Abstract
Utilization of a two-line breeding system via photoperiod-thermo sensitive male sterility has a great potential for hybrid production in wheat (Triticum aestivum L.). 337S is a novel wheat male sterile line sensitive to both short daylength/low temperature and long daylength/high temperature. Five F2 populations derived from the crosses between 337S and five common wheat varieties were developed for genetic analysis. All F1’s were highly fertile while segregation occurred in the F2 populations with a ratio of 3 fertile:1 sterile under short daylength/low temperature. It is shown that male sterility in 337S was controlled by a single recessive gene, temporarily designated as wptms3. Bulked segregant analysis (BSA) coupled with simple sequence repeat (SSR) markers was applied to map the sterile gene using one mapping population. The wptms3 gene was mapped to chromosome arm 1BS and flanked by Xgwm413 and Xgwm182 at a genetic distance of 3.2 and 23.5 cM, respectively. The accuracy and efficiency of marker-assisted selection were evaluated and proved essential for identifying homozygous recessive male sterile genotypes of the wptms3 gene in F2 generation.
Keywords: Heterosis, wptms3, Short daylength/low temperature sensitive male sterility, Molecular mapping, Triticum aestivum
1. Introduction
Heterosis is a common biological phenomenon. It has been widely utilized to improve yield and quality in many crops such as maize, rice, sorghum, oilseed rape, and cotton. The production of hybrid seeds is undertaken using the following systems: mechanical emasculation, chemical hybridizing agent (CHA), and application of male sterility (Zhou et al., 2005). Due to the laborious work of mechanical emasculation and difficulty in the right usage of CHA, the male sterile system is preferred by breeders.
It is well known that the three-line breeding system based on cytoplasmic male sterility (CMS) has made a great contribution to rice production and the discovery of a thermo-photoperiod sensitive recessive male sterile line (Shi, 1985) has resulted in the second breakthrough for hybrid rice breeding. As the photoperiod- or temperature-induced male sterile plants can be used not only as male sterile lines but also as maintainer lines, the two-line system facilitates the breeding procedure, providing an effective alternative to the CMS system for hybrid seed production. Despite unstable male sterility due to variable climatic conditions, the main advantages of the two-line system include no need to develop maintainer lines, no negative cytoplasmic effects, a wide range of genotypes as restorers, and reduced potential genetic vulnerability as caused by the three-line system.
Wheat (Triticum aestivum L.) is the most widely cultivated crop in the world and is very important for food supplies. However, breeding for hybrid wheat has encountered much more difficulty than that for rice. Since the CMS in wheat was first discovered by Kihara (1951), and the male sterile line and restorer line were developed by Wilson and Ross (1962), subsequent studies have been made to develop commercial hybrid wheat varieties. Several types of CMS such as T (Triticum timopheevii), K (Aegilops kotschyi), V (Aegilops ventricosa), and D2 (Aegilops crassa) have been identified (Murai, 2002; Chen, 2003). Utilization of these types of male sterile lines in hybrid wheat breeding has achieved limited success due to unstable male sterility, negative alloplasmic and cytoplasmic effects, and limited numbers of restoring genes (Murai and Tsunewaki, 1993; Li et al., 2006). The first long daylength-sensitive D2 type CMS wheat line was discovered by Sasakuma and Ohtsuka (1979), and thereafter a series of photoperiod-thermo sensitive male sterile lines have been identified in wheat (Tan et al., 1992; Murai and Tsunewaki, 1993; Luo et al., 1998; Murai, 1998; Xu and Yan, 1998). Most of these sterile lines are difficult to use widely for hybrid wheat production due to their requirements for extreme daylength or temperature. Recently, we identified a novel wheat male sterile line 337S, which shows good male sterility under both short daylength/low temperature and long daylength/high temperature (Guo et al., 2006a). There are two sowing windows for this line to be used as a male sterile line. Under an appropriate sowing time, it becomes fertile with the self-fertility rate >50%, and thus it can also be used as a maintainer line.
Simple sequence repeat (SSR), also called microsatellites, has been developed based on repeated DNA sequence variation and applied in many cereal crops including rice (Wu and Tanksley, 1993; Panaud et al., 1996), maize (Senior and Heun, 1993), and barley (Liu et al., 1996). In common wheat, which has a characteristic of large genomes and allopolyploidy, SSR is one of the most useful markers. The marker has the advantage of being genome-specific, codominant, and multiallelic, and thus is suitable for mapping agronomically important genes in wheat (Röder et al., 1998).
