Abstract
Propafenone, a class Ic antiarrythmic drug, inhibits growth of cultured Plasmodium falciparum. While the drug’s potency is significant, further development of propafenone as an antimalarial would require divorcing the antimalarial and cardiac activities as well as improving the pharmacokinetic profile of the drug. A small array of propafenone analogs was designed and synthesized to address the cardiac ion channel and PK liabilities. Testing of this array revealed potent inhibitors of the 3D7 (drug sensitive) and K1 (drug resistant) strains of P. falciparum that possessed significantly reduced ion channel effects and improved metabolic stability. Propafenone analogues are unusual among antimalarial leads in that they are more potent against the multi-drug resistant K1 strain of P. falciparum compared to the 3D7 strain.
Keywords: propafenone, malaria, microwave epoxide ring opening, hERG
Introduction
According to the WHO Malaria Report 2009, in 2008 there were 243 million malaria cases leading to 863,000 deaths, primarily pediatric cases in developing countries.1, 2 Globally, there is increased resistance to existing drugs,3, 4 including the more recently developed artemisinins.5 This clinical situation leads to a continuing need for novel, affordable malaria drugs that are effective against resistant strains of the parasite.
A screen of the MicroSource Spectrum and Killer compound collections6 -- containing drugs, bioactive compounds and natural products -- revealed that the class Ic anti-arrhythmic drug propafenone7, 8 (Fig. 1) was an inhibitor of the growth of both 3D7 (drug sensitive, EC50 1 µM) and W2 (drug resistant, EC50 0.2 µM) strains of Plasmodium falciparum6 in erythrocytic co-culture. Subsequent work with commercially sourced propafenone analogues revealed a consistent trend towards greater potency against the W2 strain of P. falciparum. The compounds also possessed improved potency against the drug resistant K1 strain, another multi-drug resistant strain, relative to the 3D7 strain. The limited SAR provided by the initial analog testing showed that removal of the secondary alcohol or truncation of the alkyl chain connecting the A and B rings gave a dramatic decrease in potency.6
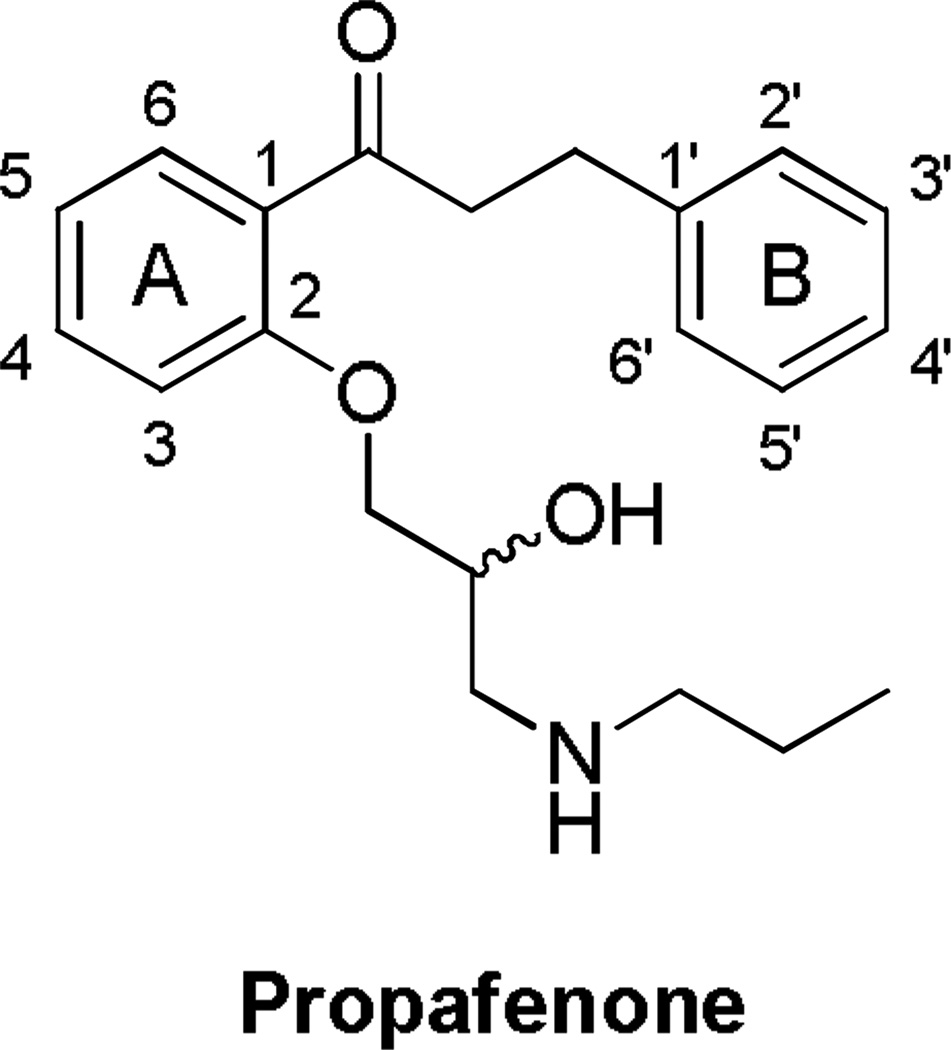
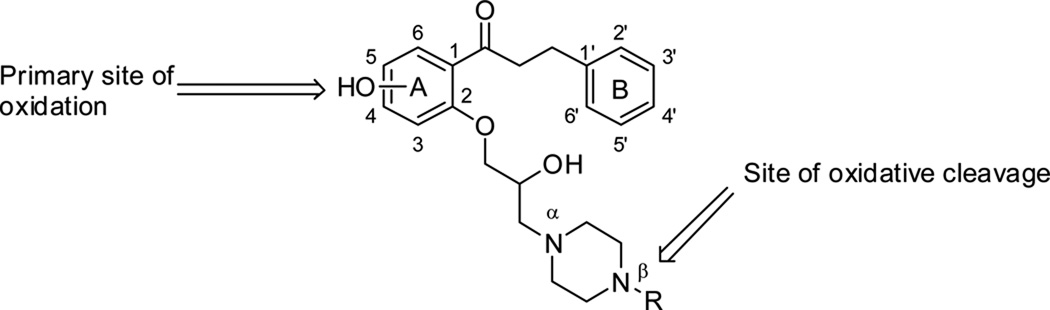
Figure 1.
Propafenone numbered according to the literature.
Propafenone (Figure 1) is a Class 1c anti-arrhythmia drug that acts on the Nav1.5 and KCNH2 (hERG) ion channels and additionally has weak β-blocking effects.8, 9 Propafenone is rapidly and extensively metabolized in 2–10 hours in 90% of patients, to two main Phase 1 metabolites, 5-hydroxypropafenone, formed by CYP2D6, and N-depropylpropafenone, formed by both CYP3A4 and CYP1A2.8 Previous studies of propafenone metabolism have revealed major species differences in the predominant metabolites produced.10, 11 Whereas humans and cynomologus monkeys show hydroxylation predominantly at the 5-position of the A-ring, rats show hydroxylation predominantly at the 4’ position of the B-ring and dogs give a mixture of both metabolites. Although propafenone is given as a racemate, the R-enantiomer is cleared faster than the S-enantiomer. Both enantiomers show equivalent potency against sodium channels, but the S-enantiomer is a more potent β-antagonist.12 Furthermore, CYP2D6*10 genotype patients, who exhibit decreased enzyme activity, have a propafenone plasma Cmax two-fold higher than individuals with the wild-type genotype and show a two-fold higher inhibitory rate of ventricular premature contractions compared with those with homozygous CYP2D6*1.13
The MMV product profile for uncomplicated malaria requires compounds with oral bioavailability, long half life, and high therapeutic index.14 Additionally, the preclinical models used to develop antimalarials require working in the mouse and those critical to assessment of cardiac risk require working in the dog. Therefore, key to development of propafenones as anti-malarials is 1) reducing or completely eliminating the cardiac effects, 2) improving the potency against both 3D7 and K1 P. falciparum, 3) increasing the elimination half life, 4) normalizing the half life across species, and 5) normalizing the half life within humans.
