Abstract
We conducted gender-stratified analyses on a systems-based candidate gene study of 53 regions involved in nicotinic response and the brain-reward pathway in two randomized clinical trials of smoking cessation treatments (placebo, bupropion, transdermal and nasal spray nicotine replacement therapy). We adjusted P-values for multiple correlated tests, and used a Bonferroni corrected α-level of 5 × 10−4 to determine system-wide significance. Four SNPs (rs12021667, rs12027267, rs6702335, rs12039988; r2>0.98) in erythrocyte membrane protein band 4.1 (EPB41) had a significant male-specific marginal association with smoking abstinence (OR=0.5; 95% CI 0.3–0.6) at end of treatment (adjusted P<6 × 10−5). rs806365 in cannabinoid receptor 1 (CNR1) had a significant male-specific gene-treatment interaction at 6-month follow-up (adjusted P=3.9 × 10−5); within males using nasal spray, rs806365 was associated with a decrease in odds of abstinence (OR=0.04; 95% CI 0.01–0.2). While the role of CNR1 in substance abuse has been well studied, we report EPB41 for the first time in the nicotine literature.
Keywords: Genetic association studies, heterogeneity, smoking cessation
INTRODUCTION
Nicotine, the addictive component of cigarettes, plays a primary role in entrapping individuals in a cycle of tobacco dependence. It acts swiftly on central nervous system receptors and stimulates the release of neurotransmitters (e.g, dopamine). These neurotransmitters are involved in the brain reward pathway, resulting in pleasurable arousal and enhanced mood.1, 2 Persistent exposure to nicotine results in the development of tolerance to its effects and a need for increased intake to achieve the same level of reward.1–3 Subsequent withdrawal symptoms (e.g., irritability, hunger, anxiety) in the absence of nicotine and their severity are tied to the extent of dependence.3–5
Earlier efforts to elucidate the variability between women and men in smoking behavior and these effects focused on smoking frequency and inhalation differences.6, 7 These data suggest that men smoke with greater frequency and inhale more deeply. However, among regular to heavy smokers, there were no gender differences in cigarette volume and inhalation patterns.7 Among smokers, nicotine blood levels after smoking one cigarette did not differ between women and men.8
Gender-specific characteristics in smoking determinants and cessation have also been characterized. Women reported greater withdrawal symptoms, including higher levels of negative affect (e.g., anxiety, nausea, depression, neuroticism) than men.9, 10 Smoking behavior in women was more strongly reinforced by external social and situational cues, and concern about the “consequences” of smoking cessation (e.g., weight gain, social isolation).9, 11 In contrast, smoking behavior in men was reinforced more by nicotine dosage12, and pharmacological treatments were more effective for men than women.10, 12, 13 Nicotine replacement therapies (e.g., nicotine patch, gum, spray) have been shown to be efficacious regardless of gender, increasing abstinence rates almost two-fold.14 However, given the greater sensitivity to the effects of nicotine in men, along with lower relapse rates following nicotine replacement therapies and bupropion,11, 15 we would anticipate a stronger pharmacogenetic effect in men.
These gender differences in response to nicotine may have a physiological basis. In a study of the effects of sex hormones on nicotine metabolism, Benowitz et al showed that females, especially those taking oral contraceptives, metabolized nicotine more quickly than men.16 Estrogen upregulates CYP2A6 activity17 and glucuronidation activity,18 two drug metabolizing activities essential for nicotine metabolism.19
Animal models focusing on gender differences in response to nicotine showed that female rodents reached a threshold tolerance with less nicotine due to greater nicotine sensitivity, and subsequently, did not discriminate as well as male rodents between varying levels of nicotine.20–22 This differential response to nicotine arises from the regulation of dopamine through ovarian, not testicular, hormones,13 with a greater increase in dopamine concentration in female rodents in response to nicotine.21 Moreover, female mice were less sensitive to the pain and anxiety reducing effects of nicotine, attributable to female sex hormones (progesterone, estradiol) and not male sex hormones (testosterone) acting as nicotinic receptor antagonists.23
These differences motivated a gender-stratified analysis to investigate genes with potentially distinct roles in addiction specific to males and females. We utilized the same phenotype and genotype data from two comparable pharmacogenetic trials of smoking cessation treatment.24, 25 As previously reported, a number of genetic variants were identified as predictors of cessation and/or therapeutic response in these studies, including the −141C Ins/Del and C957T in the dopamine D2 receptor gene (DRD2),25 CYP2A6 and CYP2B6,26–31 a VNTR and SNPs in SLC6A3,24, 32, 33 and a SNP (rs2072661) within the 3′ UTR of the nicotinic acetylcholine receptor (nAChR) β2 subunit gene (CHRNB2).24 We present gender-stratified marginal effects and gender-stratified gene-treatment interactions across both clinical trials for 1,198 SNPs in 53 gene regions, in order to identify variants of interest within each gender. SNPs with interesting effects within each gender were also assessed for differences between genders.
MATERIALS AND METHODS
Study sample and design
We analyzed subjects enrolled in two randomized clinical trials conducted by the University of Pennsylvania Transdisciplinary Tobacco Use Research Center.24, 25 The first study (Bupropion) was a double-blind randomized clinical trial comparing the efficacy of bupropion to placebo. This study population and SNPs within the candidate genes presented here were previously investigated for marginal SNP and SNP-treatment effects. 27 The second study (NRT) was an open-label randomized clinical trial comparing transdermal (patch) nicotine replacement therapy (NRT) to nicotine nasal spray NRT. The studies had similar designs, making subjects comparable for analysis. We limited our analyses to individuals who self-identified as white non-Hispanic race/ethnicity (n=411 and 378, respectively) to avoid the potential confounding and heterogeneity of effect estimates arising from differential linkage disequilibrium across ethnic groups.
In both studies, potential participants were smokers ≥ 18 years old who reported smoking ≥ 10 cigarettes per day over the prior 12 months. Through a medical and psychiatric screening, slightly different exclusion criteria were applied in each study and are detailed elsewhere.25 Treatment included 7 group behavioral counseling sessions plus study medication. Participants were instructed to quit smoking on their target quit date (TQD) which occurred 1–2 weeks after pre-quit counseling. Participants in the Bupropion study initiated treatment during the first week of the study period. Those in the NRT study began treatment at TQD. In both studies we focused on smoking abstinence at two endpoints: (1) End of treatment (EOT) assessed 8 weeks post-TQD and (2) 6-months post-TQD. For both endpoints, those self-reporting smoking abstinence for the 7 days prior to assessment were biochemically verified using saliva cotinine concentrations measured by gas-liquid chromatography.34 Subjects who self-reported abstinence over that prior week and had cotinine levels ≤ 15 ng/ml were classified as abstinent.24, 25
A candidate gene study was carried out as part of the Pharmacogenetics of Nicotine Addiction Treatment Consortium. We investigated previously genotyped SNPs within 53 genes involved in nicotine metabolism and the brain-reward pathway including nicotinic acetylcholine receptors, and dopamine candidate genes (see Supplemental Tables for complete list of genes and SNPs). Selection of the 1,528 SNPs has been described previously.24, 25, 35, 36
Among the 1,528 total SNPs, we excluded 41 SNPs with a call rate of zero and 57 SNPs with MAFs < 0.01, leaving 1,198 SNPs (118 with putative functional evidence, 1080 to capture underlying LD structure) within the 53 gene regions for association analysis and 232 ancestry informative markers (AIMs). Analysis of these AIMs using STRUCTURE37 showed negligible admixture in our self-identified white non-Hispanic study population, confirming their genetic homogeneity. Self-reported gender was verified through 41 SNPs located within X-chromosome genes. We estimated heterozygosity and excluded four individuals who self-reported male but were heterozygous for more than 30% of the X-chromosome SNPs. A complete description of the quality control procedures has been previously published.24
Imputation was carried out on selected gene regions using the haplotypes from the European populations (EUR) from the August release of the 1000 Genomes Project38 and the program IMPUTE2.39, 40 Ten regions with multiple associated SNPs or a priori evidence of association (EPB41, CHRNB2, CNR1, the CHRNA2, CHRNB3;CHRNA6 region, the ANKK1;DRD2 region, CHRNA7, the chromosome 15 nAChR CHRNA5;CHRNA3;CHRNB4 region, CHRNA4, MAPK1) were targeted and expanded up- and downstream 50 kb to encompass potential SNPs in LD with those lying in respective regions. Using the 245 SNPs across those regions, we imputed an additional 7957 SNPs. We excluded 1716 imputed SNPs with imputation certainty scores < 0.9. Based on best genotype calls, we excluded 4232 imputed SNPs with an observed MAF < 0.01, and an additional 7 with a P-value < 0.0001 from an exact test of Hardy-Weinberg proportions, leaving 2002 imputed SNPs for additional analysis. LocusZoom was used to plot P-values from the analysis of these expanded regions.41
Statistical analysis
SNP association
To estimate gender-stratified SNP associations at EOT and 6-month follow up, we pooled both studies, and used logistic regression to estimate odds ratios for marginal SNP effects and SNP × treatment interaction effects. For each SNP we tested either an additive or dominant genetic model obtained from previously reported analyses.24 The most common genotype served as the referent. We adjusted for study and treatment when estimating marginal effects, and used dummy variables indicating treatment assignment to assess within treatment effects of bupropion, patch, or spray vs. placebo. For all models we adjusted for age and the Fagerström Test for Nicotine Dependence.42 For gender-stratified marginal SNP effects we performed a 1-df LRT within each gender to identify significant SNPs. For gender-specific gene-treatment interaction effects, we performed a 3-df LRT of gene-treatment interaction terms for each gender analysis. We did not test the treatment-specific effects within each gender. Analyses were performed using the R Statistical Program.43
Correcting for correlated tests within a gene and determining system-level significance
Marginal 1-df LRT P-values were adjusted to account for the correlation and the number of tests performed across the SNPs within a gene region.24, 44 The 3-df LRT P-values for the gene-treatment interaction were adjusted using an extension of the approach from Conneely and Boehnke44 in which observed test statistics were compared to their asymptotic distribution through numerical integration. We used the correlation of individual contributions to score statistics for the 3-df tests to approximate the correlation structure. This approach performs similarly to computationally intensive permutation tests.45 P-values are reported for each SNP adjusted for the number of correlated SNPs within each gene region. These adjusted P-values achieved significance within a gene region (i.e., region-wide significance) at an α-level of 0.05. Overall significance was determined using an additional Bonferroni correction across the 53 gene regions and two genders. This gives a system-wide α-level of 0.05/(53×2)=5×10−4 to determine the significance of adjusted P-values in four independent tests: marginal effects at EOT, marginal effects at 6-month follow-up, interaction effects at EOT, and interaction effects at 6-month follow-up. As a conservative benchmark, study-wise significance across the 53 independent gene regions, two outcomes of interest, two genders, and two sets of hypotheses tested (gender-stratified marginal tests and gender-stratified SNP-treatment interactions) yields a α-level of 0.05/(53*2*2*2) = 1.1×10−4.