To date, only a few temperature, photoperiod, or photoperiod-thermo sensitive male sterile genes in wheat have been mapped (Xing et al., 2003; Cao et al., 2004). Our preliminary study showed that the photoperiod-thermo sensitive male sterility in 337S is governed by two recessive genes located on chromosomes 2B and 5B, respectively, under long daylength/high temperature (Guo et al., 2006b). In order to obtain an overall knowledge of the novel male sterile line 337S, the objective of the present study is to analyze inheritance of the short daylength/low temperature sensitive sterility in 337S and map the corresponding gene.
2. Materials and methods
2.1. Plant materials and field experimental design
The wheat line 337S was first identified at the Shayang Agricultural Research Institute as a temperature/daylength sensitive line (Rong and Cao, 1999). Based on multiple years of sowing date trials, 337S showed a high level of male sterility if sown before Sept. 30 or after Nov. 30, and it was partially fertile if sown in late October to early November (Guo et al., 2006a). Line 337S, as a female parent, was crossed with five common wheat varieties or lines (Huamai 8, Hua 12, 24I37, 24I38, 24I58) under natural conditions. All F1 hybrids were self-pollinated to generate F2 populations including the mapping population consisting of 262 F2 individuals from the cross between 337S and Huamai 8. Seeds were sown on the Experimental Station of Huazhong Agricultural University, Wuhan, China (N 30°32′ and E 114°20′). Considering wheat as an autumn-sown crop in Wuhan, an early sowing date can expose head development to a relative low temperature/short daylength. Hence, seeds of six parental lines, five F1 populations and five F2 populations were planted on Sept. 30 in 2006. The average temperature was 10.4 °C and daylength was 12.1 h during head development in 2007 (climatological data from Wuhan, Hubei Meteorological Bureau Regional Meteorological Center), which met the effect of short daylength and low temperature (Guo et al., 2006a).
2.2. DNA extraction and bulk construction
Fresh young leaves from F2 individuals of the mapping population and both parents, 337S and Huamai 8, were collected and frozen for DNA extraction. Total genomic DNA was extracted following a modified cetyltrimethylammonium bromide (CTAB) method. DNA concentration of each sample was determined using a spectrophotometer (Eppendorf BioPhotometer, Germany) and adjusted to 20 ng/µl for a working solution. Molecular markers were screened using bulked segregant analysis (BSA) (Michelmore et al., 1991). Equal amounts of DNA from 12 fertile and 12 sterile individuals were mixed to construct fertile bulk (BF) and sterile bulk (BS), respectively.
2.3. Fertility characterization
The main head of each plant was bagged at the heading stage to prevent cross pollination, and the bags were removed after 30 d (Guo et al., 2006b). Fertility evaluation was based on the percentage of fertile spikelets of the bagged main head. Plants were classified as male sterile if the self-fertility rate was less than 5% and as male fertile if the self-fertility rate was greater than or equal to 5%. A Chi-square test was performed to determine the segregation pattern of the fertility.
2.4. SSR analysis
A standard 10 μl reaction mixture contained 1× buffer, 2 mmol/L MgCl2, 1 U Taq polymerase, 0.5 μmol/L of each primer, 0.2 mmol/L of each deoxynucleotide, and 40 ng of template DNA, covered with one drop of mineral oil. Polymerase chain reaction (PCR) was performed in a thermal cycler (Bio-Rad MyCycler, USA). Amplification reactions were undertaken using the following profile: 94 °C for 3 min, then 45 cycles at 94 °C for 1 min, 1 min at either 50, 55, or 60 °C (depending on the individual primer set), 2 min at 72 °C, with a final extension step of 10 min at 72 °C (Röder et al., 1998). The PCR products were separated on a 6% polyacrylamide gel electrophoresis (PAGE) sequencing gel run at 80 W for 1.5 h. The gel was removed from the apparatus and stained using the silver-staining method (Xu et al., 2002). SSR primers were synthesized based on the primer sequences reported by Röder et al. (1998) and Holton et al. (2002). A total of 228 SSR markers distributed over all 21 wheat chromosomes were used to screen for polymorphisms between the two parental lines. The polymorphic SSR markers were selected to further screen the two DNA bulks to identify candidate markers linked to the target gene. The markers found to be polymorphic between the two bulks were selected to analyze the F2 extreme recessive class (Zhang et al., 1994) and the whole population.