With these parameters in mind, the initial medicinal chemistry plan incorporated two key elements: 1) incorporating fluorines at the 5- and 4’-positions of propafenone to modulate metabolism in order to increase and normalize the elimination half-life across species and within humans;15–17 and 2) exploring alkyl amines that have been shown in other lead series to modulate interaction with ion channels.18
Results and Discussion
Separation and testing of the propafenone enantiomers
Chiral separation of propafenone was achieved using SFC chromatography with a Chiracel OD-H column (S1 Fig 1.) and confirmed by optical rotation (R-propafenone [αD]23.7 = +2.5 (c = 0.97, CH3OH), literature value [αD]23 = +6.4, (c = 1, CH3OH) and S-propafenone [αD]23.7 = −2.5 (c = 1.03, CH3OH), literature value [αD]23 = −6.3, (c = 1, CH3OH)).19 Although the propafenone enantiomers have differing metabolism and cardiac activity, there were no significant potency differences between the propafenone enantiomers against malaria in either 3D7 or W2 strains (See S1, Table 1). With this knowledge it was decided to proceed with synthesis and testing of the propafenone analogues as racemates.
Synthetic Chemistry
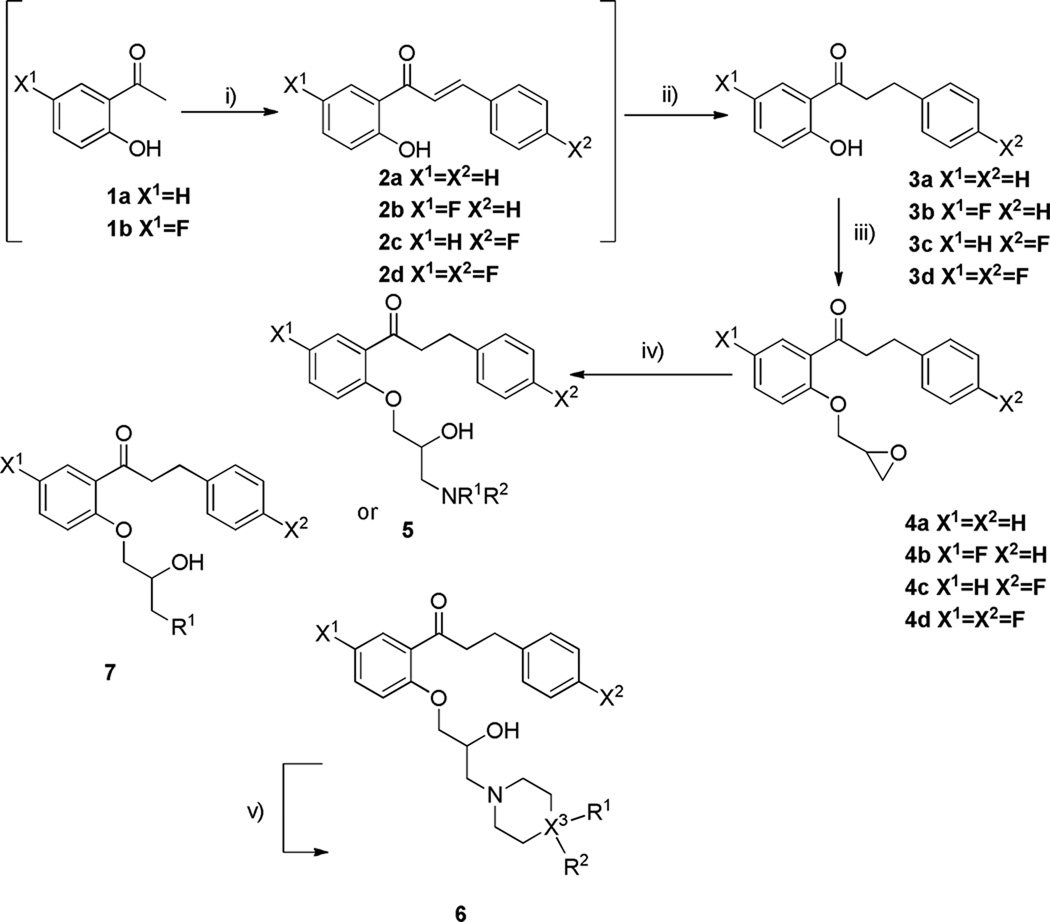
An initial set of propafenone analogues (Figure 3). was synthesized following the method of Chiba et. Al.20 The diversity in this set focused on the amine side chain of the lower part of the molecule, with most substitutions being those that had ameliorated ion channel binding in other medicinal chemistry projects.20
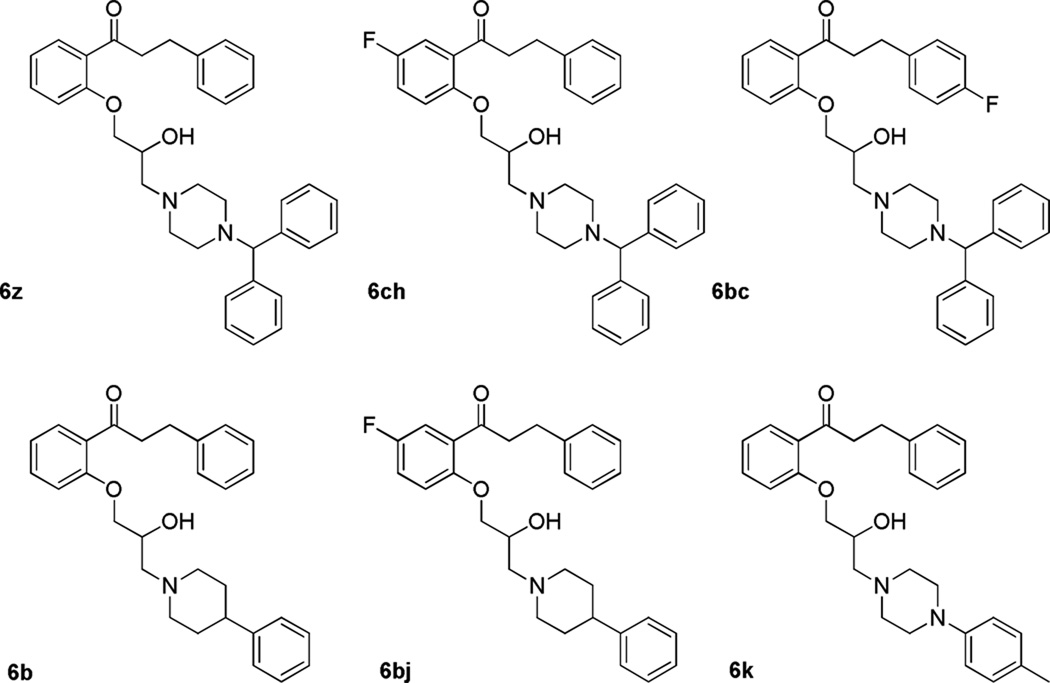
Figure 3.
