Abstract
Naturally fermented pickles harbour many lactic acid bacteria (LAB). Forty-three LAB strains with conjugated linoleic acid (CLA)-producing ability were isolated from three naturally fermented pickle brines. Of these isolates, lp15 identified as Lactobacillus plantarum by API 50 CHL system and full-length 16S rDNA sequence analysis exhibited the highest CLA-producing ability (26.1% conversion) at 48 h in de Man Rogosa Sharpe (MRS) broth in the presence of 100 µg/ml of linoleic acid (LA). Compared to other strains, L. plantarum strain lp15 showed the highest tolerance upon increased levels of LA in the medium, i.e., up to 600 µg/ml. This strain converted about 25% of LA into CLA isomers [predominantly cis-9, trans-11 CLA (9-CLA) and trans-10, cis-12 CLA (10-CLA)], of which 75% was 9-CLA. Interestingly, though the conversion rate of LA into CLA by lp15 remained stable between 100 to 600 µg/ml LA levels in the medium, it dropped sharply at 1000 µg/ml. Taken together, the lp15 strain displayed relatively high LA tolerance with higher conversion rate, which implies that this strain is a valuable candidate for enhancing the CLA content in food-sources like pickles.
Keywords: Conjugated linoleic acids, Lactobacillus plantarum, Lactic acid bacteria, Pickle, Gas chromatography
1. Introduction
Conjugated linoleic acids (CLAs) are a heterogeneous group of positional and geometric isomers of linoleic acid (LA) (MacDonald, 2000). Ever since the discovery by Ha et al. (1987), when they were investigating the carcinogenic properties of fried ground beef, CLAs have been reported to possess many important biological activities and health-promoting properties, including antiatherosclerotic, anticarcinogenic, antiadipogenic, immunoenhancing, antioxidative, hypotensive, and anti-inflammatory effects (Hayek et al., 1999; Inoue et al., 2004; Nagao and Yanagita, 2005; Bhattacharya et al., 2006; Kelley et al., 2007; Mitchell and Mcleod, 2008; Benjamin and Spener, 2009). It also has been well documented that the biological activities of CLAs are largely attributable to the separate actions of two isomers: cis-9, trans-11 CLA (9-CLA) and trans-10, cis-12 CLA (10-CLA). The isomer 9-CLA (rumenic acid), which has been considered to have the main biological activities, is mainly responsible for the anticarcinogenic effect, while 10-CLA isomer is involved in the lipid metabolism and body composition, and in some cases the health-promoting effects appear to be the synergistical interactions of these two isomers (Park et al., 1999; Pariza et al., 2001; Martin and Valeille, 2002; Bhattacharya et al., 2006; Churruca et al., 2009; Feitoza et al., 2009; Halade et al., 2009).
To date, at least 28 different CLA isomers have been identified and the major member in natural foods particularly in ruminant meat and dairy products is 9-CLA isomer, which accounts for 75%–90% of total CLAs (Kelley et al., 2007). In the case of 10-CLA, vegetable oils and partially hydrogenated oils such as shortenings and margarines are the main sources (Wall et al., 2008). The levels of CLAs in dairy products, however, are much lower than the levels needed to play a physiological role in human (Chung et al., 2008). Although substantial amounts of 9- and 10-CLAs can be produced commercially by chemical isomerization, they are contaminated with a variety of other isomers and toxic substances and cannot be used directly. Due to the high cost and difficulties in isomer purification, such synthesized CLAs are not reliable dietary sources at present (Pariza et al., 2001; Mitchell and Mcleod, 2008).
To increase the intake of CLAs, dietary and non-dietary manipulations on food products have been routinely employed (Wall et al., 2008). Supplementing the ruminant diet with plant oils or animal fats that contain either LA or linolenic acid is important and popular practices on elevating food-sourced CLAs (Donovan et al., 2000; Jones et al., 2000; Chouinard et al., 2001; Peterson et al., 2002; Lock et al., 2005). On the other hand, non-dietary methods especially CLA synthesis via food-grade bacterial strains are extensively utilized and attracting more and more world-wide attentions, which represents a more promising scheme in producing dietary CLAs (van Nieuwenhove et al., 2007; Wall et al., 2008).
Although species of propionibacterium, clostridium, and butyrivibrio possess the ability of converting LA into CLA isomers, lactic acid bacteria (LAB) strains, especially lactobacillus, are the most widely utilized sources in dietary CLA production (Jiang et al., 1998; Alonso et al., 2003; Coakley et al., 2003; Kishino et al., 2003; Ogawa et al., 2005; Peng et al., 2007; Chung et al., 2008; Zeng et al., 2009). Fermented pickles not only are world-widely enjoyed as delicious and nutritious side dishes, but also are fertile grounds for invaluable lactobacillus strains with great utilization potentials (Sanchez et al., 2004; Plengvidhya et al., 2007). The natural fermentation of Chinese sauerkraut or pickle by LAB has quite a long history in China. In order to isolate and identify lactobacillus strains possessing high abilities of CLA production from the naturally fermented Chinese pickles, we carried out this investigation and discovered one strain, termed lp15, as a prospective candidate.