2.5. Genetic mapping and evaluation of marker-assisted selection
Linkage analysis was conducted using the software MAPMAKER (Lander et al., 1987). A linkage map was constructed with a critical limit of detection (LOD) score threshold of 3.0 and the genetic distance (centimorgan, cM) was estimated using the Kosambi (1943) mapping function. The accuracy of marker-assisted selection (AMAS) and the efficiency of marker-assisted selection (EMAS) were calculated for the homozygotic genotype of the target gene in the F2 population as demonstrated by Peng et al. (2000b). In the present study, AMAS and EMAS for the male sterile gene in the F2 progeny were calculated using the following formulae.
When a single marker was used,
| AMAS=NssM/NM×100%, |
| EMAS=NssM/NssT×100%, |
where N M is total number of homozygous 337S-type marker genotype; N ssM is the number of homozygous recessive male sterile genotype out of the homozygous 337S-type marker genotype; N ssT is the total number of homozygous recessive male sterile genotypes in the population (N ssM≤N M and N ssM≤N ssT).
When two markers were used,
| AMAS=NssMc/NMc×100%, |
| EMAS=NssMc/NssT×100%, |
where N Mc is total number of 337S-type genotypes homozygous at two marker loci; N ssMc is the number of homozygous recessive male sterile genotype out of the 337S-type genotypes homozygous at two marker loci (N ssMc≤N Mc and N ssMc≤N ssT).
3. Results
3.1. Genetic analysis of male sterility
The field experiment indicated that the fertility difference between 337S and the other five common wheat varieties was significant. During anthesis, the parental line 337S exhibited apparent differences in phenotype from others, with the glume opening and the stigma exerting. The F1 progenies from the five crosses showed the same phenotype as the corresponding male parental lines whereas trait segregation occurred in the F2 generations. Fertility analysis indicated that 337S appeared completely sterile (with fertility rate equal to 0%) while the other five male parental lines and all F1’s were highly fertile (with fertility rate above 90%). Male sterility of 337S thus was controlled by recessive gene(s). The fertility rate varied from 0% to 100% and when plotted, a bimodal distribution was observed in the F2 population (Fig. 1).
Fig. 1.
Distribution of the fertility rate in the F2 population of the cross 337S (with fertility rate equal to 0%)/Huamai 8 (with fertility rate above 90%) grown under short daylength/low temperature conditions
Chi-square test showed that the fertility segregation fitted a ratio of 3:1 in all the five F2 populations. The sterility in 337S was thus governed by a single recessive gene under short daylength and low temperature (Table 1). We temporarily designated this gene wptms3. Here, wptms stands for wheat photoperiod-thermo sensitive male sterility.
Table 1.
Fertility segregation patterns of F2 populations sown on Sept. 30, 2006
| Cross | nt | ns | nf | χ2(p3:1) | P |
| 337S/Huamai 8 | 262 | 64 | 198 | 0.05 | 0.75–0.90 |
| 337S/Hua 12 | 234 | 70 | 164 | 3.01 | 0.05–0.10 |
| 337S/24I37 | 253 | 75 | 178 | 2.91 | 0.05–0.10 |
| 337S/24I38 | 246 | 55 | 191 | 0.92 | 0.25–0.50 |
| 337S/24I58 | 238 | 68 | 170 | 1.62 | 0.10–0.25 |
n t: total number of plants; n s: number of sterile plants; n f: number of fertile plants. χ 2 (p3:1): chi-square test for the ratio of 3 fertile:1 sterile; P: probability
3.2. Screening SSR markers linked to the male sterile gene
A total of 228 SSR primers were screened for polymorphism between the two parents, 337S and Huamai 8. Fourteen primer pairs failed in amplifications, and 73 primer pairs showed polymorphism between the two parents. These polymorphic SSR markers were further used to screen for polymorphisms between the sterile and fertile DNA pools. Three SSR markers, Xgwm273, Xgwm413 from chromosome 1B, and Xgwm182 from chromosome 5D, showed polymorphism between the two DNA pools. The polymorphisms were confirmed in the individuals used to construct the two gene pools. Thus, the three markers were likely to be linked with the male sterile gene. Fig. 2 indicates the amplification patterns of the primers Xgwm413 and Xgwm182, respectively.