Notably active propafenone analogues
Aldol condensation afforded hydroxychalcone compounds 2a–d under standard conditions.21 Reduction of the α,β-unsaturated carbonyl was achieved using a palladium catalyzed selective hydrogen transfer reaction with microwave heating.22 Removal of the palladium was achieved by addition of acetonitrile to the crude products and filtration through 0.2 µm syringe filters. Etherification of commercially available phenol 3a proceeded in quantitative yield giving essentially pure product, whereas 3b–d required chromatographic purification following work-up (Biotage® SP1). Aminolysis of the epoxide rings with microwave heating23 was compatible with a wide range of amine substrates leading to compound sets 5, 6, and 7.
Overall the route was high yielding with the chalcones 2 and reduced products 3 easily being produced on a multi-gram scale and purified by recrystallization. The non-fluorinated epoxide 3a was formed in almost quantitative yield and could be reacted without further purification, whereas the fluorinated products 3b–d required purification and gave yields between 75–85%. The microwave epoxide reaction to form final compound arrays 5, 6, and 7 proceeded cleanly and quickly, giving yields over 90%, with the exception of some of the hindered amines such as 30 and 31 (S1, Table 2).
All compounds were purified using normal phase chromatography, to a minimum standard of 95% purity. To improve solubility, all compounds were converted to the hydrochloride salts by dissolving in a 1.25 M solution of HCl in methanol, followed by evaporation of the solvent in vacuuo. Identity was confirmed by NMR and MS and purity by UPLC/UV/ELSD/MS (Waters Affinity).24 Testing was carried out using the purified hydrochloride salts made up as stock solutions in DMSO whose concentrations were confirmed using HPLC/MS/CLND
Antimalarial Activity
In vitro growth inhibitory activity of the propafenone analogues was tested against erythrocytic co-cultures of both the 3D7 and K1 strains of Plasmodium falciparum, using a previously described assay.25 For all compounds, concentration response curves were defined using 10-point, three-fold dilution schemes with concentrations ranging from 10 µM to 5 nM. Each experiment was performed in triplicate, and all triplicate experiments were independently replicated at least twice on different days. Data are reported as average values with standard deviations based upon all replications of the experiments (S1, Table 3). The EC50 values were confirmed independently at UCSF.
Comparison of the 3D7 and K1 activity shows a general correlation in potency across both strains (S1, Fig. 2) with many analogues showing sub-micromolar activity in both strains of the parasite. The series of propafenone analogues proved to be generally more active against the K1 strain of P. falciparum compared to the 3D7 strain. Comparing the log(K1 EC50) vs. logD(7.4) showed no correlation between the two (S1, Fig. 3), suggesting that permeability and solubility do not play major roles in determining potency in this series.
Overall, incorporation of a single fluorine at either the −5 or −4’ positions provided equivalent potency across the series for similar compounds although di-fluoro compounds appear to have decreased activity in all cases. The analogues incorporating smaller alkyl amine substituents (R1 = 3 – 8) gave poor potency compared to those with more bulky substituents. The diphenylmethylpiperazine containing analogues 6z, 6ch, and 6bc showed the greatest potency against both 3D7 and K1 P. falciparum, with EC50 ≤ 100 nM potency for both strains, in accordance with the MMV requirements for late lead compounds.14 Despite this, the relatively poor solubility and permeability of these bulky compounds along with their high molecular weight, suggests that the aromatic piperidine compounds 6b and 6bj would be more favorable as potential drug candidates (Figure 3).
Cytotoxicity Studies
The compound array was tested for growth inhibitory activity with four mammalian cell lines, HEK 293 (a human embryonic kidney cell line), Raji (a lymphoblastoid cell line derived from a Burkitt’s lymphoma), HepG2 (a liver cell line derived from a human hepatoblastoma) and BJ (immortalized normal foreskin fibroblasts). No analogues showed potent growth inhibition or cytotoxicity against any of the cell lines. This finding demonstrates a good class selectivity for the parasite, in accordance with the MMVs criteria requiring >10-fold selectivity for HepG2 relative to the parasite.14 Each experiment was performed in triplicate, and all triplicate experiments were independently replicated at least twice on different days. Data are reported as average values with standard deviations based upon all replications of the experiments (S1, Table 3).
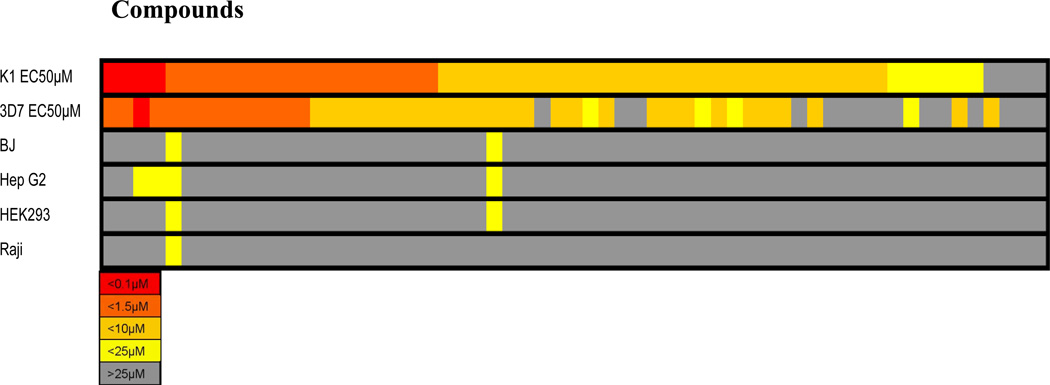
The relative potencies of the compounds in the six activity assays are shown in Figure 4 as a heat map with compound number sorted by potency against the K1 strain. This view of the data makes it clear that there is a generally good correlation between activity against the parasite strains, a reasonable range of potencies (covering 2 logs of range for EC50’s), and that there is little appreciable toxicity for mammalian cells.
Figure 4.
Heatmap Showing Relative Relationships between Growth Inhibitory Potencies against 3D7 and K1 Strains of P. falciparum and Human Cell Lines. All values represent consensus EC50 values in the respective assays.
Ion Channel Activity
One of the main objectives of the studies was to determine if the cardiac effects exhibited by the parent compound could be reduced or eliminated while retaining antimalarial potency. Since the ion channel activities of propafenone had not been established by modern methods, propafenone was screened at a fixed concentration of 10 µM against a panel of ion channels (Chantest cardiac safety panel). This study showed propafenone to be equally or more active than the controls for the hERG26, Nav1.527 and Kir6.2/SUR2Achannels28 (Table 1). While the effects on hERG and Nav1.5 were previously known, the activity against Kir6.2/SUR2A was a new finding. A consultation with cardiac toxicologists focused the program on initial optimization to eliminate the Nav1.5 and Kir6.2/SUR2A channel activities with a secondary goal of maintaining at least a 50-fold potency window against hERG. The Nav1.5 channel is encoded by the SCN5A gene and mutations in this gene are associated with long QT syndrome type 3, Brugada syndrome, primary cardiac conduction disease, and idiopathic ventricular fibrillation. It is responsible for the initial upstroke of the action potential in an electrocardiogram and is the main target for propafenone.