RESULTS
Our analyses were restricted to subjects that self-identified and were confirmed European ancestry (Table 1). Fagerström Test for Nicotine Dependence (FTND) scores for women (5.1, SD=2.2) were lower compared with men (5.6, SD=2.1). Across all treatments, at EOT, 26% of women were abstinent compared with 34% of men (OR=0.7, 95% CI 0.5–0.9). At 6-month follow-up, 19% of women were abstinent compared with 23% of men (OR=0.82, 95% CI 0.6–1.2). However, within the spray treatment arm, a slightly higher proportion of women remained abstinent (OR=1.1, 95% CI 0.5–2.3).
Table 1.
Study Characteristics
| Male N (%) |
Female N (%) |
|
|---|---|---|
| 386 | 403 | |
| Age | 46.1 ± 10.8 | 44.8 ± 12.1 |
| Treatment | ||
| Placebo | 88 (23%) | 106 (26%) |
| Bupropion | 97 (25%) | 120 (30%) |
| Patch | 107 (28%) | 81 (20%) |
| Spray | 94 (24%) | 96 (24%) |
| FTNDa | ||
| 0 | 2 (1%) | 6 (1%) |
| 1 | 13 (3%) | 22 (5%) |
| 2 | 18 (5%) | 28 (7%) |
| 3 | 32 (8%) | 37 (9%) |
| 4 | 54 (14%) | 55 (14%) |
| 5 | 63 (16%) | 70 (17%) |
| 6 | 62 (16%) | 69 (17%) |
| 7 | 68 (18%) | 58 (14%) |
| 8 | 41 (11%) | 40 (10%) |
| 9 | 26 (7%) | 14 (3%) |
| 10 | 7 (2%) | 4 (1%) |
| Mean | 5.6 ± 2.1 | 5.1 ± 2.2 |
| End of treatment - Abstinent | ||
| All treatments | 130 (34%) | 105 (26%) |
| Placebo | 24 (27%) | 18 (17%) |
| Bupropion | 35 (36%) | 35 (29%) |
| Patch | 42 (39%) | 25 (31%) |
| Spray | 29 (31%) | 27 (28%) |
| 6-month Follow-up - Abstinent | ||
| All treatments | 87 (23%) | 78 (19%) |
| Placebo | 16 (18%) | 18 (17%) |
| Bupropion | 27 (28%) | 29 (24%) |
| Patch | 26 (24%) | 11 (14%) |
| Spray | 18 (19%) | 20 (21%) |
All subjects were confirmed European ancestry.
Fagerström test for nicotine dependence
Associations with abstinence at EOT and 6-month follow-up
Gender stratified SNP associations are reported in Table 2. Gender stratified SNP × treatment associations at EOT and 6-month follow-up are reported in Tables 3 and 4, respectively. Results are presented for males and females for all SNPs with adjusted P-values < 0.05 for either gender-stratified analysis.
Table 2.
Gender stratified marginal single nucleotide polypmorphism (SNP) results for abstinence rates
| SNP | Gene Region | Chromosome | Male | Female | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAF | SNP OR (95% CT) | Observed P | Adjusted Pa | MAF | SNP OR (95% CI) | Observed P | Adjusted Pa | |||
| Significant SNPs within Males | ||||||||||
| End of treatment | ||||||||||
| rs6702335b | EPB41 | 1 | 0.39 | 0.46 (0.32–0.64) | 2.30 × 10−6 | 3.27 × 10−5 | 0.39 | 0.93 (0.67–1.30) | 6.67 × 10−1 | 1 |
| rs12021667b | EPB41 | 1 | 0.4 | 0.45 (0.32–0.64) | 2.01 × 10−6 | 3.65 × 10−5 | 0.39 | 0.96 (0.69–1.34) | 8.15 × 10−1 | 1 |
| rs12027267b | EPB41 | 1 | 0.4 | 0.46 (0.33–0.64) | 2.38 × 10−6 | 4.04 × 10−5 | 0.39 | 0.95 (0.68–1.33) | 7.62 × 10−1 | 1 |
| rs12039988b | EPB41 | 1 | 0.39 | 0.47 (0.33–0.65) | 3.40 × 10−6 | 5.77 × 10−5 | 0.39 | 1.00 (0.72–1.40) | 9.97 × 10−1 | 9.97 × 10−1 |
| rs203278b | EPB41 | 1 | 0.35 | 2.27 (1.44–3.60) | 3.30 × 10−4 | 5.57 × 10−3 | 0.34 | 0.79 (0.50–1.25) | 3.20 × 10−1 | 9.74 × 10−1 |
| rs150089 | EPB41 | 1 | 0.32 | 1.71 (1.23–2.39) | 1.48 × 10−3 | 2.20 × 10−2 | 0.31 | 0.86 (0.61–1.23) | 4.09 × 10−1 | 9.91 × 10−1 |
| rs2985322 | EPB41 | 1 | 0.32 | 1.71 (1.23–2.39) | 1.48 × 10−3 | 2.22 × 10−2 | 0.31 | 0.87 (0.61–1.24) | 4.38 × 10−1 | 9.94 × 10−1 |
| rs578776 | CHRNA3;CHRNA5 | 15 | 0.24 | 1.75 (1.21–2.54) | 2.86 × 10−3 | 2.85 × 10−2 | 0.21 | 0.94 (0.63–1.39) | 7.47 × 10−1 | 9.99 × 10−1 |
| rs4654390 | EPB41 | 1 | 0.5 | 0.62 (0.45–0.85) | 2.48 × 10−3 | 3.50 × 10−2 | 0.49 | 0.98 (0.72–1.34) | 8.97 × 10−1 | 1 |
| rs10915216 | EPB41 | 1 | 0.5 | 0.62 (0.45–0.85) | 2.68 × 10−3 | 3.72 × 10−2 | 0.49 | 0.97 (0.71–1.33) | 8.70 × 10−1 | 1 |
| 6-month Follow-up | ||||||||||
| rs6702335b | EPB41 | 1 | 0.39 | 0.46 (0.31–0.67) | 3.44 × 10−5 | 6.97 × 10−4 | 0.39 | 1.11 (0.77–1.59) | 5.87 × 10−1 | 1 |
| rs12027267b | EPB41 | 1 | 0.4 | 0.46 (0.31–0.68) | 3.89 × 10−5 | 7.95 × 10−4 | 0.39 | 1.17 (0.81–1.69) | 3.97 × 10−1 | 9.91 × 10−1 |
| rs12021667b | EPB41 | 1 | 0.4 | 0.48 (0.32–0.70) | 8.25 × 10−5 | 1.61 × 10−3 | 0.39 | 1.16 (0.80–1.66) | 4.37 × 10−1 | 9.96 × 10−1 |
| rs12039988b | EPB41 | 1 | 0.39 | 0.40 (0.24–0.66) | 2.89 × 10−4 | 5.26 × 10−3 | 0.39 | 1.53 (0.89–2.63) | 1.20 × 10−1 | 7.83 × 10−1 |
| rs2238687 | FOSB | 19 | 0.14 | 2.17 (1.33–3.55) | 2.42 × 10−3 | 1.48 × 10−2 | 0.14 | 1.57 (0.99–2.49) | 6.30 × 10−2 | 2.62 × 10−1 |
| rs11101694 | CALY | 10 | 0.13 | 0.42 (0.21–0.85) | 1.01 × 10−2 | 3.74 × 10−2 | 0.12 | 0.84 (0.44–1.59) | 5.89 × 10−1 | 5.89 × 10−1 |
| rs16837840 | EPB41 | 1 | 0.11 | 2.40 (1.38–4.17) | 2.46 × 10−3 | 3.78 × 10−2 | 0.1 | 0.70 (0.35–1.41) | 3.01 × 10−1 | 9.71 × 10−1 |
|
| ||||||||||
| Significant SNPs within Females | ||||||||||
| End of treatment | ||||||||||
| rs4809549 | CHRNA4 | 20 | 0.47 | 1.00 (0.74–1.35) | 9.96 × 10−1 | 9.96 × 10−1 | 0.48 | 0.59 (0.42–0.83) | 1.99 × 10−3 | 1.69 × 10−2 |
| rs7123797 | ANKK1 | 11 | 0.34 | 0.95 (0.62–1.46) | 8.23 × 10−1 | 1 | 0.34 | 0.46 (0.29–0.73) | 8.58 × 10−4 | 1.86 ×10−2 |
| rs4938012 | ANKK1 | 11 | 0.33 | 0.97 (0.63–1.48) | 8.74 × 10−1 | 1 | 0.33 | 0.47 (0.29–0.74) | 1.09 × 10−3 | 2.33 × 10−2 |
| rs17115439 | ANKK1 | 11 | 0.33 | 0.94 (0.61–1.44) | 7.67 × 10−1 | 1 | 0.33 | 0.47 (0.30–0.75) | 1.45 × 10−3 | 3.02 × 10−2 |
| rs4938015 | ANKK1 | 11 | 0.33 | 0.94 (0.61–1.44) | 7.67 × 10−1 | 1 | 0.33 | 0.47 (0.30–0.75) | 1.49 × 10−3 | 3.10 × 10−2 |
| 6-month Follow-up | ||||||||||
| rs3766927 | CHRNB2 | 1 | 0.31 | 0.86 (0.53–1.39) | 5.30 × 10−1 | 7.77 × 10−1 | 0.29 | 0.45 (0.26–0.76) | 2.24 × 10−3 | 1.92 × 10−2 |
| rs1127314 | CIIRNB2 | 1 | 0.31 | 0.88 (0.62–1.27) | 4.99 × 10−1 | 7.99 × 10−1 | 0.29 | 0.53 (0.34–0.82) | 2.48 × 10−3 | 2.07 × 10−2 |
| rs2834600 | CLIC6 | 21 | 0.13 | 0.89 (0.51–1.57) | 6.93 × 10−1 | 9.87 × 10−1 | 0.14 | 2.29 (1.34–3.91) | 2.86 × 10−3 | 3.08 × 10−2 |
| rs17759598 | MAPK1 | 22 | 0.17 | 0.87 (0.55–1.39) | 5.67 × 10−1 | 9.82 × 10−1 | 0.14 | 0.44 (0.23–0.83) | 5.60 × 10−3 | 3.71 × 10−2 |
| rs2131902 | CHRMB2 | 1 | 0.32 | 0.82 (0.50–1.33) | 4.20 × 10−1 | 8.24 × 10−1 | 0.3 | 0.49 (0.29–0.82) | 5.66 × 10−3 | 4.33 × 10−2 |
All models were adjusted for age, FTND, and treatment.