Fig. 2.
Amplification patterns of the SSR markers Xgwm413 (a) and Xgwm182 (b)
Lanes: P1, Huamai 8; P2, 337S; BF, fertile bulk; BS, sterile bulk; 1–12, fertile individuals; 13–24, sterile individuals. The arrows indicate the polymorphisms
3.3. Genetic mapping
In order to confirm the linkage relationship, Xgwm273, Xgwm413, and Xgwm182 combined with three neighboring markers from chromosome 1B and three from chromosome 5D, respectively, were analyzed using the recessive class consisting of 64 totally sterile plants (Zhang et al., 1994). As a result, a linkage map of 85.1 cM was constructed with a critical LOD score=3. The map consisted of six loci including four chromosome 1B marker loci, one locus amplified by a chromosome 5D marker, and the target gene. The male sterile gene wptms3 was thus mapped in an interval between Xgwm413 and Xgwm182 at a genetic distance of 3.2 and 23.5 cM, respectively, on chromosome arm 1BS (Fig. 3). According to Röder et al. (1998) and Peng et al. (1999; 2000a; 2000b), the relatively loosely linked markers, Xgwm273 and Xgwm264, were on the distal side of the map, with distances of 9.9 and 20.3 cM to the target gene wptms3, respectively, and on the proximal side, there is a marker, Xgwm11, far from the target gene.
Fig. 3.
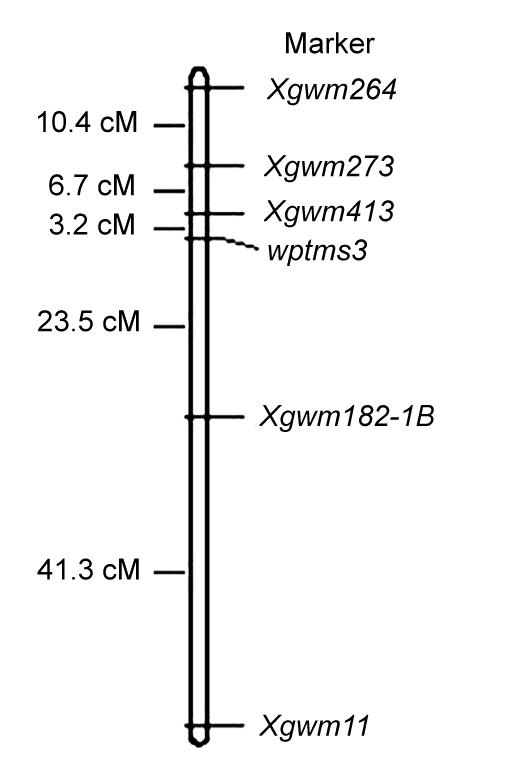
Linkage map of a chromosome 1B segment showing location of the photoperiod-thermo sensitive male sterile gene wptms3
The markers, Xgwm413 and Xgwm182, were further used to genotype the whole F2 mapping population, and the results validated the reliability of the linkage map. One-way variance analysis using marker genotypes as groups (Soller et al., 1976) showed that the fertility difference or variation between the groups was highly significant (Table 2). This again verified the linkage between markers and the target gene.
Table 2.
One-way variance analysis of the male sterility using genotypes of SSR markers as groups among F2 population (337S/Huamai 8)
| Marker | MS effect | dferror | MSerror | F | P |
| Xgwm182 | 1.41 | 260 | 0.10 | 13.78 | 0.00** |
| Xgwm413 | 1.75 | 259 | 0.11 | 16.43 | 0.00** |
| Xgwm335 | 0.12 | 259 | 0.08 | 0.64 | 0.53 |
| Xgwm371 | 0.03 | 259 | 0.76 | 0.04 | 0.96 |
| Xgwm148 | 0.68 | 259 | 0.11 | 6.32 | 0.00** |
| Xgwm374 | 2.39 | 260 | 0.10 | 23.13 | 0.00** |
| Xgwm120 | 0.72 | 259 | 1.89 | 0.38 | 0.68 |
MS: mean square; df: degree of freedom, df error value for dominant and codominant markers is 260 and 259, respectively; F: F statistic; P: probability
Significance at 0.01 probability level
Similarly, the five markers, Xgwm335 and Xgwm371 from chromosome 5B, and Xgwm148, Xgwm374, and Xgwm120 from chromosome 2B, identified to be linked with wptms1 and wptms2, respectively, in our previous study (Guo et al., 2006b), were also used to screen the whole F2 population (Table 2). The results showed that the fertility difference between the groups divided by Xgwm374 and Xgwm148 was significant, suggesting that there might be another locus on chromosome 2B, wptms2, affecting the fertility variation. This explains continuous variation in the fertility of the fertile class in Fig. 1. However, no locus on chromosome 5B was detected to significantly decorate the fertility variation.
