Abstract
MicroRNA-125b expression is modulated in macrophages in response to stimulatory cues. Here we report a functional role of miR-125b in macrophages. We found that miR-125b is enriched in macrophages compared to lymphoid cells and whole immune tissues. Enforced expression of miR-125b drives macrophages to adapt an activated morphology that is accompanied by increased co-stimulatory factor expression and elevated responsiveness to interferon gamma, while anti-miR-125b treatment decreases CD80 surface expression. To determine whether these alterations in cell signaling, gene expression and morphology have functional consequences, we examined the ability of macrophages with enhanced miR-125b expression to present antigens and found that they better stimulate T cell activation than control macrophages. Further indicating increased function, these macrophages were more effective at killing EL4 tumor cells in vitro and in vivo. Moreover, miR-125b repressed IRF4 and IRF4 knockdown in macrophages mimicked the miR-125b overexpression phenotype. In summary, our evidence suggests that miR-125b is at least partly responsible for generating the activated nature of macrophages, at least partially by reducing IRF4 levels, and potentiates the functional role of macrophages in inducing immune responses.
INTRODUCTION
The mammalian innate immune system provides a critical first line of defense against pathogens. Macrophages are key components of this system, acting to release cytokines, kill pathogens directly and present antigens to the adaptive immune system. The macrophage surface contains sensing proteins, like Toll-like receptors and interferon gamma receptor that, when engaged, lead to a rapid differentiation event termed “activation”, where the cell transforms from relative quiescence to an effector state characterized by far-heightened microbicidal ability. Macrophages also carry co-stimulatory proteins such as CD80 and CD86 for interacting with T cells, thus bridging innate immunity to adaptive immunity (1, 2).
Recently microRNAs have been shown to be important mediators of the macrophage activation process. MicroRNAs-155, 146, 147, 9 and 21 are induced by ligands of the Toll-like receptors (TLRs) (3, 4). These microRNAs, in turn, inhibit expression of protein in the inflammatory signaling cascade, thus modulating immunity through feedback regulation (3, 4). MiR-125b, a homologue of the C. elegans microRNA lin-4, has been shown to be decreased in macrophages in response to TLR4 signaling (5–7).
We and others found that miR-125b is enriched in hematopoietic stem cells and that increased miR-125b enhances hematopoietic engraftment (8, 9). Further increased levels of miR-125b cause an aggressive myeloproliferative disorder that leads to leukemia (8). Here we examine the role miR-125b plays in regulating macrophage activation. We find that macrophages express a particularly high concentration of miR-125b. When miR-125b is overexpressed in macrophages, it enhances surface activation markers both basally and in response to interferon gamma. By contrast, treatment of RAW264.7 macrophages with anti-miR-125b causes them to express decreased surface CD80 both basally and in response to interferon gamma. We demonstrate that macrophages expressing increased miR-125b become more potent stimulators of immune responses as shown by increased antigen-specific T cell activation and anti-tumor immunity. Lastly, we find that IRF4 is an important target of miR-125b in macrophages and that IRF4 knockdown mimics the miR-125b overexpression phenotype.
METHODS
Cell Culture
293T cells, RAW264.7 cells and BMMs were cultured at 37°C with 5% CO2 in DMEM supplemented with 10% FBS, 100 units/ml penicillin and 100 units/ml streptomycin. For IFNγ treatment, cells were treated overnight with 200 units/ml of recombinant mouse IFNγ (eBioscience).
Mice
C57Bl/6 and OTI Ovalbumin TCR-transgenic Balb/c mice were bred in the Caltech Office of Laboratory Animal Resources (OLAR) facility or purchased from Jackson Laboratories. The Caltech Institutional Animal Care and Use Committee (IACUC) approved all mouse experimental protocols.
Isolation of Immune Cells and Tissues
T cells and B cells were purified from the spleens of C57Bl/6 mice using magnetic beads (Miltenyi). Peritoneal macrophages were isolated four days after injecting mice with 3% Thyoglycollate.
DNA Constructs
The MG, MGP, MG-125b-1 and MGP-125b-1 vector systems have been described previously (10–12). The human miR-125b-1 sequence was also cloned into the pcDNA3 vector downstream of the CMV promoter. The IRF4 shRNA sequence was predicted and cloned into MGP as described previously (11, 13). NC1 is a negative control shRNA sequence predicted not to target any protein coding genes in the mouse genome (Invitrogen). For reporter assays, pMIR-REPORT vector (Ambion) containing Picalm and Cutl1 3’UTRs were constructed previously (10). A 3 kb region of the human IRF4 3’UTR, which includes the miR-125b putative binding site, was cloned into pMIR-REPORT downstream of luciferase. A positive control 2mer containing two tandem sites complementary to miR-125b was also cloned. Primer sequences are listed in Table S1.