SNPs above the dashed line have adjusted P-values < 0.05 within males. SNPs below the dashed line have adjusted P-values < 0.05 within females.
P-values adjusted for the number of correlated tests within respective gene regions.
Adjusted SNP × Gender Interaction P-value < 0.05.
Table 3.
Gender stratified SNP × treatment interaction results at end of treatment
| SNP | Gene Region | Gender | MAF | Placebo | Bupropion | Patch | Spray | Test of Interactiona | |
|---|---|---|---|---|---|---|---|---|---|
| SNP OR (95% CI) | SNP OR (95% CI) | SNP OR (95% CI) | SNP OR (95% CI) | Observed P | Adjusted P | ||||
| Significant SNPs within Males | |||||||||
| rs806365b | CNR1 | Male | 0.46 | 1.38 (0.44–4.30) | 2.31 (0.77–6.96) | 3.07 (1.11–8.47) | 0.17 (0.07–0.45) | 7.01 × 10−5 | 1.05 ×10−3 |
| Female | 0.44 | 0.40 (0.14–1.13) | 0.64 (0.28–1.49) | 0.67 (0.25–1.84) | 1.82 (0.63–5.24) | 2.15 × 10−1 | 7.70 × 10−1 | ||
|
| |||||||||
| rs806369b | CNR1 | Male | 0.31 | 3.53 (1.23–10.09) | 1.33 (0.57–3.10) | 3.11 (1.33–7.27) | 0.30 (0.11–0.79) | 5.56 × 10−4 | 7.08 × 10−3 |
| Female | 0.3 | 0.35 (0.11–1.06) | 0.87 (0.39–1.93) | 0.68 (0.26–1.80) | 1.83 (0.73–4.57) | 1.29 × 10−1 | 6.27 × 10−1 | ||
|
| |||||||||
| rs10107450 | CIIRNA6 | Male | 0.23 | 2.11 (0.81–5.49) | 2.43 (1.02–5.80) | 0.34 (0.14–0.80) | 0.62 (0.25–1.53) | 3.09 × 10−3 | 9.66 × 10−3 |
| Female | 0.22 | 0.69 (0.23–2.03) | 0.87 (0.39–1.95) | 0.72 (0.27–1.92) | 0.62 (0.23–1.69) | 9.61 × 10−1 | 9.61 × 10−1 | ||
|
| |||||||||
| rs7017612 | CHRNA6;CHRNB3 | Male | 0.21 | 0.91 (0.33–2.47) | 2.27 (0.96–5.34) | 0.24 (0.10–0.59) | 0.92 (0.37–2.28) | 3.94 × 10−3 | 1.16 × 10−2 |
| Female | 0.18 | 0.68 (0.22–2.13) | 0.61 (0.25–1.47) | 0.80 (0.29–2.21) | 0.43 (0.14–1.29) | 8.66 × 10−1 | 9.70 × 10−1 | ||
|
| |||||||||
| rs878887 | CRHR1 | Male | 0.22 | 2.25 (1.07–4.74) | 2.03 (0.98–4.23) | 0.40 (0.19–0.85) | 0.68 (0.31–1.50) | 1.24 × 10−3 | 1.31 × 10−2 |
| Female | 0.2 | 0.30 (0.09–1.07) | 1.07 (0.47–2.44) | 0.57 (0.23–1.39) | 0.84 (0.37–1.94) | 3.40 × 10−1 | 5.82 × 10−1 | ||
|
| |||||||||
| rs2072661 | CHRNB2 | Male | 0.26 | 0.26 (0.09–0.74) | 0.33 (0.13–0.80) | 1.30 (0.59–2.85) | 2.14 (0.87–5.24) | 2.00 × 10−3 | 1.68 × 10−2 |
| Female | 0.22 | 0.71 (0.23–2.20) | 0.54 (0.22–1.29) | 1.35 (0.51–3.57) | 0.37 (0.14–1.00) | 3.02 × 10−1 | 7.64 × 10−1 | ||
|
| |||||||||
| rs2304297 | CHRNA6 | Male | 0.24 | 2.11 (0.81–5.49) | 2.30 (0.97–5.45) | 0.44 (0.20–1.01) | 0.59 (0.24–1.48) | 1.17 × 10−2 | 2.86 × 10−2 |
| Female | 0.22 | 0.88 (0.31–2.52) | 0.96 (0.43–2.14) | 0.72 (0.27–1.92) | 0.55 (0.20–1.48) | 8.42 × 10−1 | 9.60 × 10−1 | ||
|
| |||||||||
| rs11575553 | DDC | Male | 0.1 | 2.27 (0.74–6.94) | 15.33 (3.18–74.04) | 0.67 (0.22–2.11) | 0.88 (0.32–2.42) | 1.74 × 10−3 | 2.89 × 10−2 |
| Female | 0.11 | 0.32 (0.07–1.50) | 1.59 (0.61–4.16) | 1.31 (0.42–4.12) | 0.71 (0.20–2.46) | 2.68 × 10−1 | 9.28 × 10−1 | ||
|
| |||||||||
| rs2267366 | PRKCABP | Male | 0.13 | 1.17 (0.43–3.15) | 0.56 (0.23–1.36) | 0.53 (0.21–1.38) | 4.39 (1.61–11.94) | 5.38 × 10−1 | 2.93 × 10−2 |
| Female | 0.16 | 1.39 (0.57–3.42) | 0.85 (0.41–1.77) | 0.36 (0.08–1.57) | 1.25 (0.54–2.92) | 3.41 × 10−1 | 7.06 × 10−1 | ||
|
| |||||||||
| rs1876828 | CRHR1 | Male | 0.22 | 2.19 (1.01–4.74) | 2.07 (0.98–4.38) | 0.45 (0.21–0.96) | 0.63 (0.26–1.54) | 3.34 × 10−3 | 3.20 × 10−2 |
| Female | 0.21 | 0.30 (0.09–1.08) | 0.89 (0.38–2.08) | 0.49 (0.19–1.28) | 0.90 (0.39–2.09) | 3.90 × 10−1 | 6.18 × 10−1 | ||
|
| |||||||||
| rs356165 | SNCA | Male | 0.38 | 0.86 (0.30–2.45) | 0.95 (0.39–2.30) | 2.50 (1.03–6.06) | 0.22 (0.08–0.61) | 4.54 × 10−3 | 4.27 × 10−2 |
| Female | 0.41 | 0.93 (0.29–2.95) | 2.38 (0.87–6.54) | 1.21 (0.42–3.52) | 0.88 (0.34–2.28) | 4.88 × 10−1 | 9.75 × 10−1 | ||
|
| |||||||||
| rs17690326 | CRHR1 | Male | 0.22 | 2.02 (0.96–4.23) | 1.58 (0.78–3.19) | 0.41 (0.19–0.85) | 0.67 (0.31–1.47) | 5.53 × 10−3 | 4.84 × 10−2 |
| Female | 0.2 | 0.30 (0.08–1.05) | 1.09 (0.48–2.49) | 0.48.0 (20–1.18) | 0.85 (0.37–1.94) | 2.61 × 10−1 | 4.78 × 10−1 | ||
|
| |||||||||
| Significant SNPs within Females | |||||||||
| rs950776 | CHRNB4 | Male | 0.33 | 1.65 (0.64–4.24) | 1.37 (0.56–3.31) | 0.69 (0.31–1.53) | 0.55 (0.23–1.35) | 2.62 × 10−1 | 7.68 × 10−1 |
| Female | 0.35 | 1.71 (0.58–5.02) | 0.38 (0.17–0.87) | 0.49 (0.18–1.30) | 3.61 (1.34–9.74) | 1.52 × 10−3 | 1.09 × 10−2 | ||
|
| |||||||||
| rs871058 | CHRNA5 | Male | 0.33 | 1.21 (0.47–3.11) | 0.89 (0.37–2.12) | 0.69 (0.31–1.53) | 0.41 (0.17–1.01) | 4.07 × 10−1 | 9.17 × 10−2 |
| Female | 0.37 | 1.31 (0.45–3.86) | 0.24 (0.10–0.55) | 0.59 (0.22–1.56) | 2.44 (0.95–6.28) | 1.78 × 10−3 | 1.24 × 10−1 | ||
|
| |||||||||
| rs514743 | CHRNA3;CHRNA5 | Male | 0.35 | 1.00 (0.39–2.55) | 1.16 (0.49–2.77) | 0.49 (0.21–1.09) | 0.51 (0.21–1.25) | 3.80 × 10−1 | 8.98 × 10−1 |
| Female | 0.39 | 1.07 (0.36–3.15) | 0.35 (0.15–0.81) | 0.69 (0.26–1.83) | 3.96 (1.40–11.23) | 2.79 × 10−3 | 1.87 × 10−2 | ||
|
| |||||||||
| rs9610375 | MAPK1 | Male | 0.49 | 0.78 (0.26–2.37) | 0.87 (0.33–2.29) | 0.76 (0.31–1.84) | 0.59 (0.20–1.76) | 9.65 × 10−1 | 9.65 × 10−1 |
| Female | 0.48 | 0.56 (0.19–1.64) | 0.38 (0.16–0.88) | 6.54 (1.38–30.93) | 0.66 (0.25–1.71) | 3.48 × 10−3 | 2.01 × 10−2 | ||
|
| |||||||||
| rs4275821 | CHRNA5 | Male | 0.35 | 1.25 (0.48–3.24) | 0.89 (0.37–2.12) | 0.71 (0.32–1.57) | 0.47 (0.19–1.16) | 5.14 × 10−1 | 9.67 × 10−1 |
| Female | 0.38 | 1.10 (0.37–3.23) | 0.27 (0.12–0.62) | 0.54 (0.20–1.44) | 2.58 (0.98–6.79) | 3.49 × 10−3 | 2.29 × 10−2 | ||
|
| |||||||||
| 18737865b | COMT | Male | 0.31 | 0.70 (0.34–1.44) | 1.13 (0.55–2.31) | 0.72 (0.38–1.33) | 2.11 (1.11–4.01) | 5.70 × 10−2 | 4.22 × 10−1 |
| Female | 0.28 | 1.25 (0.58–2.70) | 1.86 (0.92–3.73) | 0.64 (0.30–1.36) | 0.28 (0.12–0.67) | 3.23 × 10−3 | 3.71 × 10−2 | ||
|
| |||||||||
| rs3743075 | CHRNA3 | Male | 0.35 | 1.27 (0.49–3.25) | 1.09 (0.46–2.61) | 0.52 (0.23–1.17) | 0.51 (0.21–1.25) | 3.37 × 10−1 | 8.60 × 10−1 |
| Female | 0.38 | 1.11 (0.37–3.26) | 0.32 (0.14–0.73) | 0.50 (0.19–1.34) | 2.66 (1.01–7.02) | 6.41 × 10−3 | 3.92 × 10−2 | ||
|
| |||||||||
| rs1063311 | MAPK1 | Male | 0.46 | 0.83 (0.43–1.62) | 0.84 (0.46–1.52) | 1.37 (0.77–2.45) | 0.73 (0.36–1.46) | 4.82 × 10−1 | 8.10 × 10−1 |
| Female | 0.43 | 0.77 (0.38–1.56) | 0.61 (0.34–1.09) | 3.08 (1.36–6.98) | 0.97 (0.53–1.80) | 8.76 × 10−3 | 4.44 × 10−2 | ||
|
| |||||||||
| rs2266966 | MAPK1 | Male | 0.46 | 0.83 (0.43–1.62) | 0.80 (0.44–1.45) | 1.38 (0.77–2.45) | 0.73 (0.36–1.46) | 4.56 × 10−1 | 7.89 × 10−1 |
| Female | 0.43 | 0.81 (0.40–1.62) | 0.60 (0.33–1.07) | 3.08 (1.36–6.98) | 0.97 (0.53–1.80) | 8.72 × 10−3 | 4.44 × 10−2 | ||
All models were adjusted for age and FTND.