Table 1.
Activity of propafenone against each tested channel.
| Propafenone (10µM) | Mean % Inhibition |
|---|---|
| Cav 1.2 | 72.5 ± 6.8 |
| Cav 3.2 | 60.2 ± 4.5 |
| HCN2 | 47.8 ± 2 |
| HCN4 | 54.6 ± 12 |
| hERG | 96.9 ± 1.8 |
| Kir2.1 | 0.2 ± 0.4 |
| Kir3.1/3.4 | 46.4 ± 8.4 |
| Kir6.2/SUR2A | 89.6 ± 0.2 |
| Kv1.5 | 65.4 ± 5.4 |
| Kv4.3 | 44.1 ± 5.1 |
| KvLQT/minK | 33 ± 6.5 |
| Nav1.5 Tonic | 93.4 ± 4.5 |
| Nav1.5 Phasic | 97.1 ± 3.2 |
Three major modifications to the alkyl amine were explored: diphenyl piperidines, aromatic piperazines, and aromatic piperidines, represented by compounds 6ch, 6k, and 6b, respectively. Each of these families was chosen because similar moieties have been successfully utilized to replace simple alkyl amines in suppression of cardiac ion channel activity in other medicinal chemistry campaigns. These four particular compounds also showed good potency against both 3D7 and K1 P. falciparum strains. Compounds 6ch, 6k, and 6b were tested for inhibition of hERG, Nav1.5, and Kir6.2/SUR2A to determine if introduction of bulky aromatic piperazine and piperidine moieties reduced ion channel activity to acceptable levels. This was done using dose-response analysis of the compounds in an electrophysiological assay. All three showed greatly reduced activity against the Nav1.5 and Kir6.2/SUR2A channels compared to propafenone and gave therapeutic indices of between 23 and 895 for hERG channel activity relative to potency against K1 P. falciparum (S1, Fig. 4). Compound 6z was also tested for hERG activity as it exhibited more favorable solubility and permeability properties compared to its fluorinated counterpart 6ch. Although it shows moderate hERG channel activity it still has a reasonable TI of 26 for hERG activity relative to K1 P. falciparum.
Overall, these studies indicate that suppression of the primary propafenone cardiac targets appears reasonable and that particularly with improvements in potency one can achieve a solid therapeutic window. However, activity on hERG remains a potential liability that will have to be carefully monitored.
Microsome stability studies
In order to decrease and normalize the intrinsic clearance (Clint’) of the analogues, relative to propafenone, fluorine was incorporated at the main sites of oxidative metabolism. The 5-position of A-ring and the 4’-position of the B-ring were well known metabolic sites for the oxidation in human and rodent, respectively. It was also thought that the various amine side chains might block or reduce dealkylation, the third major metabolic event for propafenone.
Comparison of the non-fluorinated, and 4’- and 5-fluorinated diphenylpiperazine compounds 6z, 6ch, and 6bc along with the phenylpiperidine compounds 6b and 6bj, in both human liver microsomes (200 pooled mixed gender) and mouse liver microsomes (female CD9 mice) by a previously reported method29, showed that incorporation of the fluorine at the 5-position significantly decreases the Clint’ of the compounds in the human microsomes (Table 3). Analysis by MS shows oxidation on the A ring adjacent to the fluorine as the major metabolite for these compounds along with hydrolysis on one of the diphenylpiperazine rings as the minor metabolite. There was little or no sign of dealkylation at the α-amine in any of the samples. However loss of the diphenylmethyl group, in compounds 6z, 6ch, and 6bc, or methyl phenyl group, in compound 6k, was observed.
Table 3.
Multi-species liver microsome studies
| Compound | Mouse Microsomes Clint’ (mL/min/kg) |
Human Microsomes Clint’ (mL/min/kg) |
|---|---|---|
| propafenone | 45.9 | 4.4 |
| 6ch | 42.9 | 13 |
| 6z | 69.7 | 13.6 |
| 6k | 30.4 | 37.6 |
| 6b | 29.3 | 19.4 |
| 6bc | 33.7 | 4.8 |
| 6bj | 27.3 | 11.2 |
Unlike the case with the human microsomes, incorporation of the fluorine on the 4’-position did not significantly decrease the Clint’ in the mouse liver microsomes. Interestingly, only compound 6k proved to be more stable in mouse liver microsomes compared to the human liver microsomes, despite being fluorinated in the 5-position. This compound also showed little or no oxidation on the A ring. The phenyl piperidine compounds 6b and 6bj were mono or di-oxygenated on the A ring in positions 4 and 5 but showed no dealkylation (S1, Fig. 5).
Practically, these findings lead one to abandon the 4’-fluoro substitution subseries as it confers no advantage in blocking oxidative metabolism but does confer a liability in decreasing solubility and permeability. The 5-fluoro substitutions do seem to provide the potential to normalize human oxidative metabolism and potentially to increase half life.
Plasma and SGF Stability
All lead compounds used for further studies were shown to be stable for longer than 24 h in both mouse plasma and SGF (simulated gastric fluid) at pH 2 and pH 5 (S1, Table 4, S1, Fig. 6). There appear to be no issues with intrinsic stability in these matrices.
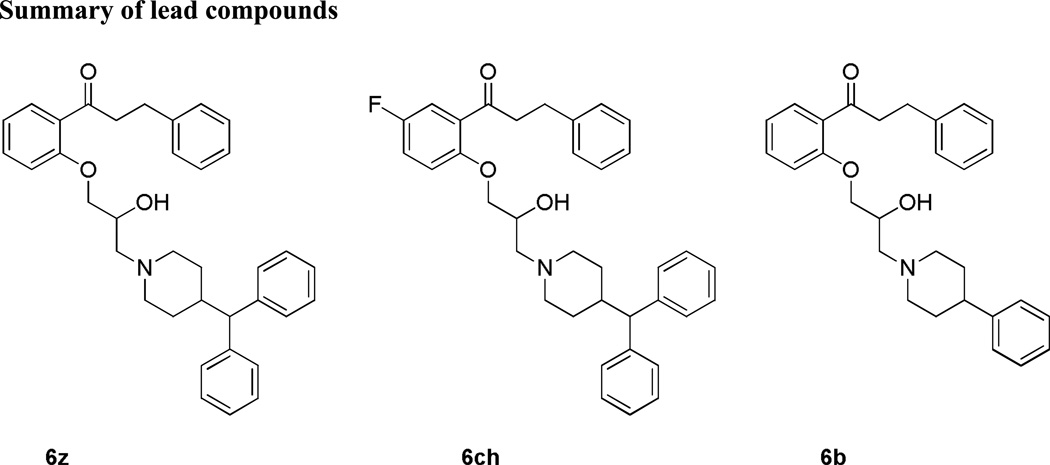
Taking into account potency against the 3D7 and K1 P. falciparum strains, solubility, cellular toxicity, stability, and ion channel activity, 3 compounds were selected for further studies (Table 4). The diphenylmethylpiperazine compound 6z exhibits greater potency but has poor solubility and permeability, a high molecular weight, and gives multiple metabolites in both the human and mouse microsomes. On the other hand, 6ch has weaker potency then the other diphenylmethylpiperazines but has a detectable permeability, high TI with respect to K1,and hERG, and a much lower Clint’ value in human microsomes. Finally, the phenylpiperidine compound 6b, exhibits weaker potency but has a lower molecular weight and is potentially more bioavailable than the other two compounds. Although 6b has the best solubility and permeability properties it also has more potent inhibition of hERG, which could lead to cardiotoxicity in vivo. Due to the fact that no one compound exhibits an optimal pattern of results across the panel of assays, all 3 compounds will be taken forward for in vivo toxicity, PK and efficacy studies (Fig 6).