Retrovirally Transduced Bone Marrow Derived Macrophages
To generate retrovirus for infecting bone marrow, 293T cells were transfected with pCL-Eco and MG, MGP, MG-125b-1 or MGP-125b-1 vectors. After 36 h, 10 ug/ml polybrene (Millipore) was added to retrovirus-containing culture supernatant, which was used to spin-infect bone marrow from C57Bl/6 mice. Cells were counted and 1 million were plated per well in a 6 well plate with 10 ng/ml M-CSF (eBioscience), and differentiated for 6 days to yield retrovirally transduced BMMs (12).
Stable Cell Lines
RAW264.7 cells were stably transduced with VSV-G-pseudotyped MGP or MGP-125b-1 retrovirus and puromycin selection was subsequently performed as described previously (11).
Electroporation of Anti-miRs
RAW264.7 cells were co-electroporated with anti-miR-125b or a mismatched control (Regulus Therapeutics) and pmaxGFP vector (Lonza) using an Amaxa Nucleofector. 36 hours post-electroporation, GFP positive cells were analyzed by FACS. Anti-miR-125b or mismatched control compound were chimeric 2′-fluoro/2′-O-methoxyethyl-modified oligonucleotides with a completely modified phosphorothioate backbone (14) (Regulus Therapeutics). The exact chemistry is available on request.
T-cell Macrophage Co-Culture
50,000 BMMs stably expressing either MG or MG-125b-1 were co-cultured with 150,000 T cells harvested from OTI Ova TCR-transgenic Balb/c mouse in a 48 well flat bottom plate in the absence or presence of ovalbumin protein. Flow cytometry and ELISAs to assess T cell activation were performed 72 hours later. ELISAs were performed with an IL-2 detection kit from eBioscience and carried out according to the manufacturer’s instructions.
EL4 Tumor Cell Experiments
500,000 BMMs stably expressing either MG or MG-125b-1 were generated as described above in 6 well plates. One million EL4-Fluc cells were added to each well supplemented with 20 ng/ml LPS. EL4-Fluc apoptosis was measured 94 hours later by staining cells in suspension with AnnexinV antibody (BD Pharmingen). For the in vivo experiments, 2 million EL4-Fluc cells were co-injected with 400,000 BMMs subcutaneously into albino C57Bl/6 mice. Mice were closely monitored over the next 12 days. Tumor luminescence was measured using a Xenogen imager. At the experimental endpoint, animals were euthanized and tumors were removed and weighed. Tumor surface area was assessed using a caliper— tumor length and width were measured in cm, and the product was taken to determine surface area.
Sequence Alignment
The miR-125b seed region and IRF4 3’UTR sequences from human (Homo sapien), mouse (Mus musculus), cat (Felis catus) and armadillo (Dasypus novemcinctus) were obtained and aligned using Targetscan (15–17).
Luciferase Reporter Assay
293T cells were co-transfected with pcDNA-125b, β-gal expression vector, and the pMIR-REPORT vectors containing 3’UTRs of Cutl1, Picalm, IRF4 or 2mer. The luciferase activity was quantified 48 hours later and normalized to β-gal activity as previously described (11, 13).
RNA Preparation and Quantification
RNA was isolated using TRIzol (Invitrogen), RNEasy (Qiagen) or miRNEasy (Qiagen) as per manufacturer’s instructions. Quantitative real-type PCR (qPCR) was conducted using a 7300 Real-time PCR machine (Applied Biosystems) or a Realplex Real-time PCR machine (Eppendorf). SYBR green was used to assay IRF4 and L32 expression. PCR with previously published primer sequences for mouse pri-miR-125b-1 and pri-miR-125b-2 were used to assay levels of miR-125b primary transcripts (18). Taqman based qPCR was conducted to assay miR-125b, miR-125a and snoRNA-202 (Applied Biosystems). Primer sequences are listed in Table S1.
Flow Cytometry
Cells were stained with the following fluorophore-conjugated antibodies: CD80, CD86, CD40 (Biolegend); MHC II (eBioscience); AnnexinV (BD Pharmingen). Cell surface receptors were measured using a FACSCalibur (Becton Dickinson) and all data was analyzed with FloJo (Treestar). Data was gated on GFP positive events when retrovirally transduced cells were analyzed.