SNPs above the dashed line have adjusted P-values < 0.05 within males. SNPs below the dashed line have adjusted P-values < 0.05 within females.
Observed P-values were obtained using a 3-df test of the gene-treatment interaction effect. P-values were adjusted for the correlation of individual contributions to score statistics for the 3-df tests within respective gene regions.
Adjusted SNP × Treatment × Gender Interaction P-value < 0.05.
Table 4.
Gender stratified SNP × treatment interaction results at 6-month follow-up
| SNP | Gene Region | Gender | MAF | Placebo | Bupropion | Patch | Spray | Test of Interactiona | |
|---|---|---|---|---|---|---|---|---|---|
| SNP OR (95% CI) | SNP OR (95% CI) | SNP OR (95% CI) | SNP OR (95% CI) | Observed P | Adjusted P | ||||
| Significant SNPs within Males | |||||||||
| rs806365b | CNR1 | Male | 0.46 | 1.06 (0.30–3.75) | 2.05 (0.62–6.78) | 2.41 (0.74–7.84) | 0.04 (0.01–0.21) | 3.76 × 10−6 | 3.86 × 10−5 |
| Female | 0.44 | 0.30 (0.11–0.87) | 0.80 (0.33–1.92) | 0.53 (0.14–1.95) | 1.32 (0.43–4.12) | 2.66 × 10−1 | 7.34 × 10−1 | ||
|
| |||||||||
| rs684513 | CHRNA5 | Male | 0.17 | 1.56 (0.51–4.74) | 0.57 (0.19–1.74) | 4.00 (1.53–10.42) | 0.11 (0.02–0.50) | 7.74 × 10−5 | 6.98 × 10−4 |
| Female | 0.15 | 1.56 (0.51–4.72) | 1.84 (0.71–4.73) | 1.19 (0.31–4.50) | 1.59 (0.55–4.59) | 9.64 × 10−1 | 9.64 × 10−1 | ||
|
| |||||||||
| rs4887069 | CHRNA3:CHRNB4 | Male | 0.2 | 1.83 (0.72–4.61) | 0.62 (0.22–1.72) | 2.63 (1.24–5.55) ) | 0.17 (0.04–0.61) | 2.83 × 10−4 | 2.30 × 10−3 |
| Female | 0.17 | 1.22 (0.46–3.27) | 0.91 (0.39–2.15) | 1.51 (0.46–5.01 | 0.69 (0.29–1.66) | 7.19 × 10−1 | 9.73 × 10−1 | ||
|
| |||||||||
| rs938682 | CHRNA3 | Male | 0.2 | 2.10 (0.79–5.57) | 0.62 (0.23–1.72) | 2.24 (1.07–4.67) | 0.17 (0.04–0.61) | 5.43 × 10−4 | 4.20 × 10−3 |
| Female | 0.16 | 1.26 (0.47–3.36) | 1.33 (0.58–3.06) | 1.08 (0.31–3.79) | 0.75 (0.32–1.78) | 7.88 × 101 | 9.48 × 10−1 | ||
|
| |||||||||
| rs637137 | CHRNA5 | Male | 0.2 | 1.85 (0.70–4.93) | 0.67 (0.24–1.86) | 2.38 (1.14–4.97) | 0.17 (0.04–0.61) | 6.30 × 10−4 | 4.81 × 10−3 |
| Female | 0.16 | 1.54 (0.60–3.98) | 1.33 (0.58–3.06) | 1.17 (0.31–4.43) | 0.73 (0.32–1.71) | 6.51 × 10−1 | 9.76 × 10−1 | ||
|
| |||||||||
| rs3743078 | CHRNA3;CHRNA5 | Male | 0.21 | 1.97 (0.81–4.81) | 0.71 (0.27–1.87) | 2.24 (1.07–4.67) | 0.17 (0.04–0.61) | 6.79 × 10−4 | 5.13 × 10−3 |
| Female | 0.16 | 1.13 (0.42–3.03) | 1.33 (0.58–3.06) | 1.17 (0.31–4.43) | 0.75 (0.32–1.78) | 8.07 × 10−1 | 9.59 × 10−1 | ||
|
| |||||||||
| rs667282 | CHRNA5 | Male | 0.2 | 1.85 (0.69–4.92) | 0.67 (0.24–1.86) | 2.32 (1.11–4.86) | 0.17 (0.04–0.61) | 7.10 × 10−4 | 5.30 × 10−3 |
| Female | 0.16 | 1.54 (0.60–3.98) | 1.33 (0.58–3.06) | 1.17 (0.31–4.43) | 0.73 (0.32–1.71) | 6.51 × 10−1 | 9.76 × 10−1 | ||
|
| |||||||||
| rs11954565 | DRD1 | Male | 0.34 | 2.77 (0.81–9.47) | 1.43 (0.57–3.59) | 0.59 (0.23–1.48) | 0.09 (0.02–0.43) | 5.83 × 10−4 | 6.66 × 10−3 |
| Female | 0.36 | 3.12 (0.84–11.64) | 1.76 (0.73–4.23) | 1.20 (0.32–4.54) | 1.11 (0.41–3.01) | 6.08 × 10−1 | 9.42 × 10−1 | ||
|
| |||||||||
| rs806369b | CNR1 | Male | 0.31 | 2.16 (0.97–4.79) | 1.31 (0.64–2.71) | 1.68 (0.82–3.42) | 0.14 (0.03–0.63) | 7.65 × 10−4 | 6.80 × 10−3 |
| Female | 0.3 | 0.55 (0.23–1.32) | 0.80 (0.43–1.49) | 0.45 (0.13–1.52) | 1.42 (0.63–3.22) | 3.14 × 10−1 | 7.88 × 10−1 | ||
|
| |||||||||
| rs6495309 | CHRNA3;CHRNB4 | Male | 0.19 | 2.19 (0.83–5.81) | 0.67 (0.24–1.86) | 1.98 (0.87–4.53) | 0.17 (0.05–0.61) | 1.37 × 10−3 | 9.26 × 10−3 |
| Female | 0.15 | 1.45 (0.54–3.86) | 0.96 (0.40–2.31) | 0.81 (0.21–3.16) | 0.80 (0.33–1.92) | 8.33 × 10−1 | 9.55 × 10−1 | ||
|
| |||||||||
| rs934778 | POMC | Male | 0.32 | 1.73 (0.54–5.53) | 2.35 (0.92–5.99) | 0.21 (0.07–0.61) | 0.85 (0.30–2.42) | 2.85 × 10−3 | 1.51 × 10−2 |
| Female | 0.34 | 0.94 (0.34–2.60) | 3.11 (1.20–8.01) | 2.45 (0.60–10.09) | 0.84 (0.31–2.29) | 1.75 × 10−1 | 4.35 × 10−1 | ||
|
| |||||||||
| rs708905b | FOSB | Male | 0.14 | 0.14 (0.02–1.17) | 2.52 (0.91–7.02) | 0.27 (0.07–0.98) | 1.29 (0.42–3.93) | 5.68 × 10−3 | 2.54 × 10−2 |
| Female | 0.13 | 0.72 (0.21–2.40) | 0.72 (0.24–2.15) | 3.14 (0.74–13.29) | 0.37 (0.10–1.39) | 1.91 × 10−1 | 4.04 × 10−1 | ||
|
| |||||||||
| rs743409b | MAPK1 | Male | 0.44 | 0.37 (0.15–0.94) | 1.22 (0.64–2.35) | 0.97 (0.52–1.80) | 2.95 (1.33–6.54) | 5.47 × 10−3 | 2.55 × 10−2 |
| Female | 0.47 | 2.10 (1.02–4.33) | 0.81 (0.46–1.42) | 1.57 (0.63–3.96) | 1.05 (0.55–2.00) | 1.84 × 10−1 | 4.72 × 10−1 | ||
|
| |||||||||
| rs2267366 | PRKCABP | Male | 0.13 | 0.61 (0.16–2.28) | 0.53 (0.20–1.41) | 0.73 (0.25–2.12) | 5.32 (1.80–15.74) | 6.33 × 10−3 | 2.64 × 10−2 |
| Female | 0.16 | 0.86 (0.33–2.29) | 0.76 (0.34–1.70) | 0.47 (0.07–3.33) | 0.56 (0.19–1.63) | 9.03 × 10−1 | 9.03 × 10−1 | ||
|
| |||||||||
| rs1892848b | MAPK1 | Male | 0.45 | 0.35 (0.14–0.90) | 1.14 (0.60–2.18) | 0.97 (0.52–1.81) | 2.79 (1.23–6.33) | 7.26 × 10−3 | 3.24 × 10−2 |
| Female | 0.47 | 2.29 (1.09–4.80) | 0.79 (0.45–1.40) | 1.57 (0.63–3.96) | 1.05 (0.55–2.00) | 1.27 × 10−1 | 3.52 × 10−1 | ||
|
| |||||||||
| rs3788332b | MAPK1 | Male | 0.45 | 0.35 (0.14–0.90) | 1.14 (0.60–2.18) | 0.97 (0.52–1.81) | 2.79 (1.23–6.33) | 7.26 × 10−3 | 3.25 × 10−2 |
| Female | 0.47 | 2.38 (1.12–5.02) | 0.77 (0.43–1.37) | 1.57 (0.63–3.92) | 1.05 (0.55–2.00) | 9.92 × 10−2 | 2.88 × 10−1 | ||
