Summary
Regulation of gene expression plays an integral role in adaptation of cells to hypoxic stress. In mammals, prolyl hydroxylases control levels of the central transcription factor hypoxia inducible factor (HIF) through regulation of HIFα subunit stability. Here, we report that the hydroxylase Ofd1 regulates the Sre1 hypoxic transcription factor in fission yeast by controlling DNA binding. Prolyl hydroxylases require oxygen as a substrate and the activity of Ofd1 regulates Sre1-dependent transcription. In the presence of oxygen, Ofd1 binds the Sre1 N-terminal transcription factor domain (Sre1N) and inhibits Sre1-dependent transcription by blocking DNA binding. In the absence of oxygen, the inhibitor Nro1 binds Ofd1, thereby releasing Sre1N and leading to activation of genes required for hypoxic growth. In contrast to the HIF system where proline hydroxylation is essential for regulation, Ofd1 inhibition of Sre1N does not require hydroxylation, and thus defines a new mechanism for hypoxic gene regulation.
Introduction
Organisms and cells adapt to changes in their environment by activating signal transduction pathways that alter gene expression. The ability to sense and respond to fluctuations in oxygen supply and hypoxic stress is essential for proper metabolic regulation and survival (Semenza, 2007). In bacteria, the FNR transcription factor utilizes the oxygen sensitivity of [4Fe-4S] clusters to sense oxygen levels. Under anaerobic conditions, the presence of [4Fe-4S] clusters promotes the dimerization of FNR which facilitates site-specific DNA binding (Green et al., 2009). Another bacterial transcription factor OxyR indirectly senses environmental oxygen by using a thiol-disulfide redox switch to detect reactive oxygen species generated by aerobic metabolism (Helmann, 2002).
In mammals, the heterodimeric transcription factor hypoxia-inducible factor (HIF) plays a key role in adaptation of cells to a hypoxic environment (Gordan and Simon, 2007; Schofield and Ratcliffe, 2005). The activity of HIF is regulated by post-translational hydroxyl modifications to the HIF-α subunit that independently control HIF-α degradation and transcriptional activity. In the presence of oxygen, a family of prolyl 4-hydroxylases, named PHD1-3, hydroxylate two proline residues in HIF-α (Ozer and Bruick, 2007). Proline hydroxylated HIF-α is recognized by the von Hippel-Lindau (VHL) E3 ubiquitin ligase, which targets HIF-α for proteasomal degradation (Kaelin, 2005). In a second mechanism of oxygen regulation, factor inhibiting HIF (FIH), an asparaginyl hydroxylase, prevents recruitment of the transcriptional activators p300/CBP by hydroxylating an asparagine residue in HIF-α (Hirota and Semenza, 2005). The HIF-α prolyl and asparaginyl hydroxylases are members of the 2-OG-Fe(II)-dependent dioxygenase family of enzymes that require oxygen as a substrate (Ozer and Bruick, 2007). Under hypoxic conditions, these 2-OG-Fe(II)-dependent dioxygenases are inhibited and fail to hydroxylate HIF-α, leading to stabilization of active HIF-α and expression of genes required for hypoxic growth. Thus, hydroxylases can act as oxygen sensors to regulate hypoxic transcription (Schofield and Ratcliffe, 2005).
The membrane-bound transcription factor Sre1 is a key regulator of hypoxic gene expression in the fission yeast Schizosaccharomyces pombe (Hughes et al., 2005; Todd et al., 2006). Yeast Sre1 is the ortholog of mammalian sterol regulatory element binding protein (SREBP) that regulates cellular cholesterol and lipid homeostasis (Espenshade and Hughes, 2007). Under low oxygen, Sre1 is proteolytically cleaved and the N-terminal transcription factor domain (Sre1N) enters the nucleus and upregulates genes essential for low oxygen growth (Hughes et al., 2005). Sre1N activates its own transcription and this positive feedback loop is required for maximal induction of Sre1N under low oxygen (Hughes and Espenshade, 2008; Todd et al., 2006). Sre1N is rapidly degraded (t1/2=7 min) through a proteasome-dependent pathway, allowing rapid down-regulation of Sre1N upon reintroduction of oxygen (Hughes and Espenshade, 2008).
Recently, we identified Ofd1, a prolyl 4-hydroxylase-like 2-OG-Fe(II) dioxygenase, and its binding partner Nro1 as regulators of Sre1N degradation (Hughes and Espenshade, 2008; Lee et al., 2009). Ofd1 consists of two domains: an N-terminal 2-OG-Fe(II) dependent dioxygenase domain (Ofd1N-REG, aa 1-254) and a C-terminal domain (Ofd1CTD, aa 255-515) that functions in Sre1N degradation. Ofd1N-REG domain is homologous to the prolyl hydroxylase domain of the HIF PHD enzymes and structural studies predict Ofd1N-REG to function as a hydroxylase (Henri et al., 2010; Kim et al., 2010). In the presence of oxygen, Ofd1 accelerates Sre1N degradation by an unknown mechanism. In the absence of oxygen, Nro1 binds to the C-terminal domain of Ofd1 (Ofd1CTD) and blocks the ability of Ofd1 to accelerate Sre1N turnover, thus, resulting in Sre1N accumulation. Unlike PHD-dependent degradation of HIF, Sre1N degradation does not require Ofd1 hydroxylase activity. Rather, the Ofd1N-REG dioxygenase domain is an oxygen sensor that regulates the inhibitory binding of Nro1 to Ofd1CTD (Hughes and Espenshade, 2008; Lee et al., 2009). Interestingly, Ofd1 and Nro1 do not contain domains related to the ubiquitin-proteasome system, so it is unclear how Ofd1 accelerates Sre1N proteasomal degradation.
To gain insight into how Ofd1 accelerates Sre1N degradation, we sought to identify components of the ubiquitin-proteasome pathway required for Sre1N degradation. These studies revealed that deletion of the E2 ubiquitin-conjugating enzyme Rhp6 or the E3 ubiquitin ligase Ubr1 prevents Sre1N degradation. Using ubr1Δ cells, we discovered a second function for Ofd1 outside of Sre1N degradation. Surprisingly, Ofd1-Nro1 cooperate to control Sre1N DNA binding in addition to regulating Sre1N stability. In the presence of oxygen, the Ofd1CTD binds Sre1N and inhibits Sre1N transcriptional activity by blocking DNA binding. In the absence of oxygen, Nro1 binds to Ofd1CTD, releasing Sre1N and allowing gene transcription required for hypoxic growth. These studies outline a new paradigm for regulation of hypoxic gene expression where the non-enzymatic domain of the Ofd1 dioxygenase controls Sre1N by inhibiting DNA binding. Thus, Ofd1-Nro1 cooperate to regulate both Sre1N transcriptional activity and stability, thereby ensuring rapid oxygen-dependent control of hypoxic gene expression.