RESULTS
MiR-125b expression is enriched in macrophages
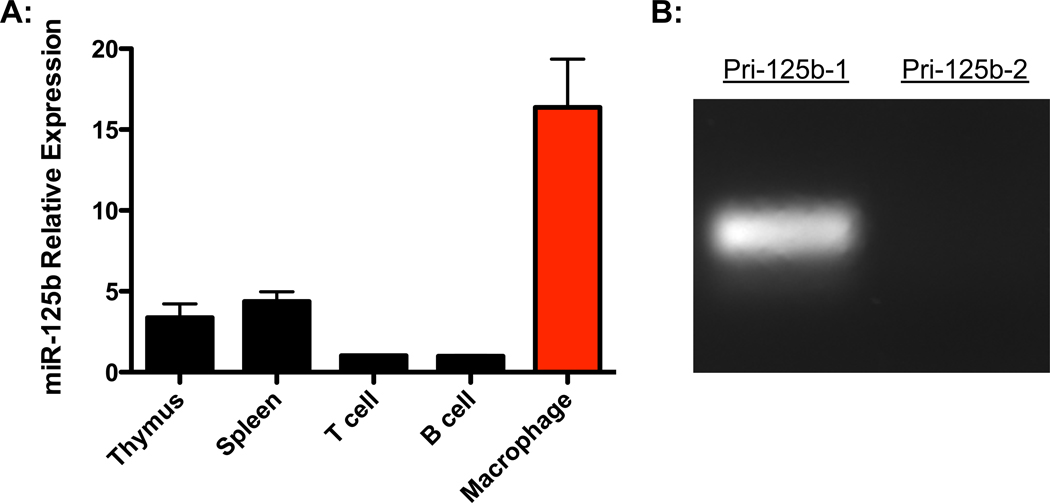
To investigate the expression of miR-125b in different immune cells and tissues, we harvested RNA from total splenocytes, thymocytes, splenic T cells, splenic B cells, and peritoneal macrophages from C57Bl/6 mice. Levels of miR-125b were assessed by reverse transcription followed by quantitative PCR. The expression of miR-125b was much higher in macrophages compared to the other immune cells and tissues (Fig. 1A). Within macrophages, miR-125b levels were also significantly higher than its homologue, miR-125a, indicating that miR-125b is the dominant isoform in these cells (Fig. S1). Also, miR-125b is expressed from two loci in the mouse genome, each encoding a different primary transcript. We performed RT-PCR for each of these primary transcripts and determined that macrophages express primarily miR-125b-1 (Fig. 1B). Because miR-125b-1 is enriched in macrophages, we set out to determine the functional role of miR-125b-1 (referred from here on as miR-125b) in these cells.
Figure 1.
MiR-125b expression is enriched in macrophages. A) Relative expression of miR-125b in immune tissues and cells assessed by quantitative PCR. Data represents the mean with SEM of 3 biological replicates per group. B) Expression of the miR-125b primary transcripts, pri-125b-1 and pri-125b-2, in bone marrow derived macrophages (BMMs). Data is representative of two independent experiments.
Enforced expression of miR-125b enhances macrophage activation status
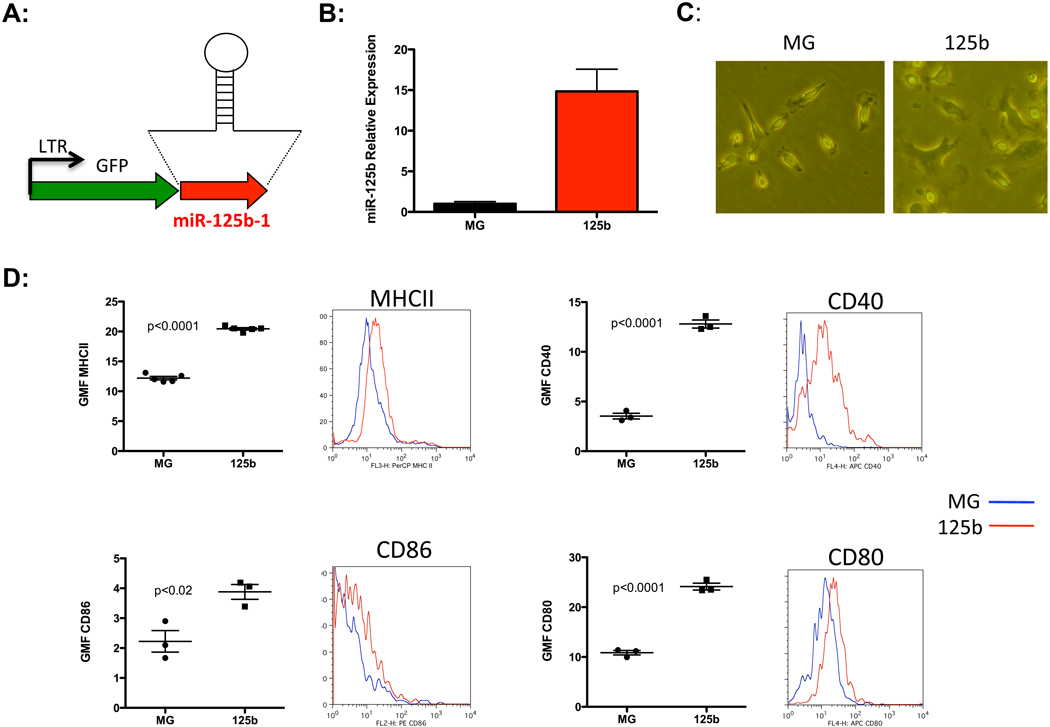
To examine the response of macrophages to miR-125b, we utilized a miR-125b over-expression system based on the MG vector (MG-miR-125b), which was originally derived from the murine stem cell virus (10) (Fig. 2A). Bone marrow cells isolated from C57Bl/6 mice were spin-infected with either MG-miR-125b or MG control vector. These cells were then differentiated into bone marrow derived macrophages (BMMs) by treatment with macrophage-colony stimulating factor (M-CSF). Using this system, miR-125b was over-expressed 15-fold above endogenous levels in BMMs (Fig. 2B). Interestingly, miR-125b over-expressing BMMs acquired a spread morphology with extensive pseudopods that resembled activated macrophages (Fig. 2C). We performed flow cytometric analyses and observed increased expression of MHCII and the co-stimulatory molecules CD40, CD86 and CD80 in these macrophages, indicating that these cells were indeed more activated (fig. 2D). Ectopic expression of miR-125b in RAW264.7 macrophages gave similar results (Fig. S2 A), further emphasizing that this microRNA promotes activation of macrophages.