2. Materials and methods
2.1. Bacteria screening and growth conditions
Three kinds of naturally fermented Chinese pickles were bought from a supermarket in Hangzhou, Zhejiang Province, China. The diluted pickle brines were coated on a culture dish with de Man Rogosa Sharpe (MRS) medium and cultured at 30 or 37 °C for 48 h in an anaerobic incubator (Model YQX-II, Shanghai CIMO Medical Instrument Co., China). Seventy-eight strains were primarily screened from the pickle brines with the gradient dilution method (Zeng et al., 2009). The culture was activated at least twice before each experiment by inoculating 1% of subcultures in MRS at 30 or 37 °C for 24 h under facultative anaerobic conditions, which were achieved by sparging nitrogen gas into the boiling medium for 5 min.
2.2. CLA determination and analysis
An emulsion of LA (purity 99%, density 0.903 g/ml; Sigma) at 100 mg/ml was prepared in 1% (v/v) polyoxyethylene sorbitan monooleate (Tween 80) by sonication for 1 min to improve its solubility and then filter-sterilized (pore size, 0.2 µm). All the strains primarily screened from the pickles were incubated with 10 ml MRS medium containing 100 µg/ml of LA in 15 mm×160 mm tubes at 30 °C for 48 h. The CLAs were extracted by using hexane/isopropanol (2:1, v/v) solution at room temperature, and the extracts were washed with distilled water and then dehydrated with anhydrous sodium sulfate. The absorption value was determined from 190 to 300 nm and the characteristic absorption peak at 233 nm indicates the presence of CLA (Barrett et al., 2007). The absorption value at 233 nm was then recorded and the CLA concentration was calculated from the standard curve constructed from standard CLAs (Sigma), while the hexane extract of MRS broth without any strain was used as blank control. The linear regression equation of the standard curve was A=0.1058C+0.1452 (A refers to the absorption value of CLA at 233 nm; C refers to the CLA concentration; R 2=0.9996). The CLA content of each sample supernatant was determined according to the equation and the strains with CLA production ability were selected.
For gas chromatography (GC), the CLA extract was recovered and dried with nitrogen gas at 70 °C in the Zymark evaporator. The dried sample was redissolved in 500 µl hexane and derivatized to methyl ester with 1 ml of 5.0% (v/v) HCl in methanol solution at 100 °C for 60 min (Jung et al., 2006). The methyl esters of CLA were analyzed using a GC system (Agilent Technologies, Series HP7890, Wilmington, Delaware, USA) equipped with an automatic injector, a flame ionization detector (FID), and an HP-88 capillary column (60 m×0.25 mm i.d.×0.20 µm film thickness). A total of 1 µl of sample was injected and the GC conditions were: injector and detector temperatures were 250 °C; oven temperature started at 90°C (5 min hold), increased to 180 °C at 10 °C/min (10 min hold), ramped to 220 °C at 5 °C/min (10 min hold), and then reached 250 °C at 5 °C/min (final 10 min hold). Hydrogen was used as the carrier gas.
2.3. Strain identification by 16S rDNA and API 50 CHL system
Biochemical strain identification was carried out according to the API 50 CHL system (BioMerieux Vitek, France). A total of 100 µl of bacterial suspension was inoculated into the 50 tubes on a clean bench and sterile liquid paraffin was added to maintain the anaerobic condition. The API 50 CHL strip was then cultured at 30 °C for 24 to 48 h. Data analysis and strain identification were performed by the Biolog microbial identification system (BMIS, Biolog Inc., Hayward, CA, USA).
Genotypical identification based on 16S rDNA was performed as described by Woo et al. (2000). Genomic DNA was extracted according to the cetyltrimethylammonium bromide (CTAB)/NaCl procedure (Wilson, 1997) and the 16S rDNA gene was amplified by polymerase chain reaction (PCR) using primers 16S5′ (AGAGTTTGATCCTGGCTCAG) and 16S3′ (GGTTACCTTGTTACGACTT) modified according to Woo et al. (2000). The PCR program is consisted of 10 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 1 min at 55 °C, and 1 min at 72 °C, and then a 5-min elongation at 72 °C. PCR products were purified using a QIAquick PCR purification kit (Qiagen Inc., Valencia, CA, USA) and sequenced commercially (Shanghai Sangon Biologic Engineering
Technology & Services Co., China). The sequence was submitted to GenBank (accession No. FJ763580) and analyzed by performing a BLAST search of nonredundant DNA sequence database (http://www.ncbi.nlm.nih.gov/).