Table 4.
Summary of Top 3 propafenones relative to propafenone
| Compound | 3D7 EC50 (µM) |
K1 EC50 (µM) |
hERG IC50 (µM) |
Avg. Pe. |
Sol. (µM) |
Microsomes Clint’(mL/min/kg) |
Human Clint’(mL/min/kg) |
|---|---|---|---|---|---|---|---|
| propafenone | 2.69 | 0.92 | 2.9 | 1517 | 69.8 | 45.9 | 4.4 |
| 6z | 0.06 | 0.1 | 2.6 | 0.8 | 0.2 | 42.9 | 13 |
| 6b | 0.09 | 0.05 | 3.5 | 490.9 | 7.27 | 29.3 | 19.4 |
| 6ch | 0.39 | 0.18 | 4.8 | 2.8 | 0.11 | 33.7 | 4.8 |
Figure 6.
Selected lead compounds
Conclusions
This paper has shown the design of a series of propafenone analogs to mitigate liabilities in the chemotype that would hinder development as antimalarials. The synthesis of this compound array proceeded rapidly and in high yield using a highly efficient microwave epoxide ring opening reaction as the central reaction of the route. Modification of the propafenone scaffold led to improved potency against both 3D7 and K1 strains of P. falciparum, with maximal potency achieved being ≤100nM (EC50), when diphenylmethyl piperazine moieties are included in the structure. There is a general correlation in the series towards increased potency against the drug resistant K1 strain of malaria.
The inclusion of bulky aromatic substituents on the amine side chain led to reduced activity in the ion channels targeted by the parent compound and gave a significant therapeutic index with respect to both strains of P. falciparum. Incorporation of fluorine at the site of primary human metabolism increased the clearance in human microsomes, but did not produce the same effect when placed at the site of primary metabolism in mouse microsomes. All compounds in the series gave, low cytotoxicity with four mammalian cell lines (BJ, Hep G2, Raji and HEK 293). All lead compounds were stable in human plasma and simulated gastric fluid at low pH. Despite successfully addressing the safety and clearance issues associated with the parent compound, the relatively poor solubility and permeability of the most potent, least cardiotoxic analogues is a concern and is currently being optimized. Overall the series shows promise for development of the compounds as novel anti-malarial therapeutics and as such 3 compounds have been selected for further investigation in vivo. Further optimization of the scaffold will focus on improvement of the solubility and permeability properties of the compounds while maintaining the potency and reduced ion channel activity. These improvements will be driven by ongoing studies of the in vivo efficacy, toxicity and PK on our current top compounds should give an insight into how these compounds perform relative to the in vitro data we have obtained so far.
Experimental Section
Chemistry
All chemicals were purchased from commercial sources and used without further purification. All glassware was pre-dried in an oven. All stirring was performed with an internal magnetic stirrer. All solvents were distilled or purified where necessary in accordance with D. D. Perrin and W. L. F. Armarego in “Purification of Laboratory Chemicals”, Pergamon Press, Fourth Edition, 1996. All chemicals were handled in accordance with the safety instructions in “Good Laboratory Practice”. Infrared spectra were recorded on a Nicolet-IR-100 with the sample in thin film (solution in CHCl3) between NaCl plates, as a Nujol Mull or as a KBr disc. Absorption maxima (νmax) are recorded in wave numbers (cm−1) and the following abbreviations are used: w, weak; m, medium; s, strong; br, broad. Proton magnetic resonance spectra were recorded on a Bruker 400 NMR spectrometer. Chemical shifts (δH) are quoted in parts per million (ppm) and are referenced to CDCl3 (δ 7.26 ppm). NMR peaks were assigned by MestReNova (5.2.2) Carbon magnetic resonance spectra were recorded on a Bruker 400 NMR spectrometer. Chemical shifts (δC) are quoted in parts per million (ppm) and are referenced to CDCl3 (δ = 77 ppm). NMR peaks were assigned by MestReNova (5.2.2). Spectra have been assigned, where possible, with use of DEPT, 1H-1H (COSY) and 1H-13C (HMQC) correlation spectra. Melting points were recorded using a Büchi-545. Optical rotations were recorded using a Jasco P-1010 at the D line of sodium (λ=589 nm) to the nearest tenth of a degree. Thin Layer Chromatography was performed on pre-coated silica gel 60 F254 plates and determined using u.v fluorescence. Flash column chromatography was performed with Merck Kieselgel 60 (239–400) mesh silica or the Biotage® SP1 flash column system. Rf values are quoted for the eluent given unless otherwise stated. Evaporation took place on a Büchi Rotavapor or in the Genevac HT series. Microwave Irradiation was carried out in a Biotage® Initiator 60
General Procedure for chalcone formation
A mixture of the acetophenone (1 eq.) and the corresponding aldehyde (1 eq.) in anhydrous ethanol (70 mL/23 mmol of acetophenone) was stirred at room temperature for 5 min. NaOH (3 eq.) was added and the reaction mixture was stirred at room temperature until completion. HCl (10%) was added to dissolve the sodium salt and the product was extracted with EtOAc and washed with brine to give the products as bright yellow solids. The product was re-crystallized from ethanol.
General procedure for selective MW enone reduction
Chalcone (1 eq.), 1,4-cyclohexene (10 eq.) and 10% Pd/C (0.1eq.) were placed in a microwave vial. Ethanol (18mL/500mg of propafenone) was added and the vial was sealed. The vial was placed in the microwave, pre-stirred for 30 s and irradiated for 10 min. at 100–300 W and 140 °C. The vial was cooled, the cap removed, and an equal volume of acetonitrile was added to the reaction mixture and the resulting mixture filtered through a 2 µM syringe filter to remove the Pd/C. The reaction was concentrated in vacuo and dissolved in ethyl acetate and washed with 10% K2CO3 solution (aq.). The product was re-crystallized from ethanol.
General procedure for epoxide formation
o-Hydroxyphenone (1 eq.) was dissolved in epichlorohydrin (30 eq.), and powdered NaOH (1.2 eq.) was added. The reaction mixture was refluxed for 18 h then allowed to cool to room temperature and concentrated in vacuuo. The yellow oil was dissolved in Et2O, washed with water, dried with MgSO4 then concentrated in vacuo to give a colorless solid. The product was purified by flash chromatography with 100% DCM.
General procedure for epoxide opening
Amines (1 eq.) were weighed into microwave vials. The epoxide (1eq.) was dissolved in ethanol (0.24 M solution) and added to the vials. Each vial was irradiated for 15 min. at 100–300 W and 160 °C. Un-reacted amine was extracted with PS-Isocyanate. Alternatively the product was purified by flash chromatography. On a large scale the products can generally be re-crystallized from ethanol.
General procedure for selective nitro reduction
The 4-nitro-phenyl piperidine compounds (1 eq.), cyclohexene (10 eq.), and 10% Pd/C (0.1 eq.) were placed in a microwave vial. Ethanol (18 mL/500 mg of starting material) was added and the vial was sealed. The vial was placed in the microwave, pre-stirred for 30 s and irradiated for 10 min. at 300 W and 140 °C. The vial was cooled, the cap removed, and an equal volume of acetonitrile was added to the reaction and the resulting mixture filtered through a 2 µM syringe filter to remove the Pd/C. The reaction was concentrated in vacuuo then dissolved in ethyl acetate and washed with 10% K2CO3 solution (aq.). The product was re-crystallized from ethanol.