3.4. Selection via single and bracketing marker(s)
When selection based on a single marker was performed, AMAS and EMAS were 82.84% and 86.85%, respectively, for Xgwm413, while they were 67.71% and 71.25% for Xgwm182 (Table 3). It is apparent that AMAS and EMAS improved when the marker loci became closer to the target gene wptms3. When the two bracketing markers were used to select male sterile plants in the F2 progeny, AMAS reached 90.10% despite the distance between the two markers extending to 26.7 cM, but EMAS slightly declined.
Table 3.
Accuracy and efficiency of marker-assisted selection of homozygous male sterile genotypes of the wptms3 gene based on a single marker and two bracketing markers in the F2 generation
| Marker | MD1 (cM) | MD2 (cM) | AMAS (%) | EMAS (%) |
| Xgwm413 | 3.2 | 82.84 | 86.85 | |
| Xgwm182 | 23.5 | 67.71 | 71.25 | |
| Xgwm413-Xgwm182 | 26.7 | 90.10 | 68.13 |
MD1: map distance of the single marker to wptms3; MD2: map distance between the two bracketing markers; AMAS: accuracy of marker-assisted selection; EMAS: efficiency of marker-assisted selection
4. Discussion
337S is the first wheat male sterile line sensitive to both short daylength/low temperature and long daylength/high temperature, providing two sowing time windows for the expression of male sterility. This male sterile line has no negative cytoplasmic effect and is controlled by recessive genes (Guo et al., 2006a). It could be widely used for hybrid wheat production because common wheat varieties are expected to carry fertility restoring genes for this male sterile line.
In the present study, the inheritance of male sterility under short daylength/low temperature was detected to be monogenic. The mapping analysis indicated that the male sterile gene wptms3 located on chromosome 1B, flanked by Xgwm413 and Xgwm182, differed from those reported by Guo et al. (2006b) and thus is a new gene. To date, several fertility restoring genes against CMS in wheat have been mapped on chromosome 1B (Ahmed et al., 2001; Li et al., 2005; Zhou et al., 2005). Therefore, there are regions on chromosome 1B related to fertility performance. The SSR marker Xgwm413 was identified to be closely linked to the male sterile gene in the present study and was found to be linked to yellow rust resistance genes in earlier reports (Peng et al., 1999; 2000a; 2000b; Ma et al., 2001). These studies indicate that genes are not randomly distributed over the genome of a species, but they are rather frequently clustered on particular chromosomes (Peng et al., 1999). The clustering of genes coding traits may be the result of the co-evolution of plant species and their adaptation to environments (Peng et al., 1999).
The microsatellite marker Xgwm182 was assigned to chromosome 5D, and sizes of the amplified bands in the two parents were 163 and 187 bp, respectively (Röder et al., 1998). However, the marker Xgwm182 was first mapped to chromosome 1B in the present study, and the amplified bands in the two parents were approximately 100 bp. Thus the Xgwm182 locus in the present study is a new marker locus and differs from that one located on chromosome 5D. Therefore, we can tentatively designate the new marker locus as Xgwm182-1B.
As analyzed above, there is a new gene controlling the fertility of 337S under short daylength/low temperature. According to the close linkage of the gene with SSR marker Xgwm413 that physically located on the terminal region of chromosome 1BS, we assume that the male sterile gene wptms3 is, in fact, physically located in a gene/marker cluster that is distal from the centromere on chromosome 1BS (Peng et al., 2000a).