2.4. Growth rate and CLA production of strain lp15 under different LA concentrations
To evaluate the growth rate of lp15 in the presence of LA, 1% inocula were added to the MRS broth supplemented with different concentrations of LA (0, 100, 200, 600, and 1 000 µg/ml), and the viable counts were determined at 0, 12, 24, 36, and 48 h using the pour plate method on the MRS agar plate which was cultured at 30 °C in an anaerobic incubator, and expressed as colony forming units (CFUs) per ml. CLA production was also determined after incubation for 48 h with different concentrations of LA (100, 200, 600, and 1 000 µg/ml) and the transformation efficiency was expressed as percentage of LA added.
2.5. Statistical analyses
All incubations were performed at least three times. Mean values and standard deviations (SDs) of the data were shown. All data were processed via software DPS 7.05.
3. Results and discussion
3.1. CLA-producing abilities of bacterial strains selected from pickles
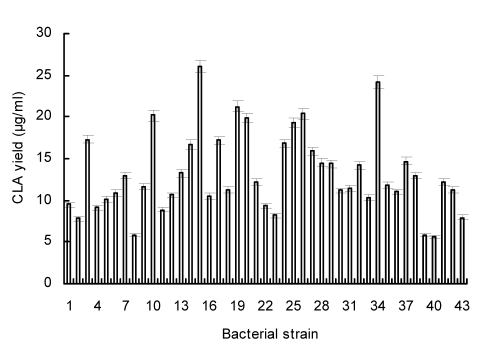
After bacterial screening, 43 LAB strains with CLA-producing abilities were isolated from three kinds of pickled vegetable brines and the CLA yields varied from 5.6 to 26.1 µg/ml (Fig. 1). Among them, five strains (designated lp10, lp15, lp19, lp26, and lp34) produced more than 20.0 µg/ml CLAs and strain lp15 exhibited the highest CLA-producing ability, with a CLA yield of 26.1 µg/ml. The corresponding conversion rate was 26.1%. The LA conversion rate of lp15 was relatively high compared with other strains, for example, Lactobacillus acidophilus CRL730 (23.8%), L. acidophilus Q42 (20.0%), Lactobacillus casei CRL87 (17%), and Lactobacillus plantarum NCUL005 (27.6%) (van Nieuwenhove et al., 2007; Zeng et al., 2009).
Fig. 1.
CLA-producing profile of 43 bacterial strains isolated from pickles grown in the MRS medium containing 100 µg/ml LA at 30 °C for 48 h
Data are shown as means±SD
3.2. Identification of strain lp15
The API system was efficient in bacterial identification, which was able to identify approximately 95.6%–98.6% of lactobacillus and streptococci species (Carmen Collado and Hernandez, 2007). Thus, the API system was employed to identify strain lp15. As shown in Table 1, lp15 could make use of 22 kinds of carbohydrates in 24 h and two other kinds in 48 h among all the 49 kinds of carbon sources tested. The carbohydrate preferences of lp15 accorded with the utilization characteristics of L. plantarum. Accordingly, strain lp15 was preliminarily classified as L. plantarum. In addition, the 16S rDNA gene of strain lp15 was sequenced in order to further confirm the classification result. The corresponding PCR product was 1 471 bp (GenBank accession No. FJ763580). The bacterial strain identification was then determined by BLAST, which revealed that the 16S rDNA sequence was most homologous to that of L. plantarum. Taken together, strain lp15 with maximum CLA-producing ability was identified as L. plantarum.
Table 1.