Results
Sre1N degradation requires the E3 ubiquitin ligase Ubr1
Proteolysis controls both the synthesis and turnover of the hypoxic transcription factor Sre1. Membrane-bound Sre1 is first activated by proteolytic cleavage to release the soluble N-terminal transcription factor domain of Sre1 (Sre1N) from the membrane. Soluble Sre1N is rapidly degraded in the presence of oxygen through a proteasome-dependent mechanism (Hughes and Espenshade, 2008). Rapid degradation of Sre1N in the presence of oxygen requires Ofd1 (t1/2=7 min in wild type and t1/2=23 min in ofd1Δ). However, the block to Sre1N degradation is incomplete in ofd1Δcells, suggesting that other proteins are required for Sre1N degradation.
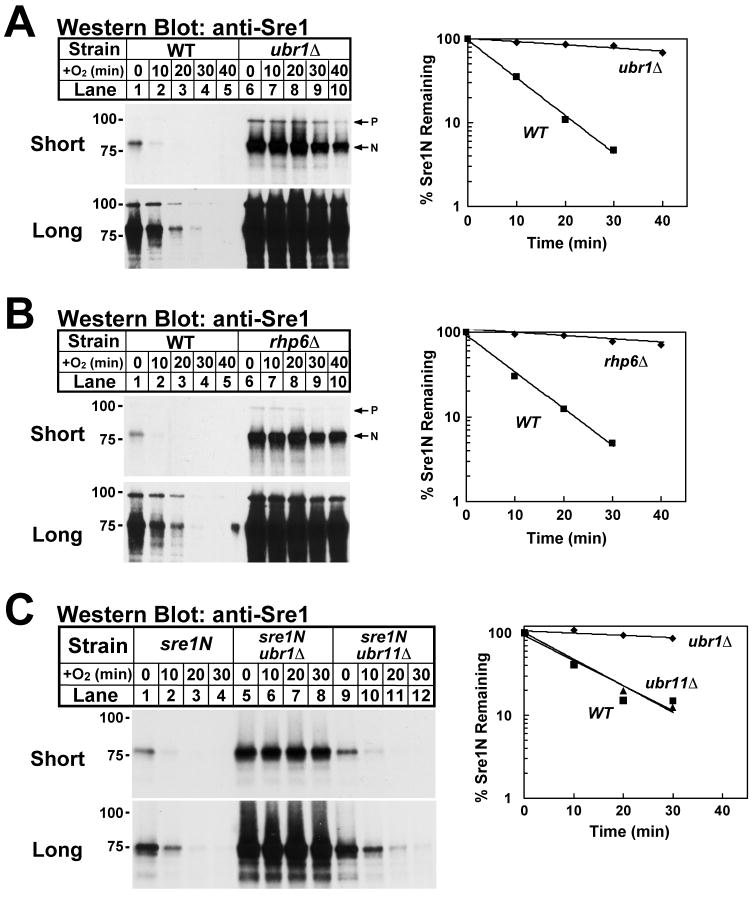
To identify ubiquitin-dependent enzymes involved in proteasome-mediated Sre1N degradation, we screened viable E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligase deletion mutants for effects on Sre1N half-life by cycloheximide chase (Hughes and Espenshade, 2008; Lee et al., 2009). We found that Sre1N degradation requires the E3 ligase Ubr1. To test the function of Ubr1 in Sre1N turnover, we cultured wild-type and ubr1Δ cells in the absence of oxygen for 6 h to accumulate Sre1N. At time t=0, cycloheximide was added to inhibit protein translation, and cells were shifted to the presence of oxygen to block Sre1 cleavage and Sre1N production. Sre1N half-life was 6.9 min in wild-type cells and 80 min in ubr1Δ, indicating that Sre1N degradation requires Ubr1 (Fig 1A). Similar studies demonstrated that Sre1N degradation required the E2 ubiquitin-conjugation enzyme Rhp6 (t1/2=6.9 min in wild type and t1/2=82 min in rhp6Δ)(Fig 1B). Consistent with results in wild-type cells, Sre1N degradation required Ubr1 in sre1N cells that express only soluble Sre1N due to insertion of a stop codon prior to the sequence encoding the first transmembrane segment of Sre1 (t1/2= 8 min in sre1N and t1/2= 103 min in sre1N ubr1Δ, average from four experiments)(Fig 1C, lanes 1-8). Sre1N degradation was not affected in cells lacking Ubr11, a highly homologous E3 ligase, demonstrating specificity for Ubr1 in Sre1N turnover (Fig 1C, lanes 9-12). Given the block to Sre1N degradation in rhp6Δ and ubr1Δ cells, we conclude that Rhp6-Ubr1 define the principal pathway for Sre1N degradation.
Figure 1. Sre1N degradation requires Rhp6 and Ubr1.
(A) Wild-type and ubr1Δ cells were grown in the absence of oxygen for 6 hours. At t=0, cycloheximide (200 μg/ml) was added and cells were shifted to the presence of oxygen. Samples were collected at the indicated times. (B) Wild-type and rhp6Δ cells were grown and processed as in (A). P denotes Sre1 precursor and N denotes the Sre1 Nuclear form. (C) sre1N, sre1N ubr1Δ and sre1N ubr11Δ cells were grown in the absence of oxygen for 3 hours. At t=0, cycloheximide (200 μg/ml) was added and cells were shifted to the presence of oxygen. Samples were collected at the indicated times. (A–C) Whole-cell extracts were subjected to western blot analysis using anti-Sre1 IgG. Both long and short exposures to film are shown. The percentage of Sre1N remaining at each time point relative to t=0 is quantified to the right. Other mutant strains tested are listed in the Strain Table (Supplemental Table S1).