With the identification of the molecular markers linked to the male sterile gene wptms3 in the present study, and wptms1 and wptms2 in our previous study, they could be very useful for developing and improving new male sterile lines via marker-assisted selection. In wheat breeding programs, we can use these linked markers to distinguish the sterile genotypes earlier, which can help to shorten the breeding time and facilitate the whole procedure. As the long daylength/high temperature induced male sterility in 337S is controlled by two complementary genes (Guo et al., 2006a; 2006b), only plants with genotype aabb are sterile and the rate of sterile plants is low in progeny. Thus, mapping of wptms3 in the present study provides a more feasible and reliable method for identifying male sterile plants in practical breeding. Furthermore, the AMAS and EMAS of selection based on single markers can be over 80%, and the AMAS of selection via two bracketing markers can exceed 90% even with a large distance between the two markers. Therefore, it is helpful for breeders to use these linked markers to identify those male sterile recessive genotypes in early generation and period with high accuracy.
Several methods discriminating fertility for genetic analysis arose in previous studies (Jia et al., 2001; Wang et al., 2003). In this study, the inflexion of a curve was used as the cut-off point between sterile and fertile individuals for the fertility analysis as this division can reflect the native change of a matter. As a result, the cut-off point was determined at the point of 5% fertile spikelets, which was consistent with the consensus in most former researches (Huang et al., 2000; Lee et al., 2005). As photoperiod-thermo sensitive male sterility is sensitive to the environment, individuals used to form the two pools in the present study were selected based on the percentage of fertile spikelets of the bagged main head while considering the other three relevant factors to ensure the reliability: heading date, characters of spikes, and the percentage of fertile spikelets of the single plant.
It is clear that the fertility of photoperiod-sensitive sterile lines is determined by the interaction of genes and environments and the mechanism is complex. Isolation, cloning, and characterization of the male sterile gene can promote molecular study of the genetic male sterile trait and improve its application in hybrid breeding. Even though three male sterile genes (wptms1, wptms2, and wptms3) have been identified in the novel wheat male sterile line 337S, the linkage between markers and target genes is not tight enough for gene cloning. Therefore, fine mapping and cloning the male sterile genes in 337S under different environments are underway and will further improve understanding of the genetic mechanism of the male sterile system in utilization of wheat heterosis.
Footnotes
Project supported by the National Basic Research Program (973) of China (Nos. 2007CB109006 and 2009CB118304), the National High-Tech R&D Program (863) of China (No. 2009AA101102), and the National Natural Science Foundation of China (Nos. 30671291 and 30971777)
References
- 1.Ahmed TA, Tsujimoto H, Sasakuma T. QTL analysis of fertility-restoration against cytoplasmic male sterility in wheat. Genes Genet Syst. 2001;76(1):33–38. doi: 10.1266/ggs.76.33. [DOI] [PubMed] [Google Scholar]
- 2.Cao SH, Guo XL, Liu DC, Zhang XQ, Zhang AM. Preliminary gene-mapping of photoperiod-thermo sensitive genic male sterility in wheat (Triticum aestivum L.) Acta Genet Sin. 2004;31(3):293–298. (in Chinese) [PubMed] [Google Scholar]
- 3.Chen QF. Improving male fertility restoration of common wheat for Triticum timopheevii cytoplasm. Plant Breeding. 2003;122(5):401–404. doi: 10.1046/j.1439-0523.2003.00875.x. [DOI] [Google Scholar]
- 4.Guo RX, Sun DF, Cheng XD, Rong DF, Li CD. Inheritance of photoperiod-sensitive male sterility in wheat. Aust J Agric Res. 2006;57(2):187–192. doi: 10.1071/AR05161. [DOI] [Google Scholar]