Test results with the API 50 CHL systems—utilization situation of 49 kinds of carbon sources by strain lp15 at 30 °C for 24 and 48 h
| Micropore | Carbon source | Utilization situation (24/48 h) |
| 0 | Water | −/− |
| 1 | Glycerol | −/w |
| 2 | Erythritol | −/− |
| 3 | D-arabinose | −/− |
| 4 | L-arabinose | −/− |
| 5 | D-ribose | +/+ |
| 6 | D-xylose | −/− |
| 7 | L-xylose | −/− |
| 8 | D-adonitol | −/− |
| 9 | Methyl-β-D-xylopyranoside | −/− |
| 10 | D-galactose | +/+ |
| 11 | D-glucose | +/+ |
| 12 | D-fructose | +/+ |
| 13 | D-mannose | +/+ |
| 14 | L-sorbose | −/− |
| 15 | L-rhamnose | −/− |
| 16 | Dulcitol | −/− |
| 17 | Inositol | −/− |
| 18 | D-mannitol | +/+ |
| 19 | D-sorbitol | −/− |
| 20 | Methyl-α-D-mannopyranoside | +/+ |
| 21 | Methyl-α-D-glucopyranoside | +/+ |
| 22 | N-acetylglucosamine | +/+ |
| 23 | Amygdaline | +/+ |
| 24 | Arbutin | +/+ |
| 25 | Esculine citrate de fer | +/+ |
| 26 | Salicine | +/+ |
| 27 | D-cellobiose | +/+ |
| 28 | D-maltose | +/+ |
| 29 | D-lactose (d′origine bovine) | −/− |
| 30 | D-melibiose | +/+ |
| 31 | D-saccharose | +/+ |
| 32 | D-trehalose | +/+ |
| 33 | Inuline | −/− |
| 34 | D-melezitose | +/+ |
| 35 | D-raffinose | −/+ |
| 36 | Amidon | −/− |
| 37 | Glycogen | −/− |
| 38 | Xylitol | −/− |
| 39 | Gentiobiose | +/+ |
| 40 | D-turanose | −/+ |
| 41 | D-lyxose | −/− |
| 42 | D-tagatose | −/− |
| 43 | D-fucose | −/− |
| 44 | L-fucose | −/− |
| 45 | D-arabitol | +/+ |
| 46 | L-arabitol | −/− |
| 47 | Potassium gluconate | +/+ |
| 48 | Potassium 2-ketogluconate | −/− |
| 49 | Potassium 5-ketogluconate | −/− |
+: utilization-positive; −: utilization-negative; w: weak utilization
Intriguingly, the other four strains, with CLA yields higher than 20 µg/ml, were also identified as L. plantarum based on API 50 CHL system (data not shown). Consistent with our results, Zeng et al. (2009) also found that L. plantarum was the predominant LAB found in Chinese pickles. The LAB, especially Lactobacillus spp. were shown to be responsible for the fermentation of multiple dietary foods, such as L. plantarum, Lactobacillus fermentum in Almagro eggplants, Lactobacillus sakei, Lactobacillus curvatus and L. plantarum in dryly fermented sausage and L. plantarum, the dominant species in naturally fermented Turkish sucuk (Aymerich et al., 2003; Sanchez et al., 2004; Drosinos et al., 2007; Kaban and Kaya, 2008). The failure of obtaining other LAB species was probably due to experimental materials used or the relatively low CLA conversion ability of other species under our experimental conditions.
3.3. Influence of LA on growth rate of lp15
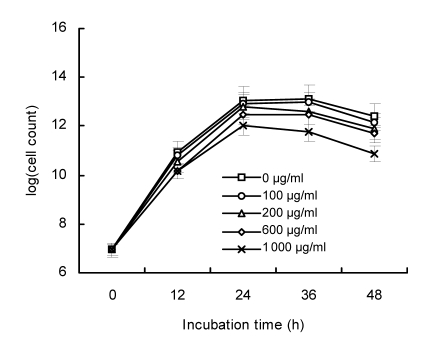
Although free LA serves as the direct precursor in CLA biosynthesis, it also inhibits bacterial growth, and the tolerance to LA varies among different strains (Jiang et al., 1998; Kim et al., 2000; van Nieuwenhove et al., 2007). To evaluate LA tolerance of L. plantarum lp15, different titers of LA were supplemented to the MRS medium (final concentrations: 0, 100, 200, 600, and 1 000 µg/ml). Cultured samples in the MRS broth containing different levels of LA were examined at regular intervals (0, 12, 24, 36, and 48 h) to determine the number of viable cells. As shown in Fig. 2, the growth rate of lp15 was hardly affected by 100 µg/ml of LA while the inhibitory effects aggravated when LA levels increased. Although previous studies found that even lower LA levels (25 µg/ml) could inhibit bacterial growth (Jiang et al., 1998), the lp15 strain was able to grow under much higher LA conditions (1 000 µg/ml), indicating a relatively high tolerance to LA (Fig. 2). It has been suggested that the conversion of free LA to CLA might function as a detoxification mechanism in bacteria and a stronger LA tolerance implies a higher CLA productivity (Jiang et al., 1998; Kim et al., 2000; Ogawa et al., 2005; Chung et al., 2008). Therefore, screening bacterial strains with high LA tolerance might be a shortcut to obtain strains that can produce more CLAs.
Fig. 2.
Viable cell counts of L. plantarum lp15 in the MRS medium supplemented with 0, 100, 200, 600, and 1 000 µg/ml LA after 0, 12, 24, 36, and 48 h of incubation at 30 °C
3.4. CLA productivity of lp15 and composition analysis
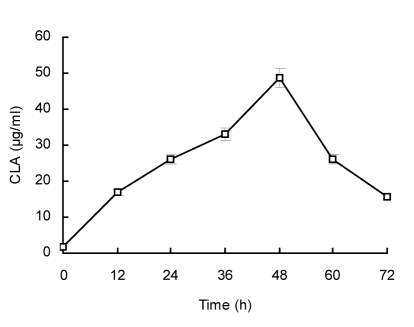
To determine the growth stage at which the CLAs were produced, we first analyzed the samples of the MRS broth supplemented with 200 µg/ml LA inoculated with lp15 after different periods (0, 12, 24, 36, 48, 60, and 72 h) of incubation at 30 °C. We found that the CLA content increased along with the fermentation progress before reaching the highest CLA production (48.7 µg/ml) after 48 h of incubation and then the CLA production declined sharply (Fig. 3). When different concentrations of LA were assayed, the similar tendency was observed (data not shown). We also discovered that most CLAs were produced at the stationary stage, similar to the findings of Lin et al. (1999) and Alonso et al. (2003).