Ofd1 functions upstream of Ubr1 in Sre1N degradation
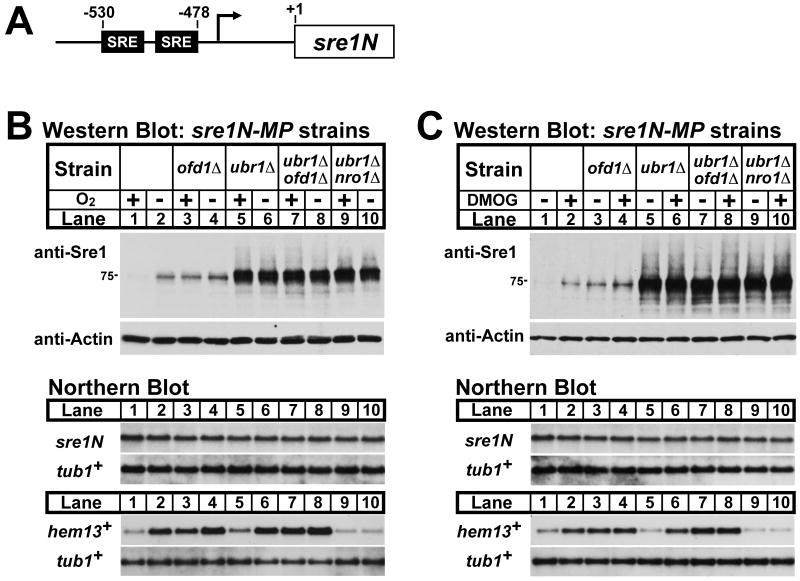
Ofd1 accelerates Sre1N degradation in the presence of oxygen by an unknown mechanism (Hughes and Espenshade, 2008). To test whether Ofd1 and Ubr1 act in the same pathway, we examined levels of Sre1N in cells lacking Ofd1, Ubr1, or both. The level of Sre1N is determined by both synthesis and degradation rates. sre1N mRNA expression is under positive feedback control whereby Sre1N binds to sterol regulatory elements (SRE) in its promoter to increase its own expression (Figure 2A). To simplify the system and focus on Sre1N degradation, we examined the functions of Ofd1 and Ubr1 in sre1N-MP cells that lack positive feedback regulation due to mutation of SRE sequences in the sre1N promoter (Hughes and Espenshade, 2008; Todd et al., 2006). As shown previously (Hughes and Espenshade, 2008), Sre1N accumulated in the absence of oxygen, and ofd1Δ cells had elevated levels of Sre1N in the presence of oxygen and lacked oxygen-dependent regulation (Fig 2B, lanes 1-4). In ubr1Δ cells, Sre1N was strongly increased in the presence of oxygen consistent with its role in Sre1N degradation. Deleting ofd1+ in sre1N-MP ubr1Δ cells had no additional effect on Sre1N (Fig 2B, lanes 5-8). Deleting nro1+, which inhibits Ofd1, also had no effect on Sre1N in sre1N-MP ubr1Δ cells (Fig 2B, lanes 9-10). In addition, oxygen did not regulate Sre1N in sre1N-MP ubr1Δ cells (Fig 2B, lanes 5-10). Together, these results indicate that Ofd1 functions upstream of Ubr1 in Sre1N degradation. Identical results were obtained when we used DMOG to inhibit Ofd1N-REG dioxygenase function (Fig 2C). As expected, all strains had equal sre1N mRNA irrespective of oxygen status due to the loss of positive feedback control (Fig 2B and 2C, lower panels). Given that sre1+ mRNA is not subject to translational control by oxygen (Sehgal et al., 2008), these data demonstrate that Ofd1 and Ubr1 function in the same pathway to control Sre1N degradation.
Figure 2. Ofd1 acts upstream of Ubr1 in Sre1N degradation.
(A) Diagram of sre1N promoter showing DNA elements required for positive feedback regulation. (B and C) sre1N-MP, sre1N-MP ofd1Δ, sre1N-MP ubr1Δ, sre1N-MP ubr1Δ ofd1Δ and sre1N-MP ubr1Δ nro1Δ cells were cultured in the presence or absence of oxygen for 3 hours (B) or with either 1% DMSO or 20 mM DMOG for 6 hours (C). Whole-cell extracts (40 μg) were subjected to western blot analysis using indicated antibodies and total RNA (5 μg) was subjected to northern analysis with the indicated 32P-labeled probes. Actin protein and α-tubulin (tub1+) RNA served as loading controls. Northern blots were stripped and reprobed for tub1+.
Ofd1 regulates Sre1N independently from degradation
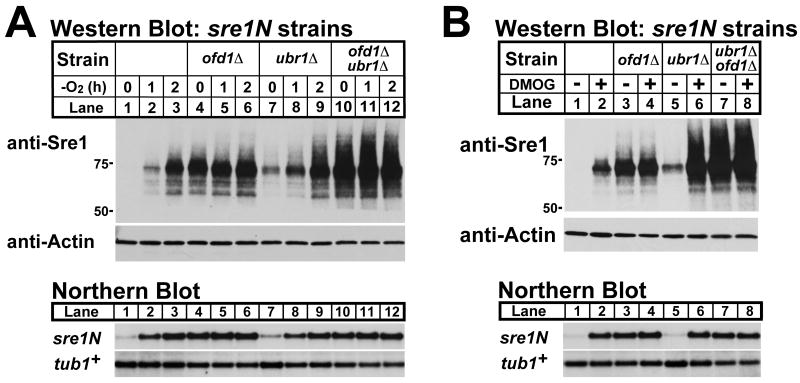
To test whether Ofd1 functions independently from its role in Ubr1 -dependent protein degradation, we examined levels of Sre1 in sre1N cells lacking Ofd1, Ubr1, or both. As reported previously, Sre1N accumulated in the absence of oxygen, while ofd1Δ cells had high levels of Sre1N in the presence of oxygen and lacked oxygen-dependent regulation (Fig 3A, lanes 1-6)(Hughes and Espenshade, 2008). In ubr1Δ cells, Sre1N was elevated in the presence of oxygen consistent with its role in Sre1N degradation, and Sre1N increased further in the absence of oxygen (Fig 3A, lanes 7-9). Importantly, deleting ofd1+ in ubr1Δ cells increased Sre1N, and ubr1Δ ofd1Δ cells lacked oxygen-dependent regulation of Sre1N (Fig 3A, lanes 10-12 and Supplemental Fig S1). If Ofd1 only functions in Sre1N degradation upstream of Ubr1, deletion of ofd1+ should have no effect on Sre1N protein levels. Thus, Ofd1 contains a degradation independent function in Sre1N regulation.
Figure 3. Ofd1 regulates Sre1N independently from degradation.
(A and B) sre1N, sre1N ofd1Δ, sre1N ubr1Δ, and sre1N ubr1Δ ofd1Δ cells were cultured in the absence of oxygen for designated time (A) or with either 1% DMSO or 20 mM DMOG for 3 h (B). Western blot and northern blot analysis were performed as described in Figure 2 and Experimental Procedures. See also Figure S1.
The Ofd1N-REG dioxygenase domain is required for oxygen-dependent regulation of Sre1N (Hughes and Espenshade, 2008; Lee et al., 2009). To test whether the function of Ofd1 in ubr1Δ cells required Ofd1N-Reg dioxygenase activity, we treated cells with the dioxygenase inhibitor dimethyloxalylglycine (DMOG). As expected from previous results, treating Sre1N cells with DMOG increased Sre1N levels in wild-type but not in ofd1Δ cells, indicating that DMOG acts through inhibition of Ofd1N-Reg (Fig 3B, lanes 1-4). Likewise, DMOG treatment increased Sre1N in ubr1Δ cells, but not in ubr1Δ ofd1Δ cells, indicating that Ofd1 regulates Sre1N in ubr1Δ cells (Fig 3B, lanes 5-8). Together, these data demonstrate that Ofd1 functions separately from Ubr1-mediated degradation to regulate Sre1N: (1) deletion of ofd1+ or inhibition of Ofd1 dioxygenase activity in ubr1Δ cells increases Sre1N; and (2) ubr1Δ cells exhibit oxygen-dependent regulation of Sre1N that requires Ofd1. These results were unexpected given that both Ofd1 and Ubr1 act in Sre1N degradation and deletion of Ubr1 blocks Sre1N degradation.