|
| |||||||||
| rs5999550b | MAPK1 | Male | 0.45 | 0.35 (0.14–0.90) | 1.14 (0.60–2.18) | 0.97 (0.52–1.81) | 2.79 (1.23–6.33) | 7.26 × 10−3 | 3.25 × 10−2 |
| Female | 0.47 | 2.29 (1.09–4.80) | 0.79 (0.45–1.40) | 1.57 (0.63–3.96) | 1.05 (0.55–2.00) | 1.27 × 10−1 | 3.52 × 10−1 | ||
|
| |||||||||
| rs11704205b | MAPK1 | Male | 0.45 | 0.35 (0.14–0.90) | 1.14 (0.60–2.18) | 0.97 (0.52–1.81) | 2.79 (1.23–6.33) | 7.26 × 10−3 | 3.26 × 10−2 |
| Female | 0.47 | 2.29 (1.09–4.80) | 0.79 (0.45–1.40) | 1.57 (0.63–3.96) | 1.05 (0.55–2.00) | 1.27 × 10−1 | 3.52 × 10−1 | ||
|
| |||||||||
| rs2797855 | DBH | Male | 0.4 | 1.91 (0.56–6.57) | 0.17 (0.06–0.45) | 0.41 (0.16–1.02) | 1.32 (0.45–3.81) | 4.14 × 10−3 | 4.74 × 10−2 |
| Female | 0.45 | 0.91 (0.29–2.84) | 1.39 (0.57–3.40) | 0.77 (0.20–2.94) | 0.31 (0.11–0.87) | 1.86 × 10−1 | 7.44 × 10−1 | ||
|
| |||||||||
| Significant SNPs within Females | |||||||||
| rs10423854b | FOSB | Male | 0.43 | 1.12 (0.35–3.61) | 1.05 (0.41–2.72) | 0.38 (0.15–0.96) | 1.78 (0.57–5.54) | 1.69 × 10−1 | 4.26 × 10−1 |
| Female | 0.41 | 0.19 (0.06–0.55) | 0.48 (0.20–1.14) | 6.33 (0.76–52.37) | 1.14 (0.40–3.19) | 2.65 × 10−3 | 1.34 × 10−2 | ||
|
| |||||||||
| rs950776 | CHRNB4 | Male | 0.33 | 1.28 (0.43–3.82) | 1.22 (0.47–3.16) | 0.75 (0.30–1.85) | 0.73 (0.26–2.06) | 7.77 × 10−1 | 8.67 × 10−1 |
| Female | 0.35 | 3.29 (1.00–10.88) | 0.41 (0.17–0.97) | 0.38 (0.10–1.44) | 2.46 (0.85–7.11) | 3.42 × 10−3 | 2.11 × 10−2 | ||
|
| |||||||||
| rs7178270 | CHRNA3;CHRNB4 | Male | 0.37 | 0.97 (0.33–2.88) | 1.45 (0.53–3.94) | 0.79 (0.31–1.97) | 1.11 (0.39–3.14) | 8.46 × 10−1 | 9.29 × 10−1 |
| Female | 0.42 | 2.83 (0.75–10.66) | 0.31 (0.13–0.75) | 0.25 (0.07–0.97) | 1.64 (0.56–4.76) | 4.24 × 103 | 2.56 × 10−2 | ||
|
| |||||||||
| rs7543174b | CHRNB2 | Male | 0.14 | 0.31 (0.06–1.48) | 2.20 (0.80–6.06) | 1.53 (0.57–4.12) | 1.71 (0.56–5.23) | 1.37 × 10−1 | 3.35 × 10−1 |
| Female | 0.18 | 4.07 (1.41–11.75) | 0.76 (0.30–1.93) | 0.15 (0.02–1.22) | 0.92 (0.31–2.69) | 7.61 × 10−3 | 3.14 × 10−2 | ||
|
| |||||||||
| rs7539745b | CHRNB2 | Male | 0.13 | 0.61 (0.16–2.37) | 3.10 (1.08–8.91) | 1.37 (0.49–3.83) | 1.81 (0.59–5.59) | 2.80 × 10−1 | 4.55 × 10−1 |
| Female | 0.15 | 4.54 (1.56–13.25) | 0.65 (0.24–1.79) | 0.19 (0.02–1.54) | 1.18 (0.40–3.49) | 7.55 × 10−3 | 3.17 × 10−2 | ||
All models were adjusted for age and FTND.
SNPs above the dashed line have adjusted P-values < 0.05 within males. SNPs below the dashed line have adjusted P-values < 0.05 within females.
Observed P-values were obtained using a 3-df test of the gene-treatment interaction effect. P-values were adjusted for the correlation of individual contributions to score statistics for the 3-df tests within respective gene regions.
Adjusted SNP × Treatment × Gender Interaction P-value < 0.05
Marginal associations
Within males at EOT, marginal associations achieving system-wide significance were observed for four SNPs (rs6702335, rs12021667, rs12027267, rs12039988; r2>0.98) in the erythrocyte membrane protein band 4.1 gene (EPB41) (P<6×10−5). Two of these SNPs (rs6702335, rs12027267) had adjusted P-values that approached system-wide significance with abstinence at 6-month follow-up (P≤8.0×10−4). Within males, these four SNPs were associated with a decrease in odds of abstinence at EOT (e.g., ORrs6702335 =0.46, 95% CI 0.32–0.64). Similar effects were observed at 6-month follow-up for those three SNPs at or approaching system-wide significance (OR rs6702335 =0.46, 95% CI 0.31–0.67). Within females at EOT and 6-month follow-up, these SNPs had no effect, and all four had region-wide significant SNP × gender interactions (adjusted P for test of heterogeneity < 0.05).
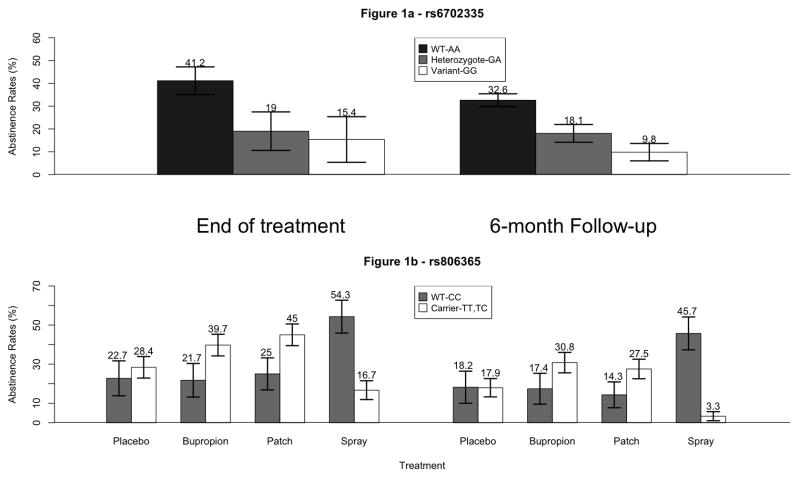
Abstinence rates within males at EOT and 6-month follow-up for each rs6702335 genotype are shown in Figure 1a. At EOT, 41% of males with two major alleles (WT-AA) were abstinent, but only 20% of heterozygotes and 15% of those with two minor alleles (Variant-GG) remained abstinent (OR=0.4, 95% CI 0.2–0.6; OR=0.25, 95% CI 0.1–0.5; respectively). At 6-month follow-up, 33% of males with two major alleles were abstinent, but only 19% of heterozygotes and 10% of those with two minor alleles remained abstinent (OR=0.4, 95% CI 0.3–0.8; OR=0.2, 95% CI 0.1–0.5; respectively).
Figure 1.
Imputation using data from the 1000 Genomes Project lends support to the association between smoking cessation at EOT and EPB41 within males (Supplemental Figure 1). For EPB41, along with the four genotyped SNPs showing association (rs6702335, rs12021667, rs12027267, rs12039988; r2>0.98), nine other imputed non-coding SNPs (rs4654328, rs2930833, rs35013556, rs7546832, rs34474391, rs10429865, rs12734106, rs35423185, rs35579088) in LD with our strongest signal (rs6702335, r2>0.80) were strongly associated with smoking abstinence. Two other SNPs lying between OPRD1 and EPB41 (rs61783628, rs1485471) were also strongly associated with smoking abstinence and in moderate LD with rs6702335 (0.6<r2<0.80). The association between rs6702335 and cessation at EOT remained the strongest. At 6-month follow-up, associations for imputed SNPs within EPB41 were similar to those at EOT.