Figure 2.
MiR-125b enhances basal macrophage activation. A) Retroviral vector design for over-expression of miR-125b-1. B) Relative expression of miR-125b in BMMs after transduction with MG or MG-miR-125b expressing vector. C) Morphology of control MG or miR-125b over-expressing BMMs. Data represent five independent experiments. D) Geometric mean fluorescence (GMF) of MHCII, CD40, CD86 and CD80 are shown. Representative plots obtained from flow cytrometric analyses are also shown for each marker. Data is the mean with SEM of 3–5 samples per group and is representative of two independent experiments.
MiR-125b increases macrophage response to interferon gamma
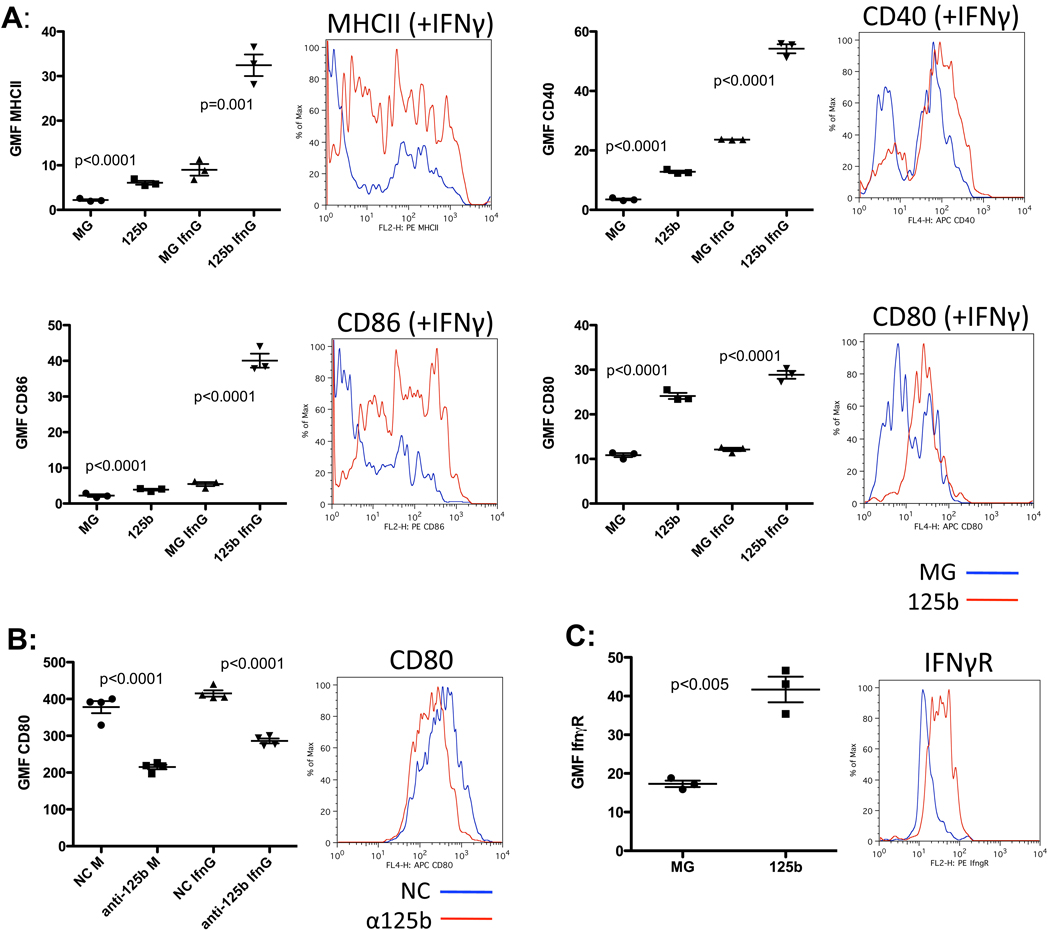
Next, we assessed the effect of miR-125b on the responsiveness of macrophages by stimulating these cells with interferon gamma (IFNγ). IFNγ treatment increased the expression of MHCII, CD40, CD86, and CD80 activation markers in control macrophages (Fig. 3A) while miR-125b over-expressing macrophages expressed significantly higher levels of these markers (Fig. 3A). Similar results were obtained in RAW264.7 macrophages with enforced miR-125b expression (Fig. S2 A).