- 5.Guo RX, Sun DF, Tan ZB, Rong DF, Li CD. Two recessive genes controlling thermophotoperiod-sensitive male sterility in wheat. Theor Appl Genet. 2006;112(7):1271–1276. doi: 10.1007/s00122-006-0228-z. [DOI] [PubMed] [Google Scholar]
- 6.Holton TA, Christopher JT, McClure L, Harker N, Henry RJ. Identification and mapping of polymorphic SSR markers from expressed gene sequences of barley and wheat. Mol Breeding. 2002;9(2):63–71. doi: 10.1023/A:1026785207878. [DOI] [Google Scholar]
- 7.Huang QY, He YQ, Jing RC, Zhu RS, Zhu YG. Mapping of the nuclear fertility restorer gene for HL cytoplasmic male sterility in rice using microsatellite markers. Chin Sci Bull. 2000;45(5):430–432. doi: 10.1007/BF02884944. [DOI] [Google Scholar]
- 8.Jia JH, Zhang DS, Li CY, Qu XP, Wang SW, Chamarerk V, Nguyen HT, Wang B. Molecular mapping of the reverse thermo-sensitive genic male-sterile gene (rtms1) in rice. Theor Appl Genet. 2001;103(4):607–612. doi: 10.1007/PL00002916. [DOI] [Google Scholar]
- 9.Kihara H. Substitution of nucleus and its effects on genome manifestations. Cytologia. 1951;16:177–193. [Google Scholar]
- 10.Kosambi DD. The estimation of map distances from recombination values. Ann Human Genet. 1943;12(1):172–175. [Google Scholar]
- 11.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1(2):174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 12.Lee DS, Chen LJ, Suh HS. Genetic characterization and fine mapping of a novel thermo-sensitive genic male-sterile gene tms6 in rice (Oryza sativa L.) Theor Appl Genet. 2005;111(7):1271–1277. doi: 10.1007/s00122-005-0044-x. [DOI] [PubMed] [Google Scholar]
- 13.Li XL, Liu LK, Hou N, Liu GQ, Liu CG. SSR and SCAR markers linked to the fertility-restoring gene for a D2-type cytoplasmic male-sterile line in wheat. Plant Breeding. 2005;124(4):413–415. doi: 10.1111/j.1439-0523.2005.01120.x. [DOI] [Google Scholar]
- 14.Li YF, Zhao CP, Zhang FT, Sun H, Sun DF. Fertility alteration in the photo-thermo-sensitive male sterile line BS20 of wheat (Triticum aestivum L.) Euphytica. 2006;151(2):207–213. doi: 10.1007/s10681-006-9141-4. [DOI] [Google Scholar]
- 15.Liu ZW, Biyashev RM, Saghai Maroof MA. Development of simple sequence repeat DNA markers and their integration into a barley linkage map. Theor Appl Genet. 1996;93(5-6):869–876. doi: 10.1007/BF00224088. [DOI] [PubMed] [Google Scholar]
- 16.Luo HB, He JM, Dai JT, Liu XL, Yang YC. Studies on the characteristics of seed production of two ecological male sterile lines in wheat. J Hunan Agric Univ. 1998;24(2):83–89. (in Chinese) [Google Scholar]
- 17.Ma JX, Zhou RH, Dong YS, Wang LF, Wang XM, Jia JZ. Molecular mapping and detection of the yellow rust resistance gene Yr26 in wheat transferred from Triticum turgidum L. using microsatellite markers. Euphytica. 2001;120(2):219–226. doi: 10.1023/A:1017510331721. [DOI] [Google Scholar]
- 18.Michelmore RW, Paran I, Kesseli V. Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. PNAS. 1991;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murai K. Two-Line System for Hybrid Wheat Production Using Photoperiod-Sensitive Cytoplasmic Male Sterility. In: Zhang A, Huang TC, editors. Proceedings of the First International Workshop on Hybrid Wheat; Beijing: China Agricultural University Press; 1998. pp. 37–39. (in Chinese) [Google Scholar]
- 20.Murai K. Comparison of two fertility restoration systems against photoperiod-sensitive cytoplasmic male sterility in wheat. Plant Breeding. 2002;121(4):363–365. doi: 10.1046/j.1439-0523.2002.720110.x. [DOI] [Google Scholar]
- 21.Murai K, Tsunewaki K. Photoperiod-sensitive cytoplasmic male sterility in wheat with Aegilops crassa cytoplasm. Euphytica. 1993;67(1-2):41–48. doi: 10.1007/BF00022723. [DOI] [Google Scholar]
- 22.Panaud O, Chen X, McCouch SR. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.) Mol Gen Genet. 1996;252(5):597–607. doi: 10.1007/BF02172406. [DOI] [PubMed] [Google Scholar]