Four SNPs (rs7123797, rs4938012, rs17115439, rs4938015; r2>0.95) in ankyrin repeat and kinase domain containing 1 (ANKK1), a gene immediately proximal of DRD2 on chromosome 11q,46 achieved region-wide significance in association with abstinence at EOT within females (OR=0.5, 95% CI 0.3–0.7, adjusted P<0.05). There was no significant SNP × gender interaction among these four SNPs (adjusted P for test of heterogeneity > 0.05).
Several SNPs located in or adjacent to nAChR subunit genes had marginal effects achieving region-wide significance. Within males, rs57877647 in the CHRNA5-CHRNA3-CHRNB4 gene cluster was associated with abstinence at EOT. Within females, rs4809549 in the nAChR α4 subunit gene (CHRNA4) was associated with abstinence at EOT, and SNPs in ADAR, immediately distal to CHRNB2 (rs3766927, rs1127314, rs2131902; r2=0.94) were associated with abstinence at 6-month follow-up. There was no significant SNP × gender interaction among these SNPs (adjusted P for test of heterogeneity > 0.05).
Gene × treatment interactions
Within males at 6-month follow-up, a gene × treatment interaction achieving system-wide significance was observed for rs806365 in the cannabinoid receptor 1 gene (CNR1) (adjusted P=3.9 × 10−5; Table 4). The odds ratio of rs806365 within the placebo arm was 1.06 (0.30–3.75) with the odds of abstinence increased more than two-fold in the bupropion and patch arms (OR=2.05, 95% CI 0.62–6.78; OR=2.41, 95% 0.74–7.84; respectively). Most notably, however, was the 25-fold decrease in odds of abstinence within the spray arm (OR=0.04, 95% CI 0.01–0.21). Estimates of effect were consistent at EOT for rs806365 within males, with an adjusted interaction P-value near system-wide significance (P=0.001).
Gene × treatment interaction analysis also identified rs806369 within CNR1. Within males, rs806369 had adjusted interaction P-values that reached region-wide significance at EOT and 6-month follow-up (adjusted Ps=0.007). These interactions were driven mostly by decreases in odds within the spray arm (OR=0.30, 95% CI 0.11–0.79; OR=0.14, 95% CI 0.03–0.63, respectively). For both cessation endpoints, rs806365 and rs806369 (r2=0.5) had region-wide significant SNP × treatment × gender interactions (adjusted P for test of heterogeneity < 0.05). Their effects within females were in the opposite direction as males with increased odds of abstinence within the spray arm for both cessation endpoints. Imputation of SNPs 50kb up- and downstream of CNR1 with 1000 Genomes Project data did not identify other SNPs with stronger SNP × treatment interactions, or SNPs in LD with our strongest signal in this region (rs806365).
Treatment stratified abstinence rates within males for rs806365 carriers and non-carriers are in Figure 1b. For both study endpoints within the bupropion and patch arms, males with at least one minor allele (Carrier-TT, TC) had a roughly two-fold increase in odds of abstinence. Within the placebo arm, those with at least one minor allele had slightly increased odds of abstinence at EOT, but this disappeared at 6-month follow-up. However, overall abstinence rates decreased in these three treatment arms from EOT to 6-month follow-up. Within the spray arm, the proportion of males abstinent remained relatively consistent for males with two major alleles (WT-CC) (54% vs. 46%); but the proportion of abstinent males with at least one minor allele decreased (17% to 3%).
Three SNPs in MAPK1 (rs9610375, rs1063311, rs2266966) achieved region-wide significant treatment interactions within females at EOT. Within males at 6-month follow-up, another five SNPs in MAPK1 (rs743409, rs1892848, rs3788332, rs5999550, rs11704205) achieved region-wide significance for SNP × treatment and SNP × treatment × gender interactions (adjusted P for test of heterogeneity < 0.05).
A number of nAChR SNPs achieved region-wide significance for gender-stratified gene × treatment interaction effects, including two that have been published previously (rs3743075,47 rs207266124). Also, within males at 6-month follow-up, rs68451348 (chromosome 15 nAChR region) had an adjusted gene × treatment interaction P-value that approached system-wide significance (P=0.0007). Within females, rs950776 had significant gene × treatment interactions at EOT and 6-month follow-up (adjusted P≤0.02). Other previously reported SNPs within the nAChR chromosome 15 region (rs1051730, rs1317286, rs578776)4, 47, 49 had null marginal and interaction effects. While two other previously reported SNPs (rs12914385, rs16969968)47, 49 were not genotyped in this study, rs16969968 is in high LD (r2>0.95) with rs1051730 and rs1317286.
DISCUSSION
Reported gender differences in smoking behavior and cessation motivated this gender-stratified analysis in two pharmacogenetic smoking cessation trials. Previous research identified SNPs and genes within the two separate clinical trials comprising this study.24–28, 30–32, 50 This analysis builds upon those study results by pooling both studies to look within genders across all four treatments across 53 candidate gene regions. Instead of focusing on estimated heterogeneity between genders, our aim was to identify SNPs with effects specific to males and/or females.
Gender-stratified SNP marginal effects revealed a male-specific association between a region of high LD within EPB41 and smoking abstinence at EOT and 6-month follow-up. Within males, we identified four non-coding SNPs in EPB41 (rs6702335, rs12021667, rs12027267, rs12039988) in strong LD (r2>0.98). rs12021667 lies in the 5′-UTR, rs12039988 in the 3′-UTR, and rs12027267 and rs6702335 both in intronic regions. These SNPs achieved system-level significance in association with smoking abstinence at EOT (adjusted P<6×10−5). At 6-month follow-up, rs6702335 and rs12027267 approached system-level significance (P≤8.0×10−4) while rs12021667 and rs12039988 achieved region-level significance (adjusted Ps=0.002 and 0.005, respectively). In males, these SNPs were associated with a more than two-fold decrease in odds of abstinence at both cessation endpoints.
Erythrocyte membrane protein band 4.1 (EPB41), known as protein 4.1R, is critical for red blood cell morphology and membrane function.51–54 This protein and its homologues have not been associated with nicotine dependence and smoking, but they were shown to stabilize the localization of dopamine receptors to the plasma membrane,55 and protein 4.1R had a specific organizational role in the arrangement of postsynaptic molecules.54 Our data suggests that genetic variation in EPB41 has gender-specific effects on smoking abstinence, potentially mediated through differential effects on the localization or function of dopamine receptors and further downstream effects on the brain reward pathway.
EPB41 is adjacent to delta opioid receptor 1 (OPRD1), which is part of the family of opioid receptor genes associated with substance abuse.56 Using HapMap SNP genotypes from the CEPH population (Release #28, NCBI Build 36)36 and Haploview,57 none of the SNPs in EPB41 that achieved system-wide significance are in LD with any SNPs in OPRD1 (r2<0.35). However, a SNP in EPB41 achieving region-wide significance (rs16837840) is in moderate LD (r2=0.6) with a SNP in OPRD1 (rs12404612). After imputation of EPB41, rs12404612 (certainty score > 0.9) had male-specific marginal associations with abstinence at EOT and 6-month follow-up (0.01 < P = 0.02 and 0.03, respectively). Interestingly, other imputed SNPs in OPRD1 with imputation certainty scores < 0.9 showed strong associations with smoking abstinence.
Gender-stratified SNP × treatment interaction analyses revealed a male-specific association between smoking abstinence and two SNPs in CNR1 in weak LD (rs806365, rs806369, r2=0.5). rs806365 lies in the 3′ flanking region of CNR1, while due to multiple CNR1 transcript variants,58 rs806369 lies either in an intron or the 5′ flanking region. For abstinence at 6-month follow-up, the male-specific interaction between rs806365 and treatment achieved system-level significance (adjusted interaction LRT P=3.9×10−5). The effect of this SNP was most prominent in males within the spray arm, where it was associated with a 25-fold decrease in odds of abstinence. Effects at EOT were similar, where rs806365 was associated with a nearly 6-fold decrease in abstinence odds for males in the spray arm (adjusted interaction LRT P=0.001). Males with two major alleles in the spray arm had the highest abstinence rates, and this remained relatively unchanged from EOT to 6-month follow-up; males with at least one minor allele had the lowest abstinence rates across all males.
The relationship between cannabinoid receptor 1 (CNR1) and addictive behaviors has been well-characterized.58–61 Along with its primary role in mediating the effects of marijuana,58 it modulates dopamine release in response to other substances.62 The CNR1 antagonist, rimonabant, was effective in suppressing smoking relapse and attenuating reward seeking behaviors during abstinence.59, 60 There was also a female-specific association between SNPs and haplotypes in CNR1 and both nicotine dependence and smoking initiation.63 Although there is no overlap of SNPs investigated in that study with our current investigation, both point to gender-specific associations of smoking behaviors with CNR1. Our analysis suggests that gene associations are not only gender-specific but treatment- and gender-specific. Of the treatments administered, the nasal spray most closely mimics the effects of smoking, introducing a sharp, rapid increase in nicotine levels.64 Since males are more responsive to nicotine dosage and pharmacological reinforcement by nicotine, they may be more sensitive to the effects of the spray. Within the spray arm, males with two major alleles for rs806365 and rs806369 had higher abstinence rates than those with at least one minor allele. Opposite and variable effects of these SNPs in females may be attributed to gender differences in the pharmacological effects of nicotine.12, 65
It is worth highlighting that for both marginal and interaction effects, no SNPs achieving region-or system-wide significance within one gender stratum achieved significance within the other gender stratum. For marginal effects, there was no overlap in gene regions between genders, with male-specific significant SNPs in EPB41, and female-specific SNPs in ANKK1 and α4 and β2 nAChR gene subunits. Across both abstinence outcomes, gene × treatment interactions identified associations across women and men in the nAChR subunit regions and MAPK1. This consistency in gene regions adds evidence of their putative roles in nicotine dependence and smoking behavior.47, 49, 66–71 These regions, along with the others without overlap, have shown prior evidence of association with nicotine dependence and smoking cessation, and require further study to validate gender-specific effects.