Fig. 3.
Total CLA produced by L. plantarum lp15 in the MRS medium supplemented with 200 µg/ml LA after 0, 12, 24, 36, 48, 60, and 72 h of incubation at 30 °C
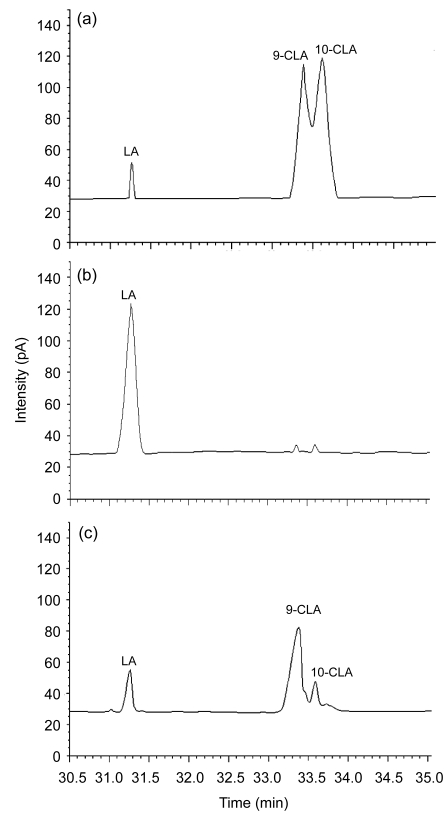
Gas chromatograms of standard CLA and LA demonstrated that the retention times of LA, 9- and 10-CLA isomers were 31.26, 33.38, and 33.62 min, respectively (Fig. 4a). The contents of major CLA isomers and the conversion rate of lp15 during fermentation with increased LA concentration are shown in Table 2. After incubation at 30 °C for 48 h in the MRS medium containing 100 to 1 000 µg/ml of LA, the total CLA yield of lp15 was 26.1, 48.7, 141.8, and 72.2 µg/ml, and the corresponding conversion rate was 26.1%, 24.4%, 23.6%, and 7.2%, respectively. The lower conversion rate at higher LA concentration can be ascribed to the growth inhibitory effect of LA. Despite a sharp drop of CLA synthesis in the medium containing 1 000 µg/ml LA, significant increases of CLA outputs were detected when increasing titers of LA (from 100 to 600 µg/ml) were supplemented (Table 2). Quantitative analysis revealed that the 9- and 10-CLA isomers comprised about 75% (w/w) and 25% (w/w) of total CLAs produced by strain lp15, respectively (Table 2 and Fig. 4c). Different from strain lp15, three quarters of total CLAs produced by L. plantarum NCUL005 were comprised of 10-CLA isomer and 9-CLA isomer accounted for only about one fourth (Zeng et al., 2009).
Fig. 4.
Gas chromatograms of LA and CLA mixture (a) and fatty acid profile of the MRS medium supplemented with 200 µg/ml of LA and L. plantarum lp15 at 0 h (b) and after 48 h of incubation (c)
Table 2.
Production of individual isomers and total CLAs by L. plantarum lp15 in the MRS broth supplemented with 100, 200, 600, and 1 000 µg/ml LA for 48 h of incubation at 30 °C
| LA (µg/ml) | 9-CLA (µg/ml) | 10-CLA (µg/ml) | Total CLA (µg/ml) | Conversion rate (%) |
| 100 | 19.9±0.1* | 6.2±0.2 | 26.1±0.3 | 26.1 |
| 200 | 37.2±0.5 | 11.5±0.3 | 48.7±0.8 | 24.4 |
| 600 | 109.2±0.2 | 32.6±0.4 | 141.8±0.6 | 23.6 |
| 1 000 | 54.8±0.8 | 17.4±0.4 | 72.2±1.2 | 7.2 |
Data are expressed as means±SD
The isolation and characterization of L. plantarum strain lp15 with a relatively high LA conversion rate and strong LA tolerance from naturally fermented pickles, not only added new bacterial strain source of producing beneficial compound CLAs, but also provided some premises for the future in production of fermented products enriched in CLAs in human diets. Next we will focus on enhancing the CLA-producing ability of this strain by optimizing the culture conditions, as well as mutagenesis or genetic engineering methods, such as whole-cell biocatalysis (Chen et al., 2011) or cell-surface displaying (Guo et al., 2010), and will try to expand its application in milk and vegetable fermentation.