Ofd1-Nro1 regulate Sre1N transcriptional activity
The sre1N promoter is subject to positive feedback regulation (Fig 2A). We hypothesized that Ofd1 may regulate Sre1N in ubr1Δ cells through control of Sre1N transcriptional activity. In sre1N-MP ubr1Δ cells, Sre1N levels remained the same despite different genetic and chemical manipulations that affect Ofd1 function (Fig 2B and 2C). This property allowed us to study the degradation-independent function of Ofd1 in cells that have equal amounts of Sre1N. First, we tested whether oxygen regulates Sre1N transcriptional activity. We cultured sre1N-MP ubr1Δ cells in the presence or absence of oxygen and assayed expression of hem13+, a direct Sre1N target gene, as a measure of Sre1N transcriptional activity (Todd et al., 2006). While sre1N-MP ubr1Δ cells contained equal amounts of Sre1N in the presence or absence of oxygen, hem13+ expression increased under low oxygen (Fig 2B, lanes 5-6, lower panel). These data suggest that the transcription factor activity of Sre1N is negatively regulated by oxygen.
Next, we tested whether this oxygen-dependent regulation required Ofd1 and Nro1 by examining sre1N-MP ubr1Δ ofd1Δ and sre1N-MP ubr1Δ nro1Δ cells. Deletion of ofd1+ increased hem13+ expression in the presence of oxygen (Fig 2B, compare lanes 5 and 7, lower panel). Conversely, deletion of nro1+, a direct inhibitor of Ofd1, blocked up-regulation of hem13+ in the absence of oxygen (Fig 2B, compare lanes 5-6 and 9-10, lower panel). In a parallel set of experiments, inhibiting Ofd1 by DMOG treatment increased hem13+ expression in the presence of oxygen and this regulation required Ofd1 and Nro1 (Fig 2C, lanes 5-10, lower panel). Under all conditions, Sre1N protein was unchanged as expected. These results suggest that Ofd1 and Nro1 cooperate to regulate the transcription factor activity of Sre1N in response to oxygen.
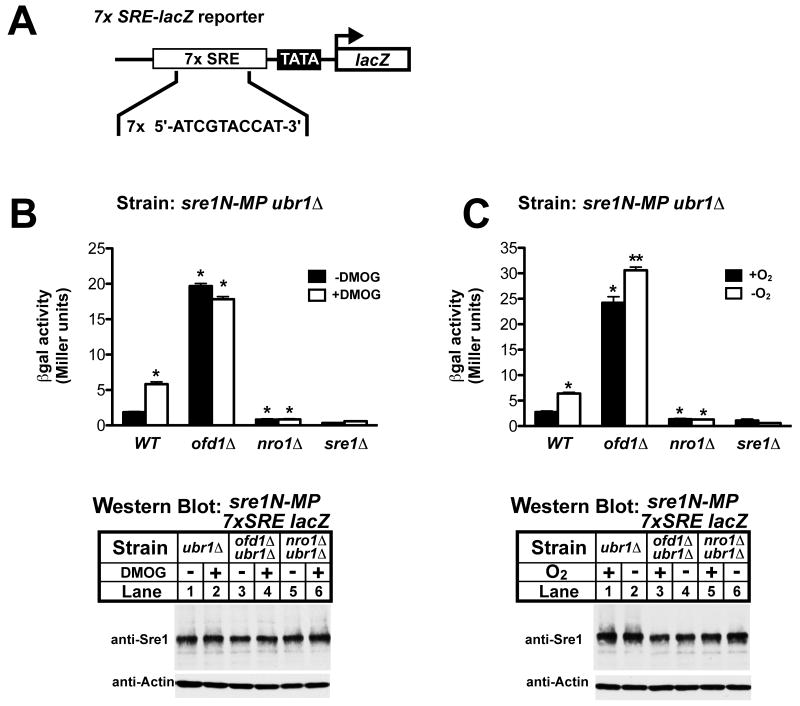
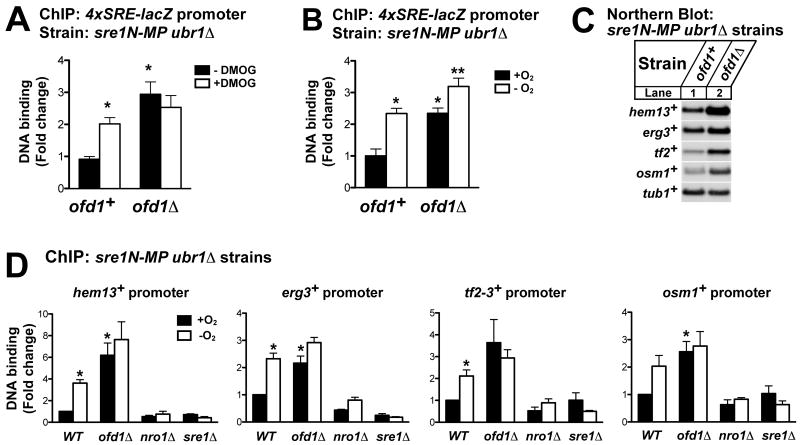
To determine whether this oxygen-dependent regulation of Sre1N transcriptional activity is a general control mechanism, we designed a chromosomally integrated lacZ reporter gene whose expression was strictly Sre1N-dependent owing to a tandem array of SRE sequences inserted upstream of a minimal promoter (Fig 4A)(Lee et al., 2009; Sehgal et al., 2007). Engineered strains, designated sre1N-MP ubr1Δ 7xSRE-lacZ, sre1N-MP ubr1Δ ofd1Δ 7xSRE-lacZ, and sre1N-MP ubr1Δ nro1Δ 7xSRE-lacZ were used to assay whether Ofd1 and Nro1 regulate Sre1N transcriptional activity. Culturing parental cells with either DMOG or in the absence of oxygen increased β-galactosidase activity significantly (Fig 4B and 4C). Consistent with these data, deletion of ofd1+ increased β-galactosidase activity about 20 fold in the presence of oxygen (Fig 4B and 4C). Low oxygen treatment significantly increased β-galactosidase activity in ofd1Δ cells, indicating that oxygen also regulates Sre1N through an Ofd1-independent mechanism (Fig 4C). Conversely, deletion of nro1+ blocked up-regulation of β-galactosidase activity under either DMOG or low oxygen treatment (Fig 4B and 4C). Cells expressed equal amounts of Sre1N in all reporter strains under different treatment conditions (Fig 4B and 4C, lower panel). Similar results were found in experiments using cells carrying the 4xSRE-lacZ reporter gene (Supplemental Fig S2). Taken together, these data demonstrate that Ofd1 inhibits Sre1N transcriptional activity in the presence of oxygen. Sre1N transcriptional activity is derepressed when Ofd1N-REG dioxygenase activity is inhibited by a chemical inhibitor or under low oxygen.