The use of 1000 Genomes Project data for imputation helped to broaden the areas for potential follow-up for the specific putatively causal variant within our top gene regions. Additionally, we imputed within the previously reported region of chromosome 15, a region linked to smoking dependence72 and other smoking related diseases.49, 73–75 For this region, imputation identified two gene regions proximal of the chromosome 15 α5-α3-β4 nAChR complex associated with smoking abstinence (Supplemental Figure 2). AGPHD1 and PSMA4 have imputed SNPs in LD (r2>0.6) with our strongest signal in the nAChR gene region (rs684513) that had comparable or more significant SNP × treatment interactions on smoking abstinence at 6-month follow-up within males (1×10−5 < P < 5×10−5). An intronic SNP in AGPHD1, rs12441354, had the strongest association (P=1.3×10−5).
Strengths and limitations of this study have been described previously,24, 25 but we highlight a few here. We did not have to address potential biases in determining smoking abstinence since information was collected from individuals in a prospective manner, and those claiming to be abstinent were subjected to biochemical verification. Similar study designs between studies suggest that subjects are comparable. However, differential responses in abstinence rates across genders and treatments could have arisen from a key difference in exclusion criteria.25 Only participants in the NRT study were excluded for drug or alcohol dependence or any subsequent treatment. Subjects in the Bupropion study may have used other substances to compensate for any adverse effects from smoking cessation.
In our analysis, we performed an adjustment of P-values that accounted for multiple correlated tests within respective gene regions. A Bonferroni-corrected P-value was then applied across the 53 gene regions and two genders. This correction was less stringent than a uniform Bonferroni-correction across all SNPs, while still accounting for independent gene regions and both genders. Thus, a SNP was significant within a respective region and gender at a P-value < 0.05, while system-wide significance was set at an α-level of 0.05/(53*2)=5×10−4.
While our determination of significance was conservative, the results require independent replication. Identified SNPs with the strongest evidence of association have not been replicated in other studies, and the region with strongest marginal SNP effects, EPB41, has not been associated with nicotine dependence or smoking previously. Confirmation from other studies, especially gender-specific results, would lend support to our findings. However, our study has identified a novel gene region, EPB41, which may be associated with smoking cessation, along with gene regions in CNR1 that may be targeted to further elucidate the etiology of gender differences in smoking behaviors.
Supplementary Material
Acknowledgments
We thank the participants of the two randomized clinical trials for their contributions to research. We acknowledge the contributions of Faith Allen, Ruth Krasnow, Huaiyu Mi, and Chris Edlund for data curation and management. This research was supported by a grant from NIDA, NCI, NIGMS and NHGRI U01 DA020830, NIDA R01 DA002277, and NCI P50 CA084735.
Footnotes
Conflict of Interest. – Drs. Swan and Conti have served as consultants for Pfizer. Dr. Lerman has served as a consultant for and/or received research support from Pfizer, AstraZeneca, Novartis, Targacept, and Glaxo SmithKline. Dr. Tyndale owns shares and participates in Nicogen Research Inc., a company focused on novel smoking cessation treatment approaches. No Nicogen funds were used in this work and no other Nicogen participants reviewed the manuscript. Dr. Tyndale has also consulted for Novartis. Dr. Benowitz is a paid consultant for several pharmaceutical companies that market smoking cessation medications and has been a paid expert witness against tobacco companies in litigation related to nicotine addiction. This research was not supported by industry funds.
References
- 1.Benowitz NL. Nicotine addiction. N Engl J Med. 2010;362(24):2295–2303. doi: 10.1056/NEJMra0809890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hogg RC, Bertrand D. Neuroscience. What genes tell us about nicotine addiction. Science. 2004;306(5698):983–985. doi: 10.1126/science.1106030. [DOI] [PubMed] [Google Scholar]
- 3.Zbikowski SM, Swan GE, McClure JB. Cigarette smoking and nicotine dependence. Med Clin North Am. 2004;88(6):1453–1465. x. doi: 10.1016/j.mcna.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Berrettini WH, Lerman CE. Pharmacotherapy and pharmacogenetics of nicotine dependence. Am J Psychiatry. 2005;162(8):1441–1451. doi: 10.1176/appi.ajp.162.8.1441. [DOI] [PubMed] [Google Scholar]
- 5.Ray R, Schnoll RA, Lerman C. Nicotine dependence: biology, behavior, and treatment. Annu Rev Med. 2009;60:247–260. doi: 10.1146/annurev.med.60.041707.160511. [DOI] [PubMed] [Google Scholar]
- 6.Russell MA, Feyerabend C, Cole PV. Plasma nicotine levels after cigarette smoking and chewing nicotine gum. Br Med J. 1976;1(6017):1043–1046. doi: 10.1136/bmj.1.6017.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell MA, Wilson C, Taylor C, Baker CD. Smoking habits of men and women. Br Med J. 1980;281(6232):17–20. doi: 10.1136/bmj.281.6232.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell MA, Jarvis M, Iyer R, Feyerabend C. Relation of nicotine yield of cigarettes to blood nicotine concentrations in smokers. Br Med J. 1980;280(6219):972–976. doi: 10.1136/bmj.280.6219.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogle JM, Curtin JJ. Sex differences in negative affective response during nicotine withdrawal. Psychophysiology. 2006;43(4):344–356. doi: 10.1111/j.1469-8986.2006.00406.x. [DOI] [PubMed] [Google Scholar]
- 10.Schnoll RA, Patterson F. Sex heterogeneity in pharmacogenetic smoking cessation clinical trials. Drug Alcohol Depend. 2009;104 (Suppl 1):S94–99. doi: 10.1016/j.drugalcdep.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgess DJ, Fu SS, Noorbaloochi S, Clothier BA, Ricards J, Widome R, et al. Employment, gender, and smoking cessation outcomes in low-income smokers using nicotine replacement therapy. Nicotine Tob Res. 2009;11(12):1439–1447. doi: 10.1093/ntr/ntp158. [DOI] [PubMed] [Google Scholar]
- 12.Perkins KA, Jacobs L, Sanders M, Caggiula AR. Sex differences in the subjective and reinforcing effects of cigarette nicotine dose. Psychopharmacology (Berl) 2002;163(2):194–201. doi: 10.1007/s00213-002-1168-1. [DOI] [PubMed] [Google Scholar]
- 13.Pogun S, Yararbas G. Sex differences in nicotine action. Handb Exp Pharmacol. 2009;(192):261–291. doi: 10.1007/978-3-540-69248-5_10. [DOI] [PubMed] [Google Scholar]
- 14.Munafo MR, Shields AE, Berrettini WH, Patterson F, Lerman C. Pharmacogenetics and nicotine addiction treatment. Pharmacogenomics. 2005;6(3):211–223. doi: 10.1517/14622416.6.3.211. [DOI] [PubMed] [Google Scholar]
- 15.Perkins KA, Scott J. Sex differences in long-term smoking cessation rates due to nicotine patch. Nicotine Tob Res. 2008;10(7):1245–1250. doi: 10.1080/14622200802097506. [DOI] [PubMed] [Google Scholar]
- 16.Benowitz NL, Lessov-Schlaggar CN, Swan GE, Jacob P., 3rd Female sex and oral contraceptive use accelerate nicotine metabolism. Clin Pharmacol Ther. 2006;79(5):480–488. doi: 10.1016/j.clpt.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Higashi E, Fukami T, Itoh M, Kyo S, Inoue M, Yokoi T, et al. Human CYP2A6 is induced by estrogen via estrogen receptor. Drug Metab Dispos. 2007;35(10):1935–1941. doi: 10.1124/dmd.107.016568. [DOI] [PubMed] [Google Scholar]
- 18.Harrington WR, Sengupta S, Katzenellenbogen BS. Estrogen regulation of the glucuronidation enzyme UGT2B15 in estrogen receptor-positive breast cancer cells. Endocrinology. 2006;147(8):3843–3850. doi: 10.1210/en.2006-0358. [DOI] [PubMed] [Google Scholar]
- 19.Benowitz NL, Hukkanen J, Jacob P., 3rd Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol. 2009;(192):29–60. doi: 10.1007/978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatchell PC, Collins AC. Influences of genotype and sex on behavioral tolerance to nicotine in mice. Pharmacol Biochem Behav. 1977;6(1):25–30. doi: 10.1016/0091-3057(77)90156-3. [DOI] [PubMed] [Google Scholar]
- 21.Isiegas C, Mague SD, Blendy JA. Sex differences in response to nicotine in C57Bl/6:129SvEv mice. Nicotine Tob Res. 2009;11(7):851–858. doi: 10.1093/ntr/ntp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lajtha A, Sershen H. Nicotine: alcohol reward interactions. Neurochem Res. 2010;35(8):1248–1258. doi: 10.1007/s11064-010-0181-8. [DOI] [PubMed] [Google Scholar]
- 23.Damaj MI. Influence of gender and sex hormones on nicotine acute pharmacological effects in mice. J Pharmacol Exp Ther. 2001;296(1):132–140. [PubMed] [Google Scholar]
- 24.Conti DV, Lee W, Li D, Liu J, Van Den Berg D, Thomas PD, et al. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17(18):2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lerman C, Jepson C, Wileyto EP, Epstein LH, Rukstalis M, Patterson F, et al. Role of functional genetic variation in the dopamine D2 receptor (DRD2) in response to bupropion and nicotine replacement therapy for tobacco dependence: results of two randomized clinical trials. Neuropsychopharmacology. 2006;31(1):231–242. doi: 10.1038/sj.npp.1300861. [DOI] [PubMed] [Google Scholar]
- 26.Lee AM, Jepson C, Hoffmann E, Epstein L, Hawk LW, Lerman C, et al. CYP2B6 genotype alters abstinence rates in a bupropion smoking cessation trial. Biol Psychiatry. 2007;62(6):635–641. doi: 10.1016/j.biopsych.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Strasser AA, Malaiyandi V, Hoffmann E, Tyndale RF, Lerman C. An association of CYP2A6 genotype and smoking topography. Nicotine Tob Res. 2007;9(4):511–518. doi: 10.1080/14622200701239605. [DOI] [PubMed] [Google Scholar]