Figure 3.
MiR-125b increases macrophage response to IFNγ. A) Surface expression of MHCII, CD40, CD86, and CD80 in response to media alone or IFNγ. A representative flow cytometric plot of the IFNγ treated samples is shown for each factor. B) Raw264.7 macrophages electroporated with control (NC) or anti-miR-125b were subjected to flow cytometry for the surface expression of CD80. A representative FACS plot of the media-treated samples shown. C) Surface expression of IFNγR in control (MG) versus miR-125b over-expressing macrophages. A representative FACS plot is shown. All data shown represents the mean expressed with SEM of three samples per group and is representative of two independent experiments.
To examine whether reducing the concentration of miR-125b had an effect inverse to that of overexpression. RAW264.7 macrophages were treated with synthetic antisense oligonucleotides (anti-miRs) and surface CD80 levels were monitored as an indication of the activation status of the cells. Anti-miR125b did cause a reduction of both basal and IFNγ induced levels of CD80 compared to cells treated with a control anti-miR (Fig3 B). Thus, miR-125b appears to control CD80 expression in macrophages under normal, physiological conditions.
A likely reason for the heightened response to IFNγ in miR-125b-treated cells could be an increased expression of the interferon gamma receptor (IFNγR). Indeed, miR-125b over-expressing BMMs (Fig. 3C) and RAW264.7 macrophages (Fig. S2 B) had significantly higher levels of the receptor. Thus, in addition to potentiating macrophage activation, miR-125b promotes enhanced macrophage responsiveness to IFNγ and increases surface expression of its cognate receptor.
MiR-125b enhances macrophage-mediated function
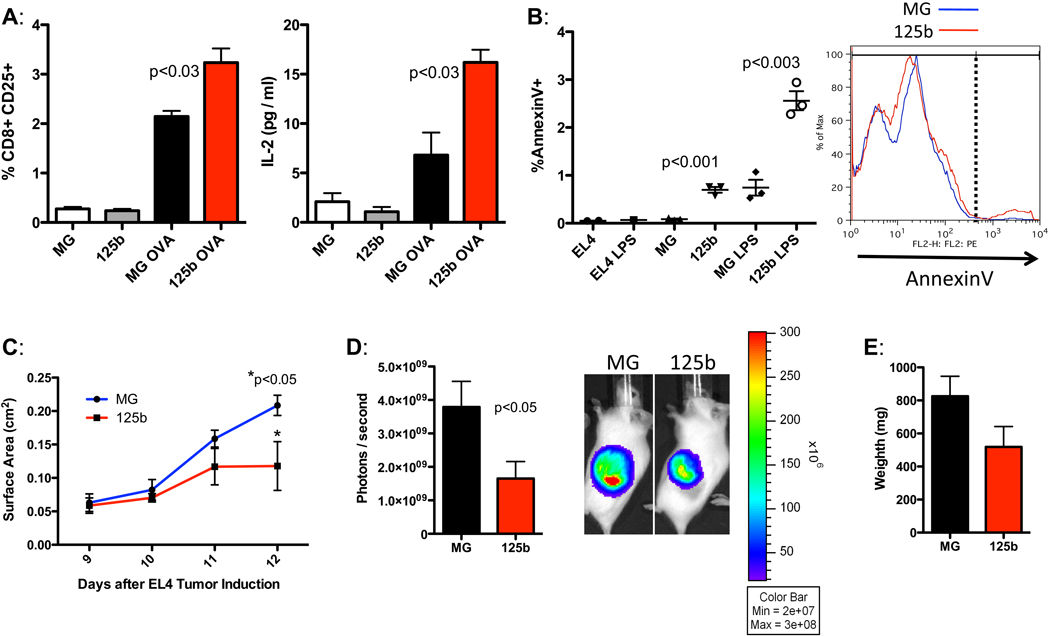
Because we found that miR-125b drove macrophages to adopt an elevated activation status and become more responsive to stimulatory cues, we examined whether miR-125b would also potentiate macrophage-mediated immune function. To this end, we investigated whether miR-125b would increase the ability of macrophages to present antigens and induce activation of T cells. MiR-125b over-expressing macrophages were co-cultured with transgenic T cells that express a chicken ovalbumin-specific T cell receptor (OT1) in the presence of ovalbumin. Indeed, compared to control macrophages, miR-125b over-expressing cells were more effective at inducing T cell activation, which was indicated by increased CD25 expression and IL-2 secretion by the T cells in response to ovalbumin (Fig. 4A). Thus, enforced expression of miR-125b led to an elevated ability of macrophages to act as effective antigen presenting cells for stimulation of T cell responses.