General procedure for HCl salt formation
The propafenone analogues were dissolved in 1.25 M solution of HCl in methanol then evaporated to dryness to form HCl salts.
All compounds were purified using normal phase chromatography, to a minimum standard of 95% purity. To improve solubility, all compounds were converted to the hydrochloride salts by dissolving in a 1.25 M solution of HCl in methanol, followed by evaporation of the solvent in vacuo. Purity was confirmed both by NMR and by UPLC/UV/ELSD/MS (Waters Affinity).24 Testing was carried out using the purified hydrochloride salts.
Growth of parasites and IC50 determinations
Two P. falciparum strains were used in this study and were provided by the MR4 Unit of the American Type Culture Collection (ATCC, Manassas, VA). Those two strains were the chloroquine sensitive strain 3D7 and the chloroquine resistant strain K1. Asynchronous parasites were maintained in culture based on the method of Trager30. Parasites were grown in presence of fresh group O-positive erythrocytes (Lifeblood Memphis, TN) in Petri dishes at a hematocrit of 4–6% in media consisted of RPMI 1640 supplemented with 0.5% AlbuMAX II, 25 mM HEPES, 25 mM NaHCO3 (pH 7.3), 100 µg/mL hypoxanthine, and 5 µg/mL gentamycin. Cultures were incubated at 37° C in a gas mixture of 90% N2, 5% O2, 5% CO2. For IC50 determinations, 20 µL of RPMI 1640 with 5 µg/mL gentamycin were dispensed per well in an assay plate (Corning 8807BC 384-well microtiter plate). 40 nL of each compound, previously serial diluted in a separate assay plate (Corning 3657 384-well white polypropylene plate), were dispensed in the assay plate followed by 20 µL of a synchronized culture suspension (1% rings, 10% hematocrit) were added per well thus giving a final hematocrit and parasitemia of 5% and 1%, respectively. Assay plates were incubated for 72 h and the parasitemia were determined by a method previously described.31 Briefly, 10 µL of the development solution (10X Sybr Green I, 0.5% v/v triton, 0.5 mg/ml saponin, in RPMI) was added per well, assay plates were shaken for 30 s, incubated in the dark for 4 h, then read with the Envision spectrophotometer at Ex/Em 485nm/535nm. EC50s were calculated with the RISE (Robust Investigation of Screening Experiments) in-house protocol.
Toxicity
BJ, HEK293, Hep G2, and Raji cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and were cultured according to recommendations. Cell culture media were purchased from ATCC. Cells were routinely tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza). Exponentially growing cells were plated in Corning 384-well white custom assay plates, and incubated overnight at 37 °C in a humidified incubator with atmosphere controlled at 5% CO2 and 100% humidity. DMSO inhibitor stock solutions were added the following day to a final concentration of 25 µM, 0.25% DMSO and then diluted 1/3 for a total of ten testing concentrations. Cytotoxicity was determined following a 72 h incubation using Promega Cell Titer Glo Reagent according to the manufacturer’s recommendation. Luminescence was measured on an Envision plate reader (Perkin Elmer)
Ion Channel Activity Panel Screen by Chantest
The in vitro effects of one test article propafenone on 12 cardiac ion channels that are responsible for major components of the cardiac action potential were evaluated at room temperature using the PatchXpress 7000A (Molecular Devices), an automatic parallel patch clamp system. Propafenone was evaluated at 10 µM in two cells (n ≥ 2) for each channel. The duration of exposure was 5 minutes. The effects were evaluated using IonWorks Quattro system (MDS-AT). In case of maximal blocking effect less then 50% the IC50 value was not calculated.
Ion Channel Activity of hERG, hNav1.5 and Kir6.2/SUR2A
The channels tested were as follows: Cloned hERG potassium channels (encoded by the KCNH2 gene and expressed in CHO cells), responsible for IKr cloned hNav1.5 sodium channel (encoded by the human SCN5A gene and expressed in CHO cells), responsible for INa, fast sodium current. Cloned Kir6.2/SUR2A potassium channels (expressed by the human KCNJ11 and SUR2A genes and co-expressed in HEK293 cells), responsible for the ATP-sensitive current, IKATP.
The in vitro effects of four test articles were evaluated using these cells at room temperature using the PatchXpress 7000A (Molecular Devices), an automatic parallel patch clamp system. Each test article was evaluated at 0.2, 0.6, 1.6, and 5 µM with each concentration tested in two cells (n ≥ 2). The duration of exposure to each test article concentration was 5 minutes.
Solubility
Solubility assays were carried out on a Biomek FX lab automation workstation (Beckman Coulter, Inc., Fullerton, CA) using µSOL Evolution software (pION Inc., Woburn, MA). The detailed method is described as follows: 10 µL of compound stock was added to 190 µL 1-propanol to make a reference stock plate. 5 µL from this reference stock plate was mixed with 70 µL 1-propanol and 75 µL phosphate buffered saline (PBS, pH 7.4 and 4) to make the reference plate, and the UV spectrum (250 nm – 500 nm) of the reference plate was read. 6 µL of 10 mM test compound stock was added to 600 µL PBS in a 96-well storage plate and mixed. The storage plate was sealed and incubated at room temperature for 18 hours. The suspension was then filtered through a 96-well filter plate (pION Inc., Woburn, MA). 75 µL of filtrate was mixed with 75 µL 1-propanol to make the sample plate, and the UV spectrum of the sample plate was read. Calculation was carried out by µSOL Evolution software based on the AUC (area under curve) of UV spectrum of the sample plate and the reference plate. All compounds were tested in triplicate.
Permeability assay
The Parallel Artificial membrane Permeability Assay (PAMPA) was conducted on a Biomek FX lab automation workstation (Beckman Coulter, Inc., Fullerton, CA) using the PAMPA Evolution software (pION Inc., Woburn, MA). The detailed method is described as follows: 3 µL 10 µM test compound stock was mixed with 600 µL of SSB (system solution buffer, pH 7.4 or 4, pION Inc., Woburn, MA) to make diluted test compound. 150 µL of diluted test compound was transferred to a UV plate (pION Inc., Woburn, MA) and the UV spectrum was read as the reference plate. The membrane on pre-loaded PAMPA sandwich (pION Inc., Woburn, MA) was painted with 4 µL GIT lipid (pION Inc., Woburn, MA). The acceptor chamber was then filled with 200 µL ASB (acceptor solution buffer, pION Inc., Woburn, MA), and the donor chamber was filled with 180 µL diluted test compound. The PAMPA sandwich was assembled, placed on the Gut-box and stirred for 30 minutes. The aqueous Boundary Layer was set to 40 µm for stirring. The UV spectrum (250–500 nm) of the donor and the acceptor were read. The permeability coefficient was calculated using PAMPA Evolution software (pION Inc., Woburn, MA) based on the AUC of the reference plate, the donor plate, and the acceptor plate. All compounds were tested in triplicate.