Figure 4. Ofd1-Nro1 regulate oxygen-dependent Sre1N transcriptional activity.
(A) Diagram of the integrated 7xSRE-lacZ reporter gene in sre1N-MP ubr1Δ strain. (B and C) sre1N-MP ubr1Δ, sre1N-MP ubr1Δ ofd1Δ, sre1N-MP ubr1Δ nro1Δ, and ubr1Δ sre1Δ cells with 7xSRE-lacZ reporter integrated were cultured in rich medium with either 1% DMSO or 20 mM DMOG for 6 hours (B) or with or without oxygen for 3 hours (C). Cells were assayed for β-galactosidase activity as described in Experimental Procedures. Data are the mean of six replicates and error bars indicate standard error. * Indicates significant difference from sre1N-MP ubr1Δ (WT) control samples, either −DMOG or + oxygen (P value <0.0003). ** Indicates significant difference from ofd1Δ + oxygen samples (P value = 0.001). Whole-cell extracts (40 μg) were subjected to western blot analysis using indicated antibodies. See also Figure S2.
Ofd1-Nro1 regulate Sre1N DNA binding
Next, we tested whether Ofd1 regulates Sre1N transcriptional activity by controlling Sre1N binding to promoter DNA. sre1N-MP ubr1Δ 4xSRE-lacZ and sre1N-MP ubr1Δ ofd1Δ 4xSRE-lacZ cells were cultured in the absence or presence of DMOG, and we assayed Sre1N binding to the lacZ promoter by chromatin immunoprecipitation (ChIP). Inhibiting Ofd1 by DMOG treatment significantly increased Sre1N DNA binding in ofd1+ cells (Fig 5A). Deletion of ofd1+ increased Sre1N DNA binding >3-fold and DMOG regulation was lost in these cells because DMOG signals through Ofd1 (Fig 5A). Consistent with the DMOG results, low oxygen also increased Sre1N DNA binding and this regulation required Ofd1 (Fig 5B). These data suggest that Ofd1 inhibits Sre1N DNA binding in the presence of oxygen.
Figure 5. Oxygen regulates Sre1N DNA binding through Ofd1.
(A) sre1N-MP ubr1Δ 4xSRE-lacZ and sre1N-MP ubr1Δ ofd1Δ 4xSRE-lacZ cells were grown with either 1% DMSO or 20 mM DMOG for 6 hours and subjected to chromatin immunoprecipitation using anti-Sre1 IgG or rabbit IgG. Binding of Sre1N to lacZ promoter regions was assayed by real-time PCR. Bound DNA was expressed as the fold change normalized to ofd1+ treated with 1% DMSO. Recovery of Sre1N bound DNA ranged from 0.35% to 1.1% of input gDNA for untreated sre1N-MP ubr1Δ cells. (B) sre1N-MP ubr1Δ 4xSRE-lacZ and sre1N-MP ubr1Δ ofd1Δ 4xSRE-lacZ cells were grown in the presence or absence of oxygen for 3 hours and subjected to chromatin immunoprecipitation as in (A). Bound DNA was expressed as the fold change normalized to ofd1+ grown with oxygen. Recovery of Sre1N bound DNA ranged from 0.22% to 0.72% of input gDNA for sre1N-MP ubr1Δ cells grown plus oxygen. For (A) and (B), data are the mean of 4 biological replicates, and error bars denote standard error among biological replicates. Non-specific DNA binding to rabbit IgG ranged from 0.0022% to 0.005% of input DNA and is not shown. (C) sre1N-MP ubr1Δ and sre1N-MP ubr1Δ ofd1Δ cells were cultured in rich medium to exponential phase. Total RNA (5 μg) was subjected to Northern blot analysis with the indicated 32P-labeled probes. (D) sre1N-MP ubr1Δ, sre1N-MP ubr1Δ ofd1Δ, sre1N-MP ubr1Δ nro1Δ, and sre1N-MP ubr1Δ sre1Δ cells were grown in the presence or absence of oxygen for 3 hours and subjected to chromatin immunoprecipitation using anti-Sre1 IgG. Binding of Sre1 to different promoter regions was assayed by real-time PCR. Bound DNA was expressed as the fold change normalized to sre1N-MP ubr1Δ cells (WT) grown with oxygen for each primer pair. Recovery of Sre1N bound DNA relative to input gDNA for sre1N-MP ubr1Δ cells grown plus oxygen was: 0.1% for hem13+; 0.22% for erg3+; 0.019% for tf2-3+; and 0.079% for osm1+. Data are the mean of 3 biological replicates, and error bars denote standard error among biological replicates. * Indicates significant difference from sre1N-MP ubr1Δ (WT) plus oxygen samples (P value <0.02). ** Indicates significant difference from ofd1Δ plus oxygen sample (P value = 0.04).
To test whether Ofd1 regulates Sre1N DNA binding to promoters of physiological target genes, we assayed Sre1N binding to four Sre1N target gene promoters involved in different biological processes: hem13+, erg3+, osm1+ and tf2+. In each case, Sre1N target gene expression increased in ofd1Δ cells grown in the presence of oxygen (Fig 5C), despite cells expressing equal amounts of Sre1N (Fig 2B, lanes 5-8). To test whether Ofd1-Nro1 control expression of these genes by regulating Sre1N binding to promoter DNA, we measured Sre1N DNA binding by ChIP in the parental stain sre1N-MP ubr1Δ and the indicated mutants in the presence or absence of oxygen. Low oxygen increased Sre1N binding 2- to 3-fold at each target gene promoter in the parental cells (Fig 5D). Deleting ofd1+ increased Sre1N promoter binding 2 to 6-fold and led to a loss of oxygen regulation. Conversely, deleting nro1+, which leads to highly active Ofd1, decreased Sre1N DNA binding to background levels seen in sre1Δ cells (Fig 5D). Collectively, these results demonstrate that Ofd1-Nro1 regulate oxygen-dependent Sre1N transcriptional activity by controlling Sre1N binding to DNA.