Figure 4.
MiR-125b enhances macrophage function. A) BMMs expressing the vectors MG or MG-125b were co-cultured with ovalbumin-specific OT1 T cells with or without ovalbumin for 72 hours. The percent CD8+CD25+ T cells are shown in the left panel. Concentration of IL-2 (pg/ml) produced by the T cells in the supernatant is shown in the right panel. Data represents the mean with SEM of 3 biological replicates per group. B) The percent AnnexinV+ EL4-Fluc cells after 94 hours of co-culture with control or miR-125 over-expressing macrophages in the presence of media alone or lipopolysaccharide (LPS). A representative flow cytometric plot of the LPS-treated group is shown. Data is expressed in mean with SEM of 1–3 experimental samples per group. C–E) EL4-Fluc cells were subcutaneously co-injected with LPS-activated control or miR-125b over-expressing macrophages into albino C57Bl/6 mice. Tumor surface area in cm2 was monitored from day 9–12 (C). The relative intensity of luminescence (D) and weight (E) of the EL4 tumors were measured on day 12. Data represents the mean plotted with SEM of eight mice per group. Representative of two independent experiments.
In addition to serving as antigen presenting cells, another major function of macrophages is to eliminate aberrant cells, such as tumor cells. We therefore assessed whether miR-125b-stimulated macrophages were more effective at killing tumor cells. We used the EL4-Fluc thymoma tumor line (19), which was engineered to express luciferase, and co-cultured these cells with either control macrophages or macrophages over-expressing miR-125b. Consistent with augmented function, miR-125b expressing macrophages were better at inducing apoptosis of EL4-Fluc cells (Fig. 4B). Macrophages exposed to LPS gained the ability to induce apoptosis of EL4-Fluc cells, with miR-125b-overexpressing macrophages having superior effectiveness (Fig. 4B). To test whether miR-125b levels in macrophages affect tumor killing in vivo, we subcutaneously co-injected into mice equal numbers of LPS-activated control or LPS-activated miR-125b overexpressing macrophages with EL4-Fluc cells and tracked growth of the resulting tumor by measuring tumor surface area over time. Since EL4-Fluc cells were engineered to express luciferase, we also monitored tumor growth by measuring luminescence in vivo. Consistent with our in vitro data, macrophages with miR-125b ectopic expression suppressed the ability of EL4 cells to expand in vivo (Fig. 4C). At the endpoint of the experiment on day 12, animals injected with MG-125b macrophages had smaller EL4-derived tumors that were significantly less luminescent than those injected with control macrophages (Fig. 4 D–E). Thus, miR-125b expression in macrophages appears to aid them in preventing the expansion of tumorigenic cells, further demonstrating that miR-125b enhances macrophage function.
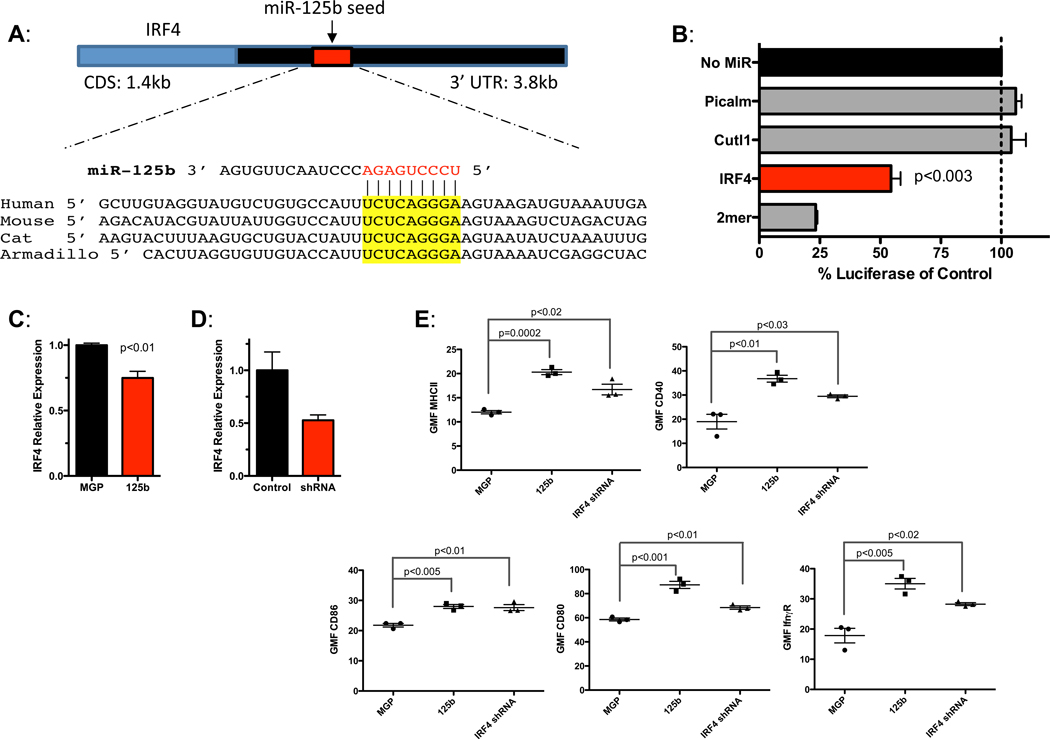
IRF4 is a miR-125b target in macrophages
To identify targets regulated by miR-125b that modulate macrophage activation, we used TargetScan 5.1 to identify transcripts in the mouse genome that contain conserved putative miR-125b binding sites in their 3’ untranslated regions (UTRs). Among these genes, the 3’UTR of IRF4 harbored a conserved miR-125b binding site (Fig. 5A) and has been previously validated as a miR-125b target in B cell lines (20–23). We found that miR-125b indeed represses via the 3’UTR of IRF4 (Fig. 5B) and that miR-125b inhibits IRF4 expression in macrophages (Fig. 5C). Next, using the MGP retroviral vector system (11), we knocked down the expression of IRF4 using RNAi (Fig. 5D) and examined the effect in macrophages. MGP-125b led to a 6-fold increase in miR-125b in BMMs. Similar to miR-125b overexpression, decreased IRF4 expression resulted in increased surface expression of MHCII, CD40, CD86, CD80 and IFNγR (Fig 5E). Thus IRF4 knockdown in macrophages enhances activation, mimicking the miR-125b overexpression phenotype. This data is consistent with previous reports demonstrating that IRF4 is a negative regulator of macrophage pro-inflammatory pathways.(21, 24) Collectively, our data suggests that IRF4 is a primary target of miR-125b in regulating macrophage activation.