4. Conclusions
Forty-three LAB strains with CLA-producing ability were isolated from three kinds of naturally fermented pickles. Of these isolates, lp15 identified as L. plantarum exhibited the highest CLA yield. Strain lp15 also displayed a relatively high LA tolerance and its growth was almost unaffected in the presence of high levels of LA (600 µg/ml). Under our culture conditions, the maximum amount of CLAs (48.7 µg/ml) produced by strain lp15 was detected at 48 h of incubation in the MRS broth containing 200 µg/ml LA. The total CLA production of strain lp15 supplemented with different concentrations of LA (100, 200, 600, and 1 000 µg/ml) ranged from 26.1 to 141.8 µg/ml in which the two CLA isomers (9-CLA and 10-CLA) were detected and more than 75% (w/w) of these isomers was 9-CLA. The corresponding conversion rate varied from 26.1% to 7.2% and a relatively high conversion rate was observed when lower LA was supplemented. The L. plantarum strain lp15 showing robust LA tolerance and high conversion rate is a valuable candidate that can be explored to enhance the CLA contents in fermented food products.
Footnotes
Project (No. 2007AA100402) supported by the National High-Tech R&D Program (863) of China
References
- 1.Alonso L, Cuesta EP, Gilliand SE. Production of free conjugated linoleic acid by Lactobacillus acidophilus and Lactobacillus casei of human intestinal origin. J Dairy Sci. 2003;86(6):1941–1946. doi: 10.3168/jds.S0022-0302(03)73781-3. [DOI] [PubMed] [Google Scholar]
- 2.Aymerich T, Martın B, Garriga M, Hugas M. Microbial quality and direct PCR identification of lactic acid bacteria and nonpathogenic staphylococci from artisanal low-acid sausages. Appl Environ Microbiol. 2003;69(8):4583–4594. doi: 10.1128/AEM.69.8.4583-4594.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett E, Ross RP, Fitzgerald GF, Stanton C. Rapid screening method for analyzing the conjugated linoleic acid production capabilities of bacterial cultures. Appl Environ Microbiol. 2007;73(7):2333–2337. doi: 10.1128/AEM.01855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjamin S, Spener F. Conjugated linoleic acids as functional food: an insight into their health benefits. Nutr Metab. 2009;6:36. doi: 10.1186/1743-7075-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya A, Banu J, Rahman M, Causey J, Fernandes G. Biological effects of conjugated linoleic acids in health and disease. J Nutr Biochem. 2006;17(12):789–810. doi: 10.1016/j.jnutbio.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Carmen Collado M, Hernandez M. Identification and differentiation of Lactobacillus, Streptococcus and Bifidobacterium species in fermented milk products with bifidobacteria. Microbiol Res. 2007;162(1):86–92. doi: 10.1016/j.micres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Chen ML, Guo Q, Wang RZ, Xu J, Zhou CW, Ruan H, He GQ. Construction of the yeast whole-cell Rhizopus oryzae lipase biocatalyst with high activity. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2011;12(7):545–551. doi: 10.1631/jzus.B1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chouinard PY, Corneau L, Butler WR, Chilliard Y, Drackley JK, Bauman DE. Effect of dietary lipid source on conjugated linoleic acid concentrations in milk fat. J Dairy Sci. 2001;84(3):680–690. doi: 10.3168/jds.S0022-0302(01)74522-5. [DOI] [PubMed] [Google Scholar]
- 9.Chung SH, Kim IH, Park HG, Kang HS, Yoon CS, Jeong HY, Choi NJ, Kwon EG, Kim YJ. Synthesis of conjugated linoleic acid by human-derived Bifidobacterium breve LMC 017: utilization as a functional starter culture for milk fermentation. J Agric Food Chem. 2008;56(9):3311–3316. doi: 10.1021/jf0730789. [DOI] [PubMed] [Google Scholar]
- 10.Churruca I, Fernandez-Quintela A, Portillo MP. Conjugated linoleic acid isomers: differences in metabolism and biological effects. Biofactors. 2009;35(1):105–111. doi: 10.1002/biof.13. [DOI] [PubMed] [Google Scholar]