Low oxygen rapidly stimulates Sre1N DNA binding
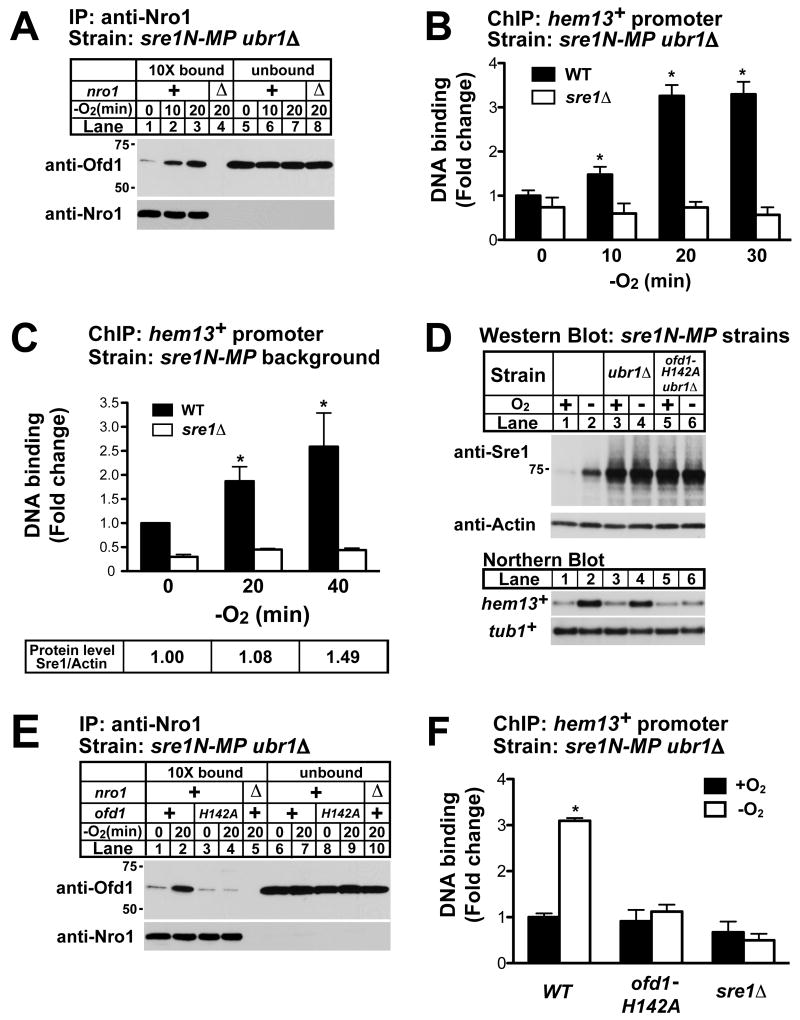
Activity of the Ofd1N-REG dioxygenase controls direct binding of Nro1 to Ofd1 and thereby Ofd1 regulation of Sre1N (Lee et al., 2009). Low oxygen and DMOG inhibit the Ofd1N-REG dioxygenase, inducing Nro1 binding to the Ofd1CTD and blocking Ofd1CTD regulation of Sre1N. Our new data suggest a model in which Ofd1-Nro1 regulate Sre1N transcriptional activity by Ofd1CTD-mediated inhibition of Sre1N DNA binding. To begin to test this model, we examined the kinetics of Ofd1-Nro1 binding and Sre1N DNA binding under low oxygen. Ofd1-Nro1 binding was assayed in sre1N-MP ubr1Δ cells by co-immunoprecipitation using anti-Nro1 antibody. Ofd1 binding to Nro1 increased rapidly when cells were shifted to low oxygen with changes occurring after 10 minutes (Fig 6A, lanes 1-3). To determine whether Ofd1-Nro1 binding correlates with Sre1N DNA binding, we measured Sre1N DNA binding under low oxygen to the hem13+ promoter by ChIP. Sre1N DNA binding increased 10 minutes after shifting to low oxygen and maximal DNA binding occurred at 20 minutes (Fig 6B). Sre1N DNA binding was specific as sre1Δ cells showed background ChIP signal using anti-Sre1 antibody. Consistent with the model for Sre1N regulation, Ofd1-Nro1 binding correlates with Sre1N DNA binding.
Figure 6. Low oxygen rapidly stimulates Sre1N DNA binding.
(A) sre1N-MP ubr1Δ and sre1N-MP ubr1Δ nro1Δ cells were grown in the absence of oxygen for the indicated times and were treated with 0.5 mM DSP crosslinker in PBS for 5 min. Detergent-solubilized whole-cell extracts were subjected to immunoprecipitation with anti-Nro1 IgG. Bound (10-fold overloaded) and unbound fractions were analyzed by western blot using anti-Nro1–HRP and anti-Ofd1–HRP antibodies. (B) sre1N-MP ubr1Δ and sre1N-MP ubr1Δ sre1Δ cells were grown in the absence of oxygen for the indicated times and subjected to chromatin immunoprecipitation using anti-Sre1 IgG. Sre1N promoter occupancy was assayed by real-time PCR using hem13+ primer pair. Bound DNA was expressed as the fold change normalized to sre1N-MP ubr1Δ cells grown with oxygen. Recovery of Sre1N bound DNA to input gDNA in sre1N-MP ubr1Δ samples ranged from 0.12% to 0.39%. Data are the mean of six biological replicates, and error bars denote standard error among biological replicates. (C) Upper panel: sre1N-MP 7xSRE lacZ cells were grown in the absence of oxygen for the indicated times and subjected to chromatin immunoprecipitation using anti-Sre1 IgG, and western blot analysis using anti-Sre1 and anti-Actin antibodies. Sre1N promoter occupancy was assayed by real-time PCR using hem13+ primer pair. Bound DNA was expressed as the fold change normalized to sre1N-MP cells grown with oxygen. Recovery of Sre1N bound DNA to input gDNA in sre1N-MP samples ranged from 0.04% to 0.26 %. Lower panel: Sre1N protein levels were quantified and normalized to actin. Sre1N / Actin ratio was expressed as the fold change normalized to samples grown with oxygen. Data are the mean of four biological replicates for sre1N-MP and two biological replicates for control sre1Δ cells. Error bars denote standard error among biological replicates. * Indicates significant difference from samples grown in the presence of oxygen (P value < 0.05). See also Figure S3. (D) sre1N-MP, sre1N-MP ubr1Δ, and sre1N-MP ubr1Δ ofd1-H142A cells were grown in the presence or absence of oxygen for 3 hours. Western blot and northern blot analysis were performed as described in Figure 2 and Experimental Procedures. (E) sre1N-MP ubr1Δ, sre1N-MP ubr1Δ ofd1-H142A, and sre1N-MP ubr1Δ nro1Δ cells were grown with or without oxygen for 20 minutes and then were treated with 0.5 mM DSP crosslinker in PBS for 5 min. Immunoprecipitation and western blot were performed as described in (A).(F) sre1N-MP ubr1Δ, sre1N-MP ubr1Δ ofd1-H142A, and sre1N-MP ubr1Δ sre1Δ cells were grown with or without oxygen for 20 minutes. Cells were subjected to chromatin immunoprecipitation using anti-Sre1 IgG. Promoter occupancy by Sre1N was assayed by real-time PCR using hem13+ primer pair. Bound DNA was expressed as the fold change normalized to sre1N-MP ubr1Δ cells (WT) grown with oxygen. Recovery of Sre1N bound DNA in sre1N-MP ubr1Δ samples ranged from 0.14% to 0.5% of input gDNA. Data are the mean of four biological replicates, and error bars denote standard error among biological replicates. * Indicates significant difference from sre1N-MP ubr1Δ (WT) plus oxygen samples (P value < 0.02).