Figure 5.
IRF4 is a target of miR-125b in macrophages. A) IRF4 contains a conserved miR-125b target site. B) Luciferase reporters carrying the 3’UTR of IRF4, Picalm (negative control), Cutl1 (negative control) or 2mer (positive control) were co-transfected into 293T cells with β-gal reporter and +/− miR-125b. The relative luciferase activity of each reporter in the presence of miR-125b is shown relative to the no microRNA control. C) RAW264.7 macrophages were transduced with either a control (MGP) or miR-125b expressing vector, or with D) control (NC1) or IRF4 shRNA expressing vector. RNA was harvested and L32-normalized IRF4 levels were determined by qPCR. E) BMMs expressing MGP, MGP-125b, or shRNA against IRF4 were measured for surface expression of the activation markers MHCII, CD40, CD86, CD80 and IFNγR. Geometric Mean Fluorescence (GMF) measured by flow cytometry is shown. All data represents the mean with SEM of 3 samples per group and is representative of two independent experiments.
DISCUSSION
In this study, we demonstrate that miR-125b is enriched in macrophages and that further elevation of miR-125b promotes greater activation, IFNγ response, and immune function in these cells. We also performed loss-of-function studies using synthetic antisense oligonucleotides designed to inhibit miR-125b, and found that this anti-miR-125b compound effectively decreased CD80 levels, supporting a physiological role for miR-125b in macrophage activation. Other groups have reported that miR-125b levels decrease in macrophages 3 hours post-inflammatory stimulation (5, 7). Thus, decreasing miR-125b may serve as a natural mechanism to limit the inflammatory response. In our studies, miR-125b overexpression also promoted the ability of macrophages to present antigen and induce T cell activation, demonstrating that miR-125b can enhance the macrophage’s role in mediating adaptive immunity. The increase in activated T cells would in theory result in more secretion of IFNγ, which in turn would further magnify the activation status of miR-125b-expressing macrophages. In this way, by affecting macrophage function alone, miR-125b could amplify both innate and adaptive immune responses by orchestrating positive feedback loops between macrophages and T cells.
In B cells, miR-125b inhibits differentiation of germinal center B cells into plasma cells, and does so via repression of the transcription factors IRF4 and BLIMP1 (23, 25). Recently, IRF4 was shown in B cell lines to regulate levels of BIC/miR-155, an interaction that might be important in leukemic transformation (26). IRF4, a member of the Interferon Response Factor family of transcription factors, also has important functions in macrophages where it acts as an inhibitor of the inflammatory response (21, 24). We demonstrate here that IRF4 is a target of miR-125b and that IRF4 knockdown mimics the miR-125b mediated activation phenotype in macrophages. Thus, miR-125b’s ability to potentiate macrophage activation is consistent with previously demonstrated roles of its target, IRF4, to inhibit pro-inflammatory macrophage polarization (27).
MiR-125b is upregulated in certain leukemias and downregulated in many non-hematopoietic solid cancers (12, 28–38). We have shown here that increased miR-125b expression in tumor macrophages slows tumor growth. Our data suggests that supplementing tumor macrophages with miR-125b may be a useful strategy for treating certain cancers. Future studies should aim to better understand the physiological and pathological mechanisms underlying control of miR-125b expression in macrophages.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Lili Yang for providing us with Ovalbumin TCR-transgenic mice and the EL4-Fluc cell line.