- 23.Peng JH, Fahima T, Röder MS, Li YC, Dahan A, Grama A, Ronin YI, Korol AB, Nevo E. Microsatellite tagging of the stripe-rust resistance gene YrH52 derived from wild emmer wheat, Triticum dicoccoides, and suggestive negative crossover interference in chromosome 1B. Theor Appl Genet. 1999;98(6-7):862–872. doi: 10.1007/s001220051145. [DOI] [Google Scholar]
- 24.Peng JH, Fahima T, Röder MS, Huang QY, Dahan A, Li YC, Grama A, Nevo E. High-density molecular map of chromosome region harboring stripe-rust resistance genes YrH52 and Yr15 derived from wild emmer wheat, Triticum dicoccoides . Genetica. 2000;109(3):199–210. doi: 10.1023/A:1017573726512. [DOI] [PubMed] [Google Scholar]
- 25.Peng JH, Fahima T, Röder MS, Li YC, Grama A, Nevo E. Microsatellite high-density mapping of the stripe rust resistance gene YrH52 region on chromosome 1B and evaluation of its marker-assisted selection in the F2 generation in wild emmer wheat. New Phytol. 2000;146(1):141–154. doi: 10.1046/j.1469-8137.2000.00617.x. [DOI] [Google Scholar]
- 26.Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149(4):2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rong DF, Cao WM. The fertility characters of a novel thermo-photoperiod sensitive male sterile line 337S. Trit Crops. 1999;19(1):20–24. (in Chinese) [Google Scholar]
- 28.Sasakuma T, Ohtsuka I. Cytoplasmic effects of Aegilops species having D genome in wheat. I. Cytoplasmic differentiation among five species regarding pistilody induction. Seiken Ziho. 1979;27:59–65. [Google Scholar]
- 29.Senior ML, Heun M. Mapping maize microsatellites and polymerase chain reaction confirmation of the targeted repeats using a CT primer. Genome. 1993;36(5):884–889. doi: 10.1139/g93-116. [DOI] [PubMed] [Google Scholar]
- 30.Shi MS. The discovery and study of the photosensitive recessive male-sterile rice (Oryza sativa L. Japonica) Sci Agric Sin. 1985;(2):44–48. (in Chinese) [Google Scholar]
- 31.Soller M, Brody T, Genizi A. On the power of experimental designs for the detection of linkage between marker loci and quantitative loci in crosses between inbred lines. Theor Appl Genet. 1976;47(1):35–39. doi: 10.1007/BF00277402. [DOI] [PubMed] [Google Scholar]
- 32.Tan CH, Yu GD, Yang PF, Zhang ZH, Pan Y, Zheng J. Preliminary study on sterility of thermo-photo-sensitive genetic male sterile wheat in Chongqing. Southwest China J Agric Sci. 1992;5(4):1–6. (in Chinese) [Google Scholar]
- 33.Wang YG, Xing QH, Deng QY, Liang FS, Yuan LP, Weng ML, Wang B. Fine mapping of the rice thermo-sensitive genic male-sterile gene tms5 . Theor Appl Genet. 2003;107(5):917–921. doi: 10.1007/s00122-003-1327-8. [DOI] [PubMed] [Google Scholar]
- 34.Wilson JA, Ross WM. Male sterility interaction of the Triticum aestivum nucleus and Triticum timopheevi cytoplasm. Wheat Inf Serv. 1962;14:29–30. [Google Scholar]
- 35.Wu KS, Tanksley SD. Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol Gen Genet. 1993;241(1-2):225–235. doi: 10.1007/BF00280220. [DOI] [PubMed] [Google Scholar]
- 36.Xing QH, Ru ZG, Zhou CJ, Xue X, Liang CY, Yang DE, Jin DM, Wang B. Genetic analysis, molecular tagging and mapping of the thermo-sensitive genic male-sterile gene (wtms1) in wheat. Theor Appl Genet. 2003;107(8):1500–1504. doi: 10.1007/s00122-003-1385-y. [DOI] [PubMed] [Google Scholar]
- 37.Xu NY, Yan JQ. Studies on photoperiod-sensitive cytoplasmic male sterility in wheat. J Wuhan Bot Res. 1998;16(2):97–105. (in Chinese) [Google Scholar]
- 38.Xu SB, Tao YF, Yang ZQ, Chu JY. A simple and rapid method used for silver staining and gel preservation. Hereditas. 2002;24(3):335–336. (in Chinese) [PubMed] [Google Scholar]
- 39.Zhang QF, Shen BZ, Dai XK, Mei MH, Saghai MAM, Li ZB. Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male sterility in rice. PNAS. 1994;91(18):8675–8679. doi: 10.1073/pnas.91.18.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou WC, Kolb FL, Domier LL, Wang SW. SSR markers associated with fertility restoration genes against Triticum timopheevii cytoplasm in Triticum aestivum . Euphytica. 2005;141(1-2):33–40. doi: 10.1007/s10681-005-5067-5. [DOI] [Google Scholar]