- 11.Coakley M, Ross RP, Nordgren M, Fitzgerald G, Devery R, Stanton C. Conjugated linoleic acid biosynthesis by human-derived Bifidobacterium species. J Appl Microbiol. 2003;94(1):138–145. doi: 10.1046/j.1365-2672.2003.01814.x. [DOI] [PubMed] [Google Scholar]
- 12.Donovan DC, Schingoethe DJ, Baer RJ, Ryali J, Hippen AR, Franklin ST. Influence of dietary fish oil on conjugated linoleic acid and other fatty acids in milk fat from lactating dairy cows. J Dairy Sci. 2000;83(11):2620–2628. doi: 10.3168/jds.S0022-0302(00)75155-1. [DOI] [PubMed] [Google Scholar]
- 13.Drosinos EH, Paramithiotis S, Kolovos G, Tsikouras I, Metaxopoulos I. Phenotypic and technological diversity of lactic acid bacteria and staphylococci isolated from traditionally fermented sausages in southern Greece. Food Microbiol. 2007;24(3):260–270. doi: 10.1016/j.fm.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Feitoza AB, Pereira AF, da Costa NF, Ribeiro BG. Conjugated linoleic acid (CLA): effect modulation of body composition and lipid profile. Nutr Hosp. 2009;24(4):422–428. [PubMed] [Google Scholar]
- 15.Guo Q, Zhang W, Ma LL, Chen QH, Zhang HB, Ruan H, He GQ. A food industrial arming yeast expressing β-1,3-1,4-glucanase with enhanced thermal stability. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2010;11(1):41–51. doi: 10.1631/jzus.B0900185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha YL, Grimm NK, Pariza MW. Anticarcinogens from fried ground beef: heat-altered derivatives of linoleic acid. Carcinogenesis. 1987;8(12):1881–1887. doi: 10.1093/carcin/8.12.1881. [DOI] [PubMed] [Google Scholar]
- 17.Halade GV, Rahman MM, Fernandes G. Effect of CLA isomers and their mixture on aging C57BI/6J mice. Eur J Nutr. 2009;48(7):409–418. doi: 10.1007/s00394-009-0029-7. [DOI] [PubMed] [Google Scholar]
- 18.Hayek MG, Han SN, Wu DY, Watkins BA, Meydani M, Dorsey JL, Smith DE, Meydani SN. Dietary conjugated linoleic acid influences the immune response of young and old C57BL/6NCrlBR mice. J Nutr. 1999;129(1):32–38. doi: 10.1093/jn/129.1.32. [DOI] [PubMed] [Google Scholar]
- 19.Inoue N, Nagao K, Hirata J, Wang YM, Yanagita T. Conjugated linoleic acid prevents the development of essential hypertension in spontaneously hypertensive rats. Biochem Biophys Res Commun. 2004;323(2):679–684. doi: 10.1016/j.bbrc.2004.08.139. [DOI] [PubMed] [Google Scholar]
- 20.Jiang J, Bjorck L, Fonden R. Production of conjugated linoleic acid by dairy starter cultures. J Appl Microbiol. 1998;85(1):95–102. doi: 10.1046/j.1365-2672.1998.00481.x. [DOI] [PubMed] [Google Scholar]
- 21.Jones DF, Weiss WP, Palmquist DL. Short communication: influence of dietary tallow and fish oil on milk fat composition. J Dairy Sci. 2000;83(9):2024–2026. doi: 10.3168/jds.S0022-0302(00)75082-X. [DOI] [PubMed] [Google Scholar]
- 22.Jung MY, Kim GB, Jang ES, Jung YK, Park SY, Lee BH. Technical note: improved extraction method with hexane for gas chromatographic analysis of conjugated linoleic acids. J Dairy Sci. 2006;89(1):90–94. doi: 10.3168/jds.S0022-0302(06)72072-0. [DOI] [PubMed] [Google Scholar]
- 23.Kaban G, Kaya M. Identification of lactic acid bacteria and gram-positive catalase-positive cocci isolated from naturally fermented sausage (Sucuk) J Food Sci. 2008;73(8):M385–M388. doi: 10.1111/j.1750-3841.2008.00906.x. [DOI] [PubMed] [Google Scholar]
- 24.Kelley NS, Hubbard NE, Erickson KL. Conjugated linoleic acid isomers and cancer. J Nutr. 2007;137(12):2599–2607. doi: 10.1093/jn/137.12.2599. [DOI] [PubMed] [Google Scholar]
- 25.Kim YJ, Liu RH, Bond DR, Russell JB. Effect of linoleic acid concentration on conjugated linoleic acid production by Butyrivibrio fibrisolvens A38. Appl Environ Microbiol. 2000;66(12):5226–5230. doi: 10.1128/AEM.66.12.5226-5230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kishino S, Ogawa J, Ando A, Iwashita T, Fujita T, Kawashima H, Shimizu S. Structural analysis of conjugated linoleic acid produced by Lactobacillus plantarum, and factors affecting isomer production. Biosci Biotechnol Biochem. 2003;67(1):179–182. doi: 10.1271/bbb.67.179. [DOI] [PubMed] [Google Scholar]