Liver microsomal stability
0.633 mL of mouse liver microsomes (20 mg/mL, female CD9 mice, Fisher Scientific, #NC9567486) or human liver microsomes (20 mg/mL, 200 pooled mixed gender, Fisher Scientific #50-722-552) was mixed with 0.051 mL of 0.5 M EDTA solution and 19.316 mL potassium phosphate buffer (0.1M, pH 7.4, 37°C) to make 20 mL of liver microsome solution. 1 part of 10 mM DMSO compound stock was mixed with 4 part of acetonitrile to make 2 mM diluted compound stock in DMSO and acetonitrile. 29.1 µL of the diluted compound stock was added to 2.3 mL of liver microsomal solution and vortexed to make a microsomal solution with compound. 180 µL of the microsomal solutions with different compounds were dispensed into respective rows of a 96-well storage plate (pION Inc., MA, #110323). For 0 hour time point, 450 µL pre-cooled (4 °C) internal standard (10 µM warfarin in methanol) was added to the first three columns before the reaction starts. 1.25 mL of microsome assay solution A (Fisher Scientific, #NC9255727) was combined with 0.25 mL of solution B (Fisher Scientific, #NC9016235) in 3.5 mL of potassium phosphate buffer (0.1 M, pH 7.4). 45 µL of this A+B solution was added to each well of the 96-well storage plate (reaction plate). Liquid in the first 3 columns was moved to another storage plate (quenched plate). The reaction plate was then sealed and incubated at 37 °C, shaken at a speed of 60 rpm. The solutions were sampled at 0.5 hr, 1 hr, and 2 hr time points. At each time point, 450 µL of pre-cooled internal standard was added to 3 rows in the reaction plate, and the liquid was then transferred to the quenched plate. The quenched plate was then centrifuged (model 5810R, Eppendorf, Westbury, NY) at 4000 rpm for 20 minutes. 200 µL supernatant was then transferred to a 96-well plate and analyzed by UPLC-MS (Waters Inc., Milford, MA). The compounds and internal standard were detected by SIR. The log peak area ratio (compound peak area / internal standard peak area) was plotted against time and the slope was determined to calculate the elimination rate constant [k = (−2.303) * slope]. The half life (hour) was calculated as t (1/2) = 0.693 / k.
Supplementary Material
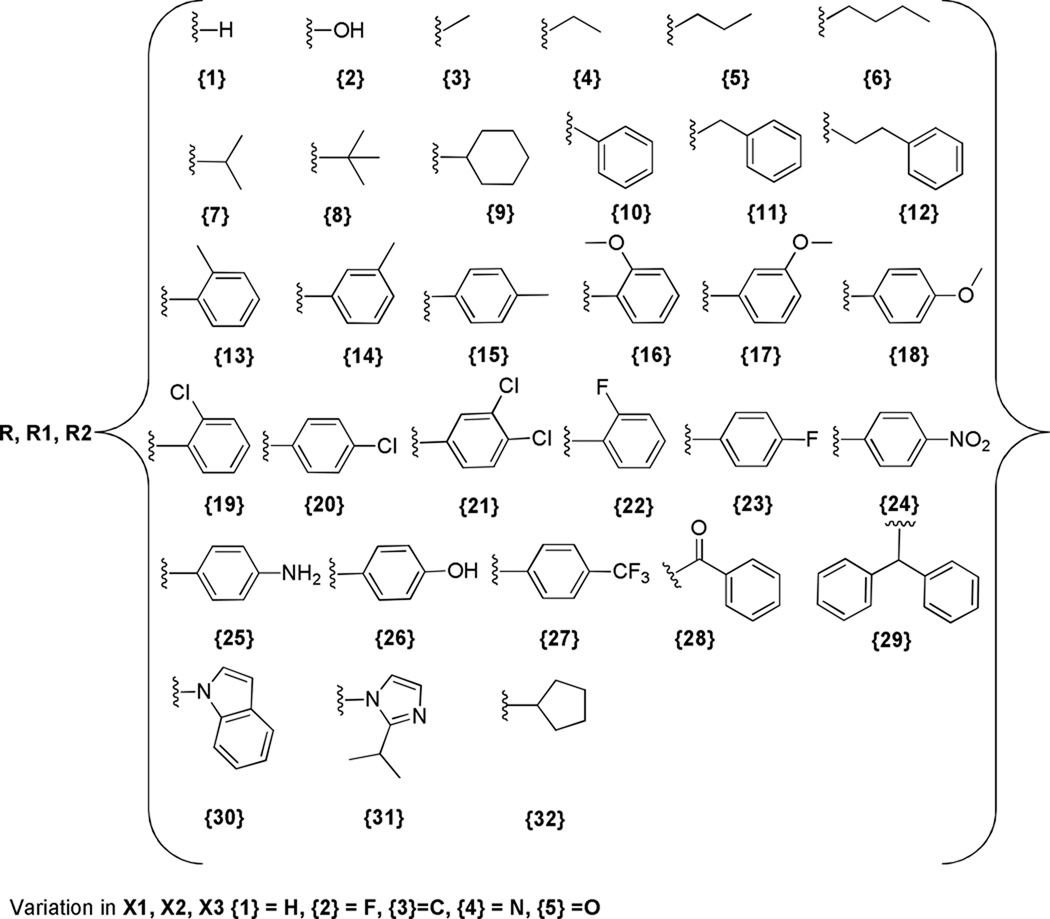
Figure 2.
Diversity Set for X and R groups in Compound Arrays 1 through 7.
Figure 5.
Number and naming convention for the piperazine propafenones
Scheme 1.
General synthetic scheme for the propafenone analoguesa
aReagents and conditions: i) aldehyde (1 eq.), NaOH, EtOH ii) 10% Pd/C (0.1 eq.), 1,4-cyclohexadiene (10 eq.), ethanol, µW (100–300W, 160 °C, 15 min.) iii) epi-chlorohydrin, NaOH, reflux, 18 h.; iv) amine (1 eq.), µW (100–300W, 160 °C, 15 min.) v) 10% Pd/C (0.1 eq.), cyclohexene (10 eq.), EtOH, µW (100–300 W, 140 °C, 10 min.).
Table 2.
Summary of ion channel activity with the three major scaffold types
| EC50 (µM) | IC50 (µM) | ||||||
|---|---|---|---|---|---|---|---|
| hNav1.5 | |||||||
| Compound | 3D7 | K1 | hERG | tonic | phasic | Kir6.2/SUR2A | TI (hERG IC50/ K1 EC50) |
| propafenone | 2.69 | 0.92 | 2.88 | 3.3 | >5 | 2.2 | 3 |
| 6z | 0.06 | 0.10 | 2.6 | N/A | N/A | N/A | 26 |
| 6ch | 0.13 | 0.005 | 4.84 | >5 | >5 | >5 | 895 |
| 6k | 0.39 | 0.18 | 4.31 | >5 | >5 | >5 | 23 |
| 6b | 0.09 | 0.05 | 3.46 | >5 | >5 | >5 | 65 |
Acknowledgements
This work was supported by funding from ALSAC, St. Jude Children’s Research Hospital and the NIH (AI075517). The authors would like to thank Rui Chen, Mettler Toledo, for assistance with the separation of the propafenone isomers, Anang Shelat and the St. Jude Children’s Research Hospital Chemical Biology Cheminformatics Group for development and assistance with the RISE (Robust Investigation of Screening Experiments) protocol, and Jimmy Cui and the St. Jude Children’s Research Hospital Chemical Biology High Throughput Screening Group for assistance with the automated screening procedures.