Next, we examined the effect of Ofd1 control on Sre1N DNA binding in cells with an intact Sre1N degradation pathway. Since Ofd1 controls both Sre1N DNA binding and Sre1N degradation, we assayed oxygen-dependent Sre1N DNA binding in sre1N-MP cells to minimize effects on Sre1N protein due to changes in sre1N transcription (Supplemental Fig S3A). In sre1N-MP cells, Sre1N DNA binding increased after 20 minutes in the absence of oxygen, while Sre1N protein remained unchanged (Fig 6C and Supplemental Fig 3B). Sre1N DNA binding increased 2.5-fold after 40 minutes at which time Sre1N protein increased slightly due to Ofd1-dependent changes in protein degradation (Fig 6C)(Hughes and Espenshade, 2008). Thus, low oxygen induces Sre1N DNA binding prior to changes in Ofd1-dependent protein stability. Given that sre1N-MP cells express ∼250-fold less Sre1N than sre1N-MP ubr1Δ cells (Fig 2B, lanes 1 and 5), these experiments demonstrate that Ofd1 controls Sre1N DNA binding across a broad range of transcription factor concentration.
Ofd1 inhibition of Sre1N DNA binding does not require dioxygenase activity
Previous studies showed that mutation of the iron-coordinating residues (H142A) in the Ofd1N-REG dioxygenase domain disrupts oxygen-dependent regulation of Sre1N due to the failure of the Ofd1-H142A mutant to bind Nro1 under low oxygen. Consequently, Ofd1-H142A down regulates Sre1N protein both in the presence and absence of oxygen (Hughes and Espenshade, 2008; Lee et al., 2009). To test whether Ofd1N-REG dioxygenase is also required for proper regulation of Sre1N DNA binding, we measured target gene expression, Ofd1-Nro1 binding and Sre1N DNA binding in ofd1+ or ofd1-H142A iron-coordinating mutant cells. From previous results, we expected that loss of Ofd1-Nro1 binding under low oxygen would abolish oxygen-dependent Sre1N DNA binding in Ofd1 mutant cells. We first measured hem13+ expression in sre1N-MP ubr1Δ and sre1N-MP ubr1Δ ofd1-H142A cells. Expression of hem13+ increased under low oxygen in ofd1+ cells, but failed to increase under low oxygen in the ofd1-H142A mutant (Fig 6D, lanes 3-6, lower panel). These cells express equal amounts of Sre1N and Ofd1 protein (Fig 6D, lanes 3-6 and Fig 6E, lanes 6-9). Unlike wild-type Ofd1, the Ofd1-H142A mutant failed to bind Nro1 under low oxygen (Fig 6E, compare lanes 2 and 4). Consistent with the failure of Ofd1-H142A to bind Nro1 and up-regulate hem13+ expression, Sre1N DNA binding did not increase under low oxygen in ofd1-H142A cells (Fig 6F). Taken together, these data support a model in which formation of the Ofd1-Nro1 complex under low oxygen activates Sre1-dependent transcription by increasing Sre1N DNA binding. In addition, Ofd1 inhibition of Sre1N DNA binding does not require Ofd1N-REG hydroxylase activity because the Ofd1-H142A iron-coordinating mutant maximally inhibits Sre1N DNA binding.
Ofd1CTD binds Sre1N
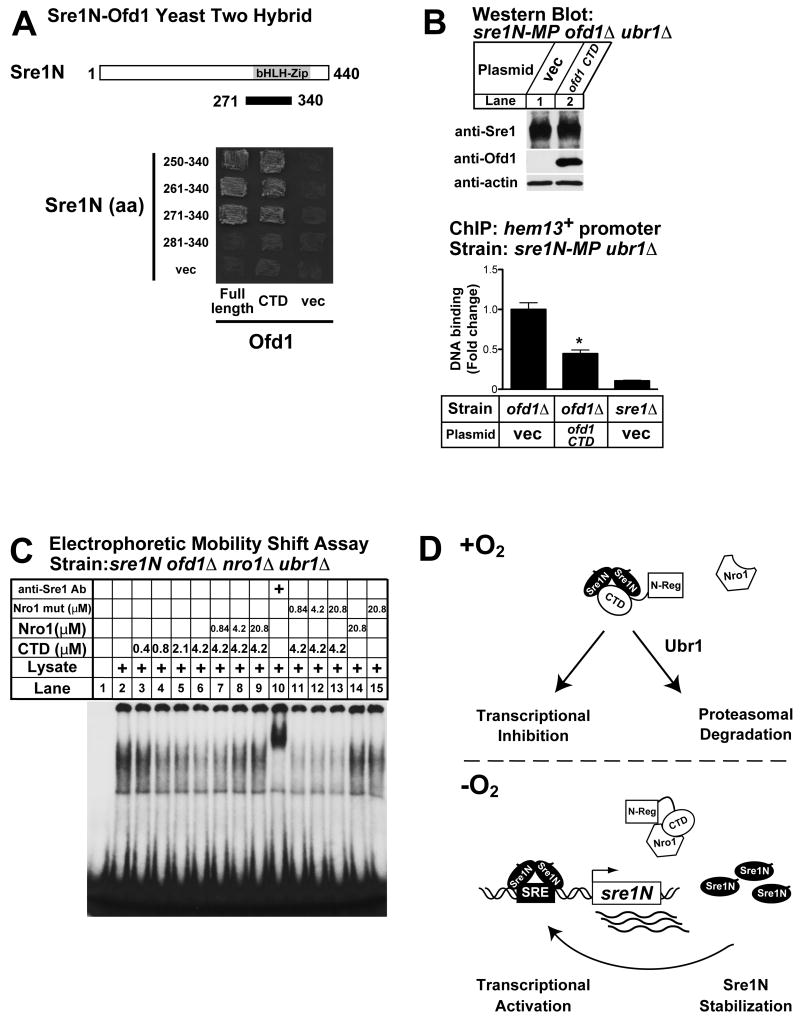
Our data suggest that Ofd1CTD inhibits Sre1N DNA binding. To determine if Ofd1CTD acts directly on Sre1N, we assayed Ofd1 binding to Sre1N. We performed yeast two-hybrid experiments between Ofd1 or Ofd1CTD (aa 255-515) and different truncations of Sre1N. Cells expressing Sre1N and full-length Ofd1 or Ofd1CTD, but not empty vector, grew well on selective medium suggesting a strong interaction between the Ofd1CTD and Sre1N (Fig 7A). Cells containing Ofd1N-REG (aa 1-254) and Sre1N (aa 250-340) plasmids showed no growth (data not shown). The minimal Ofd1CTD binding site mapped to amino acids 271-340 of Sre1N, which spans the basic helix-loop-helix DNA binding domain of the transcription factor (Fig 7A). These data suggest that Ofd1CTD binds to the bHLH domain of Sre1N.
Figure 7. Ofd1 CTD binds Sre1N and inhibits DNA binding.