Abbreviations used in this paper
- miR
microRNA
- BMM
bone marrow-derived macrophage
- IRF4
IFN regulatory factor 4
- UTR
untranslated region
- Cutl1
Cut-like homeobox 1
- Picalm
phosphatidylinositol binding clathrin assembly protein
- BLIMP1
B lymphocyte-induced maturation protein 1
Footnotes
A.A.C. was supported by the National Science Foundation Graduate Research Fellowship Program, and by the Paul and Daisy Soros Fellowship for New Americans. A.Y.S. was supported by Award 1F32 CA139883-01A1 from the National Institutes of Health. N.S. was supported by the Caltech Amgen Scholars program. R.M.O. was supported by Award K99HL102228 from the National Heart, Lung and Blood Institute. This work was supported in part by National Institutes of Health Grant 1RO1AI079243-01.
REFERENCES
- 1.Murphy KP, Travers P, Walport M, Janeway C. Janeway's immunobiology. New York: Garland Science; 2008. [Google Scholar]
- 2.Kindt TJ, Goldsby RA, Osborne BA, Kuby J. Kuby immunology. New York: W.H. Freeman; 2007. [Google Scholar]
- 3.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 4.O'Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 5.Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 7.Murphy AJ, Guyre PM, Pioli PA. Estradiol suppresses NF-kappa B activation through coordinated regulation of let-7a and miR-125b in primary human macrophages. J Immunol. 2010;184:5029–5037. doi: 10.4049/jimmunol.0903463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci U S A. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao DS, O'Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, Bhat B, Swayze EE, Bennett CF, Esau C. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37:70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Baltimore D. Long-term in vivo provision of antigen-specific T cell immunity by programming hematopoietic stem cells. Proc Natl Acad Sci U S A. 2005;102:4518–4523. doi: 10.1073/pnas.0500600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gururajan M, Haga CL, Das S, Leu CM, Hodson D, Josson S, Turner M, Cooper MD. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol. 2010;22:583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, Matsuyama T, Taniguchi T, Honda K. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc Natl Acad Sci U S A. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honma K, Udono H, Kohno T, Yamamoto K, Ogawa A, Takemori T, Kumatori A, Suzuki S, Matsuyama T, Yui K. Interferon regulatory factor 4 negatively regulates the production of proinflammatory cytokines by macrophages in response to LPS. Proc Natl Acad Sci U S A. 2005;102:16001–16006. doi: 10.1073/pnas.0504226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honma K, Udono H, Kohno T, Yamamoto K, Ogawa A, Takemori T, Kumatori A, Suzuki S, Matsuyama T, Yui K. Interferon regulatory factor 4 negatively regulates the production of proinflammatory cytokines by macrophages in response to LPS. Proc Natl Acad Sci U S A. 2005;102:16001–16006. doi: 10.1073/pnas.0504226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gururajan M, Haga CL, Das S, Leu CM, Hodson D, Josson S, Turner M, Cooper MD. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol. 2010;22:583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Toomey NL, Diaz LA, Walker G, Ramos JC, Barber GN, Ning S. Oncogenic IRFs Provide a Survival Advantage for Epstein-Barr Virus- or Human T-Cell Leukemia Virus Type 1-Transformed Cells through Induction of BIC Expression. J Virol. 2011;85:8328–8337. doi: 10.1128/JVI.00570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 28.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci U S A. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, Lippert E, Talmant P, Lafage-Pochitaloff M, Leroux D, Gervais C, Viguie F, Lai JL, Terre C, Beverlo B, Sambani C, Hagemeijer A, Marynen P, Delsol G, Dastugue N, Mecucci C, Brousset P. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klusmann JH, Li Z, Bohmer K, Maroz A, Koch ML, Emmrich S, Godinho FJ, Orkin SH, Reinhardt D. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24:478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gefen N, Binder V, Zaliova M, Linka Y, Morrow M, Novosel A, Edry L, Hertzberg L, Shomron N, Williams O, Trka J, Borkhardt A, Izraeli S. Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia. 2010;24:89–96. doi: 10.1038/leu.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 33.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 34.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 35.Guan Y, Yao H, Zheng Z, Qiu G, Sun K. MiR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int J Cancer. 2011;128:2274–2283. doi: 10.1002/ijc.25575. [DOI] [PubMed] [Google Scholar]
- 36.Glud M, Rossing M, Hother C, Holst L, Hastrup N, Nielsen FC, Gniadecki R, Drzewiecki KT. Downregulation of miR-125b in metastatic cutaneous malignant melanoma. Melanoma Res. 2010;20:479–484. doi: 10.1097/CMR.0b013e32833e32a1. [DOI] [PubMed] [Google Scholar]
- 37.Henson BJ, Bhattacharjee S, O'Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48:569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.