- 27.Lin TY, Lin CW, Lee CH. Conjugated linoleic acid concentration as affected by lactic cultures and added linoleic. Food Chem. 1999;67(1):1–5. doi: 10.1016/S0308-8146(99)00077-1. [DOI] [Google Scholar]
- 28.Lock AL, Bauman DE, Garnsworthy PC. Short communication: effect of production variables on the cis-9, trans-11 conjugated linoleic acid content of cows’ milk. J Dairy Sci. 2005;88(8):2714–2717. doi: 10.3168/jds.S0022-0302(05)72950-7. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald HB. Conjugated linoleic acid and disease prevention: a review of current knowledge. J Am Coll Nutr. 2000;19(2):111S–118S. doi: 10.1080/07315724.2000.10718082. [DOI] [PubMed] [Google Scholar]
- 30.Martin JC, Valeille K. Conjugated linoleic acids: all the same or to everyone its own function? Reprod Nutr Dev. 2002;42(6):525–536. doi: 10.1051/rnd:2002042. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell PL, Mcleod RS. Conjugated linoleic acid and atherosclerosis: studies in animal models. Biochem Cell Biol. 2008;86(4):293–301. doi: 10.1139/O08-070. [DOI] [PubMed] [Google Scholar]
- 32.Nagao K, Yanagita T. Conjugated fatty acids in food and their health benefits. J Biosci Bioeng. 2005;100(2):152–157. doi: 10.1263/jbb.100.152. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa J, Kishino S, Ando A, Sugimoto S, Mihara K, Shimizu S. Production of conjugated fatty acids by lactic acid bacteria. J Biosci Bioeng. 2005;100(4):355–364. doi: 10.1263/jbb.100.355. [DOI] [PubMed] [Google Scholar]
- 34.Pariza MW, Park Y, Cook ME. The biologically active isomers of conjugated linoleic acid. Prog Lipid Res. 2001;40(4):283–298. doi: 10.1016/S0163-7827(01)00008-X. [DOI] [PubMed] [Google Scholar]
- 35.Park Y, Storkson JM, Albright KJ, Liu W, Pariza MW. Evidence that the trans-10, cis-12 isomer of conjugated linoleic acid induces body composition changes in mice. Lipids. 1999;34(3):235–241. doi: 10.1007/s11745-999-0358-8. [DOI] [PubMed] [Google Scholar]
- 36.Peng SS, Deng MD, Grund AD, Rosson RA. Purification and characterization of a membrane-bound linoleic acid isomerase from Clostridium sporogenes . Enzyme Microb Technol. 2007;40(4):831–839. doi: 10.1016/j.enzmictec.2006.06.020. [DOI] [Google Scholar]
- 37.Peterson DG, Kelsey JA, Bauman DE. Analysis of variation in cis-9, trans-11 conjugated linoleic acid (CLA) in milk fat of dairy cows. J Dairy Sci. 2002;85(9):2164–2172. doi: 10.3168/jds.S0022-0302(02)74295-1. [DOI] [PubMed] [Google Scholar]
- 38.Plengvidhya V, Breidt F, Lu Z, Fleming HP. DNA fingerprinting of lactic acid bacteria in sauerkraut fermentations. Appl Environ Microbiol. 2007;73(23):7697–7702. doi: 10.1128/AEM.01342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez I, Sesena S, Palop LL. Polyphasic study of the genetic diversity of lactobacilli associated with ‘Almagro’ eggplants spontaneous fermentation, based on combined numerical analysis of randomly amplified polymorphic DNA and pulsed-field gel electrophoresis patterns. J Appl Microbiol. 2004;97(2):446–458. doi: 10.1111/j.1365-2672.2004.02324.x. [DOI] [PubMed] [Google Scholar]
- 40.van Nieuwenhove CP, Oliszewski R, Gonzalez SN, Perez Chaia AB. Conjugated linoleic acid conversion by dairy bacteria cultured in MRS broth and buffalo milk. Lett Appl Microbiol. 2007;44(5):467–474. doi: 10.1111/j.1472-765X.2007.02135.x. [DOI] [PubMed] [Google Scholar]
- 41.Wall R, Ross RP, Fitzgerald GF, Stanton C. Microbial congjugated linoleic acid production—a novel probiotic trait? FST Bull. 2008;4(8):87–97. [Google Scholar]
- 42.Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 1997;27:2.4.1–2.4.5. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- 43.Woo PCY, Leung PKL, Leung KW, Yuen KY. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient. Mol Path. 2000;53(4):211–215. doi: 10.1136/mp.53.4.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng Z, Lin J, Gong D. Identification of lactic acid bacterial strains with high conjugated linoleic acid-producing ability from natural sauerkraut fermentations. J Food Sci. 2009;74(4):M154–M158. doi: 10.1111/j.1750-3841.2009.01123.x. [DOI] [PubMed] [Google Scholar]