Abbreviations
- WHO
World Health Organization
- MMV
Medicines for Malaria Venture
- HTS
High Throughput Screening
- SFC
Super Critical Fluid chromatography
- SAR
Structure Activity Relationship
- hERG
human Ether-à-go-go Related Gene
- TI
Therapeutic Index
- Clint’
Intrinsic Clearance
Footnotes
Supporting Information: Experimental procedures, tabulated activity data, and characterization data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.WHO. World Malaria Report 2009. http://whqlibdoc.who.int/publications/2009/9789241563901_eng.pdf.
- 2.Snow R, Guerra C, Noor A, Myint H, Hay S. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sidhu A, Verdier-Pinard D, Fidock D. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olliaro P. Drug resistance hampers our capacity to roll back malaria. Clin Infect Dis. 2005;41 Suppl 4:S247–S257. doi: 10.1086/430785. [DOI] [PubMed] [Google Scholar]
- 5.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisman J, Liou A, Shelat A, Cohen F, Guy R, DeRisi J. Searching for new antimalarial therapeutics amongst known drugs. Chemical Biology & Drug Design. 2006:409–416. doi: 10.1111/j.1747-0285.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck OA, Witt E, Hochrein H. Influence of New Antiarrhythmic Agent Propafenone on Cardiac Conduction. Zeitschrift Fur Kardiologie. 1975;64:179–187. [PubMed] [Google Scholar]
- 8.Abbott Laboratories. Rythmol ® SR. http://www.accessdata.fda.gov/drugsatfda_docs/label/2003/21416_rythmol_lbl.pdf.
- 9.Barbey J. Clinical-Pharmacology And Beta-Blocking Efficacy Of Propafenone. Journal Of Cardiovascular Pharmacology. 1991:S41–S43. [PubMed] [Google Scholar]
- 10.Tan W, Li Q, McKay G, Semple HA. Identification and determination of phase I metabolites of propafenone in rat liver perfusate. Journal of Pharmaceutical and Biomedical Analysis. 1998;16:991–1003. doi: 10.1016/s0731-7085(97)00055-1. [DOI] [PubMed] [Google Scholar]
- 11.Reder-Hilz B, Ullrich M, Ringel M, Hewitt N, Utesch D, Oesch F, Hengstler JG. Metabolism of propafenone and verapamil by cryopreserved human, rat, mouse and dog hepatocytes: comparison with metabolism in vivo. Naunyn-Schmiedebergs Archives of Pharmacology. 2004;369:408–417. doi: 10.1007/s00210-004-0875-z. [DOI] [PubMed] [Google Scholar]
- 12.Burnett D, Gal J, Zahniser N, Nies A. Propafenone Interacts Stereoselectively With Beta-1-Adrenergic And Beta-2-Adrenergic Receptors. Journal Of Cardiovascular Pharmacology. 1988:615–619. doi: 10.1097/00005344-198811000-00017. [DOI] [PubMed] [Google Scholar]
- 13.Cai W, Xu J, Chen B, Zhang F, Huang Y, Zhang Y. Effect of CYP2D6*10 genotype on propafenone pharmacodynamics in Chinese patients with ventricular arrhythmia. Acta Pharmacol Sin. 2002;23:1040–1044. [PubMed] [Google Scholar]
- 14.MMV. MMV Compound Progression Criteria. http://www.mmv.org/sites/default/files/uploads/docs/essential_info_for_scientists/Compound_progression_criteria.pdf. [Google Scholar]
- 15.Bohm HJ, Banner D, Bendels S, Kansy M, Kuhn B, Muller K, Obst-Sander U, Stahl M. Fluorine in medicinal chemistry. Chembiochem. 2004;5:637–643. doi: 10.1002/cbic.200301023. [DOI] [PubMed] [Google Scholar]
- 16.Purser S, Moore PR, Swallow S, Gouverneur V. Fluorine in medicinal chemistry. Chemical Society Reviews. 2008;37:320–330. doi: 10.1039/b610213c. [DOI] [PubMed] [Google Scholar]
- 17.Shah P, Westwell AD. The role of fluorine in medicinal chemistry. Journal of Enzyme Inhibition and Medicinal Chemistry. 2007;22:527–540. doi: 10.1080/14756360701425014. [DOI] [PubMed] [Google Scholar]
- 18.Schmid D, Staudacher DL, Loew HG, Spieckermann PG, Ecker GF, Kopp S, Chiba P. A subset of highly effective propafenone-type multidrug resistance modulators lacks effects on cardiac action potential and mechanical twitch parameters of rat papillary muscles. J Pharmacol Exp Ther. 2003;307:589–596. doi: 10.1124/jpet.103.052993. [DOI] [PubMed] [Google Scholar]
- 19.Gottfried B, Bernd W. Racemattrennung von Propafenon und Diprafenon, Konfiguration der Propafenon-Enantiomeren. Liebigs Annalen der Chemie. 1987;1987:561–563. [Google Scholar]
- 20.Chiba P, Burghofer S, Richter E, Tell B, Moser A, Ecker G. Synthesis, Pharmacological Activity, and Structure-Activity-Relationships of a Series of Propafenone-Related Modulators of Multidrug-Resistance. Journal of Medicinal Chemistry. 1995;38:2789–2793. doi: 10.1021/jm00014a031. [DOI] [PubMed] [Google Scholar]
- 21.Powers DG, Casebier DS, Fokas D, Ryan WJ, Troth JR, Coffen DL. Automated parallel synthesis of chalcone-based screening libraries. Tetrahedron. 1998;54:4085–4096. [Google Scholar]
- 22.Quinn JF, Razzano DA, Golden KC, Gregg BT. 1,4-Cyclohexadiene with Pd/C as a rapid, safe transfer hydrogenation system with microwave heating. Tetrahedron Letters. 2008;49:6137–6140. [Google Scholar]
- 23.Lindstrom UM, Olofsson B, Somfai P. Microwave-assisted aminolysis of vinylepoxides. Tetrahedron Letters. 1999;40:9273–9276. [Google Scholar]
- 24.Lemoff A, Yan B. Dual detection approach to a more accurate measure of relative purity in high-throughput characterization of compound collections. J Comb Chem. 2008;10:746–751. doi: 10.1021/cc800100g. [DOI] [PubMed] [Google Scholar]
- 25.Madrid P, Wilson N, DeRisi J, Guy R. Parallel synthesis and antimalarial screening of a 4-aminoquinoline library. J Comb Chem. 2004;6:437–442. doi: 10.1021/cc0340473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 27.Catterall W, Goldin A, Waxman S. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 28.Kubo Y, Adelman J, Clapham D, Jan L, Karschin A, Kurachi Y, Lazdunski M, Nichols C, Seino S, Vandenberg C. International Union of Pharmacology. LIV. Nomenclature and molecular relationships of inwardly rectifying potassium channels. Pharmacol Rev. 2005;57:509–526. doi: 10.1124/pr.57.4.11. [DOI] [PubMed] [Google Scholar]
- 29.Di L, Kerns E, Li S, Petusky S. High throughput microsomal stability assay for insoluble compounds. Int J Pharm. 2006;317:54–60. doi: 10.1016/j.ijpharm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Trager W, Jensen J. Human malaria parasites in continuous culture. 1976. J Parasitol. 2005;91:484–486. doi: 10.1645/0022-3395(2005)091[0484:HMPICC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 31.Smilkstein M, Sriwilaijaroen N, Kelly J, Wilairat P, Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.