- 28.Malaiyandi V, Goodz SD, Sellers EM, Tyndale RF. CYP2A6 genotype, phenotype, and the use of nicotine metabolites as biomarkers during ad libitum smoking. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1812–1819. doi: 10.1158/1055-9965.EPI-05-0723. [DOI] [PubMed] [Google Scholar]
- 29.Yamanaka H, Nakajima M, Fukami T, Sakai H, Nakamura A, Katoh M, et al. CYP2A6 AND CYP2B6 are involved in nornicotine formation from nicotine in humans: interindividual differences in these contributions. Drug Metab Dispos. 2005;33(12):1811–1818. doi: 10.1124/dmd.105.006254. [DOI] [PubMed] [Google Scholar]
- 30.Lerman C, Tyndale R, Patterson F, Wileyto EP, Shields PG, Pinto A, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther. 2006;79 (6):600–608. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Patterson F, Schnoll RA, Wileyto EP, Pinto A, Epstein LH, Shields PG, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther. 2008;84(3):320–325. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 32.Lerman C, Shields PG, Wileyto EP, Audrain J, Hawk LH, Jr, Pinto A, et al. Effects of dopamine transporter and receptor polymorphisms on smoking cessation in a bupropion clinical trial. Health Psychol. 2003;22(5):541–548. doi: 10.1037/0278-6133.22.5.541. [DOI] [PubMed] [Google Scholar]
- 33.Bergen AW, Conti DV, Van Den Berg D, Lee W, Liu J, Li D, et al. Dopamine genes and nicotine dependence in treatment-seeking and community smokers. Neuropsychopharmacology. 2009;34 (10):2252–2264. doi: 10.1038/npp.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feyerabend C, Russell MA. A rapid gas-liquid chromatographic method for the determination of cotinine and nicotine in biological fluids. J Pharm Pharmacol. 1990;42(6):450–452. doi: 10.1111/j.2042-7158.1990.tb06592.x. [DOI] [PubMed] [Google Scholar]
- 35.Edlund CK, Lee WH, Li D, Van Den Berg DJ, Conti DV. Snagger: a user-friendly program for incorporating additional information for tagSNP selection. BMC Bioinformatics. 2008;9:174. doi: 10.1186/1471-2105-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15(11):1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67(1):170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 41.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86(9):1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 43.Team RDC. R. A Language and Environment for Statistical Computing. 2010. [Google Scholar]
- 44.Conneely KN, Boehnke M. So Many Correlated Tests, So Little Time! Rapid Adjustment of P Values for Multiple Correlated Tests. Am J Hum Genet. 2007;81(6) doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li D. In: Multiple degree of freedom p-value adjustment using correlation of score statistics. Lee W, editor. Los Angeles, CA: 2010. [Google Scholar]
- 46.Perkins KA, Lerman C, Grottenthaler A, Ciccocioppo MM, Milanak M, Conklin CA, et al. Dopamine and opioid gene variants are associated with increased smoking reward and reinforcement owing to negative mood. Behav Pharmacol. 2008;19(5–6):641–649. doi: 10.1097/FBP.0b013e32830c367c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keskitalo K, Broms U, Heliovaara M, Ripatti S, Surakka I, Perola M, et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum Mol Genet. 2009;18(20):4007–4012. doi: 10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amos CI, Gorlov IP, Dong Q, Wu X, Zhang H, Lu EY, et al. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. J Natl Cancer Inst. 2010;102(15):1199–1205. doi: 10.1093/jnci/djq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le Marchand L, Derby KS, Murphy SE, Hecht SS, Hatsukami D, Carmella SG, et al. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68(22):9137–9140. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee AM, Jepson C, Shields PG, Benowitz N, Lerman C, Tyndale RF. CYP2B6 genotype does not alter nicotine metabolism, plasma levels, or abstinence with nicotine replacement therapy. Cancer Epidemiol Biomarkers Prev. 2007;16(6):1312–1314. doi: 10.1158/1055-9965.EPI-07-0188. [DOI] [PubMed] [Google Scholar]
- 51.Han BG, Nunomura W, Takakuwa Y, Mohandas N, Jap BK. Protein 4.1R core domain structure and insights into regulation of cytoskeletal organization. Nat Struct Biol. 2000;7(10):871–875. doi: 10.1038/82819. [DOI] [PubMed] [Google Scholar]
- 52.Shiffer KA, Goodman SR. Protein 4.1: its association with the human erythrocyte membrane. Proc Natl Acad Sci U S A. 1984;81(14):4404–4408. doi: 10.1073/pnas.81.14.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran YK, Bogler O, Gorse KM, Wieland I, Green MR, Newsham IF. A novel member of the NF2/ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res. 1999;59 (1):35–43. [PubMed] [Google Scholar]
- 54.Scott C, Keating L, Bellamy M, Baines AJ. Protein 4.1 in forebrain postsynaptic density preparations: enrichment of 4.1 gene products and detection of 4.1R binding proteins. Eur J Biochem. 2001;268(4):1084–1094. doi: 10.1046/j.1432-1327.2001.01968.x. [DOI] [PubMed] [Google Scholar]
- 55.Binda AV, Kabbani N, Lin R, Levenson R. D2 and D3 dopamine receptor cell surface localization mediated by interaction with protein 4.1N. Mol Pharmacol. 2002;62(3):507–513. doi: 10.1124/mol.62.3.507. [DOI] [PubMed] [Google Scholar]
- 56.Gelernter J, Gueorguieva R, Kranzler HR, Zhang H, Cramer J, Rosenheck R, et al. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: results from the VA Cooperative Study. Alcohol Clin Exp Res. 2007;31(4):555–563. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- 57.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 58.Zhang PW, Ishiguro H, Ohtsuki T, Hess J, Carillo F, Walther D, et al. Human cannabinoid receptor 1: 5′ exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Mol Psychiatry. 2004;9(10):916–931. doi: 10.1038/sj.mp.4001560. [DOI] [PubMed] [Google Scholar]
- 59.De Vries TJ, Schoffelmeer AN. Cannabinoid CB1 receptors control conditioned drug seeking. Trends Pharmacol Sci. 2005;26(8):420–426. doi: 10.1016/j.tips.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 60.Forget B, Hamon M, Thiebot MH. Cannabinoid CB1 receptors are involved in motivational effects of nicotine in rats. Psychopharmacology (Berl) 2005;181(4):722–734. doi: 10.1007/s00213-005-0015-6. [DOI] [PubMed] [Google Scholar]
- 61.Tanda G, Goldberg SR. Cannabinoids: reward, dependence, and underlying neurochemical mechanisms--a review of recent preclinical data. Psychopharmacology (Berl) 2003;169(2):115–134. doi: 10.1007/s00213-003-1485-z. [DOI] [PubMed] [Google Scholar]
- 62.Mascia MS, Obinu MC, Ledent C, Parmentier M, Bohme GA, Imperato A, et al. Lack of morphine-induced dopamine release in the nucleus accumbens of cannabinoid CB(1) receptor knockout mice. Eur J Pharmacol. 1999;383(3):R1–2. doi: 10.1016/s0014-2999(99)00656-1. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Williamson VS, An SS, Hettema JM, Aggen SH, Neale MC, et al. Cannabinoid receptor 1 gene association with nicotine dependence. Arch Gen Psychiatry. 2008;65(7):816–824. doi: 10.1001/archpsyc.65.7.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lerman C, Kaufmann V, Rukstalis M, Patterson F, Perkins K, Audrain-McGovern J, et al. Individualizing nicotine replacement therapy for the treatment of tobacco dependence: a randomized trial. Ann Intern Med. 2004;140(6):426–433. doi: 10.7326/0003-4819-140-6-200403160-00009. [DOI] [PubMed] [Google Scholar]
- 65.Robinson JD, Cinciripini PM, Tiffany ST, Carter BL, Lam CY, Wetter DW. Gender differences in affective response to acute nicotine administration and deprivation. Addict Behav. 2007;32(3):543–561. doi: 10.1016/j.addbeh.2006.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baker TB, Weiss RB, Bolt D, von Niederhausern A, Fiore MC, Dunn DM, et al. Human neuronal acetylcholine receptor A5-A3-B4 haplotypes are associated with multiple nicotine dependence phenotypes. Nicotine Tob Res. 2009;11(7):785–796. doi: 10.1093/ntr/ntp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marubio LM, Gardier AM, Durier S, David D, Klink R, Arroyo-Jimenez MM, et al. Effects of nicotine in the dopaminergic system of mice lacking the alpha4 subunit of neuronal nicotinic acetylcholine receptors. Eur J Neurosci. 2003;17(7):1329–1337. doi: 10.1046/j.1460-9568.2003.02564.x. [DOI] [PubMed] [Google Scholar]
- 68.Picciotto MR, Zoli M, Rimondini R, Lena C, Marubio LM, Pich EM, et al. Acetylcholine receptors containing the beta2 subunit are involved in the reinforcing properties of nicotine. Nature. 1998;391(6663):173–177. doi: 10.1038/34413. [DOI] [PubMed] [Google Scholar]
- 69.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wada T, Naito M, Kenmochi H, Tsuneki H, Sasaoka T. Chronic nicotine exposure enhances insulin-induced mitogenic signaling via up-regulation of alpha7 nicotinic receptors in isolated rat aortic smooth muscle cells. Endocrinology. 2007;148(2):790–799. doi: 10.1210/en.2006-0907. [DOI] [PubMed] [Google Scholar]
- 71.Chen RJ, Ho YS, Guo HR, Wang YJ. Long-term nicotine exposure-induced chemoresistance is mediated by activation of Stat3 and downregulation of ERK1/2 via nAChR and beta-adrenoceptors in human bladder cancer cells. Toxicol Sci. 2010;115(1):118–130. doi: 10.1093/toxsci/kfq028. [DOI] [PubMed] [Google Scholar]
- 72.Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(4):453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gago-Dominguez M, Jiang X, Conti DV, Castelao JE, Stern MC, Cortessis VK, et al. Genetic variations on chromosomes 5p15 and 15q25 and bladder cancer risk: findings from the Los Angeles-Shanghai bladder case-control study. Carcinogenesis. 2011;32(2):197–202. doi: 10.1093/carcin/bgq233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.