(A) Yeast two-hybrid assay showing the in vivo interaction between Sre1N and Ofd1CTD. Cells containing an ofd1CTD plasmid were transformed with plasmids expressing different truncation mutants of Sre1N or empty vector. Cells were plated on selection medium. (B) sre1N-MP ubr1Δ ofd1Δ cells containing empty vector or ofd1CTD plasmid were cultured to exponential phase. Whole-cell extracts (40 μg) were subjected to western blot analysis using indicated antibodies (upper panels). sre1N-MP ubr1Δ ofd1Δ cells containing empty vector or ofd1CTD, and sre1N-MP ubr1Δ sre1Δ cells containing empty vector were subjected to chromatin immunoprecipitation using anti-Sre1 IgG. Promoter occupancy by Sre1N was assayed by real-time PCR using hem13+ primer pair. Bound DNA was expressed as the fold change normalized to sre1N-MP ubr1Δ ofd1Δ containing vector. Recovery of Sre1N bound DNA in sre1N-MP ubr1Δ ofd1Δ samples ranged from 0.24% to 0.94% of input gDNA. Data are the mean of six biological replicates for sre1N-MP ubr1Δ ofd1Δ containing ofd1CTD plasmid and four biological replicates for others. Error bars denote standard error among biological replicates. * Indicates significant difference from sre1N-MP ubr1Δ ofd1Δ control samples (P value = 2.1 × 10-5). (C) Extracts from sre1N ofd1Δ nro1Δ ubr1Δ cells were mixed with Sre1N specific 32P-labeled DNA probes in the presence of indicated concentrations of Ofd1CTD, Nro1 or Nro1 (aa 59-393) and DNA binding was assayed. Monoclonal antibody to Sre1N (4 μg) was added to the reaction in lane 10. See also Figure S4. (D) Model for oxygen-dependent regulation of Sre1N DNA binding. The dioxygenase domain of Ofd1 (N-Reg) regulates oxygen-dependent binding of Nro1 to Ofd1. In the presence of oxygen, Nro1 does not bind Ofd1CTD, allowing Ofd1CTD to bind Sre1N, leading to Sre1N degradation and transcriptional inhibition. In the absence of oxygen, Nro1 binds Ofd1CTD, thereby allowing Sre1N to bind DNA and activate its own expression.
Ofd1CTD blocks Sre1N DNA binding
Given that Ofd1CTD binds Sre1N, we next tested whether overexpression of Ofd1CTD is sufficient to block Sre1N DNA binding. We cultured sre1N-MP ubr1Δ ofd1Δ cells containing empty vector or Ofd1CTD plasmid in the presence of oxygen and assayed Sre1N DNA binding by ChIP. While Ofd1CTD expression did not affect Sre1N levels, Sre1N binding to the hem13+ promoter decreased in cells expressing Ofd1CTD (Fig 7B). To test whether Ofd1CTD can block Sre1N DNA binding in vitro, we established an electrophoretic mobility shift assay using extract from sre1N ofd1Δ nro1Δ ubr1Δ cells. Incubation of cell extract with a DNA probe containing two tandem SRE sequences resulted in formation of a specific Sre1N-DNA complex, as shown by the ability of Sre1N monoclonal antibody to supershift the Sre1N-DNA complex (Fig 7C, lanes 2 and 10). Sre1N DNA binding was specific insomuch as increasing concentrations of cold, wild-type probe, but not mutant probe, blocked binding (Supplemental Fig S4).
Next, we tested the effect of Ofd1CTD on Sre1N DNA binding. Addition of purified recombinant Ofd1CTD inhibited Sre1N DNA binding (Fig 7C, lanes 3-6). Importantly, Ofd1 inhibition of Sre1N DNA binding was reversed by addition of purified Nro1 (Fig 7C, lanes 7-9). Addition of Nro1 (aa 59-393), a mutant that does not bind Ofd1CTD, had no effect on Ofd1CTD inhibition of Sre1N DNA binding (Fig 7C, lanes 11-13)(Yeh et al., 2011). Importantly, the effect of Nro1 on Sre1N DNA binding required the presence of Ofd1CTD since addition of Nro1 alone did not increase Sre1N DNA binding (Fig 7C, lane 14). Collectively, these results demonstrate that (1) Ofd1CTD binds Sre1N and inhibits DNA binding and (2) that Nro1 regulates Sre1N by competing for binding to Ofd1CTD.
Discussion
In this study, we describe a new mechanism for control of hypoxic gene expression in which oxygen controls DNA binding of the Sre1 transcription factor through Ofd1, a 2-OG-Fe(II)-dependent dioxygenase. Our model for this regulatory mechanism is outlined in Figure 7D. In the presence of oxygen, Ofd1N-REG is active, and Nro1 is unable to bind and inhibit Ofd1CTD. Consequently, Ofd1CTD binds Sre1N leading to both transcriptional inhibition and proteasomal degradation. In the absence of oxygen, Nro1 binds Ofd1CTD, and accumulating free Sre1N can activate expression of itself and other target genes. In this model, the Ofd1CTD binds Sre1N and prevents DNA binding, while the Ofd1N-REG dioxygenase domain is an oxygen sensor and confers oxygen-dependent regulation to this interaction. This is in contrast to the HIF system in which protein hydroxylation is essential for regulation by either the PHDs or FIH.
Combining two cooperative regulatory functions in Ofd1 ensures a coordinated response to changes in oxygen concentration. These two mechanisms act together with positive feedback regulation of the sre1N promoter to rapidly increase the Sre1N transcription factor under low oxygen (Supplemental Fig S3A, lanes 6-10). In addition, Ofd1 controls Sre1N DNA binding across a wide concentration range of transcription factor (Fig 5D and 6C), consistent its ability to inhibit basal levels of Sre1N and accumulated Sre1N upon reintroduction of oxygen (Hughes and Espenshade, 2008).
Multiple independent lines of evidence support this model (Fig 7D). First, Ofd1 regulates Sre1N levels in ubr1Δ cells where Sre1N degradation is blocked, demonstrating a second function for Ofd1 (Fig 3). Second, in sre1N-MP ubr1Δ cells Ofd1-Nro1 control Sre1N target gene expression without affecting Sre1N protein level (Figs 2, 4, and 5C). Third, Ofd1-Nro1 regulate Sre1N target gene expression by controlling its DNA binding (Fig 4 and 5). Fourth, under low oxygen formation of Ofd1-Nro1 protein complex triggers Sre1N DNA binding (Fig 6A and 6B). Fifth, an Ofd1 iron-binding mutant lacking hydroxylase activity suppresses DNA binding and Sre1N target gene expression (Fig 6D-F). Sixth, Ofd1CTD binds Sre1N to block DNA binding, and Nro1 competes with Sre1N for binding to Ofd1CTD (Fig 7). In summary, these data demonstrate that the oxygen-dependent regulation of Sre1N DNA binding involves stoichiometric binding between Ofd1CTD and Sre1N, rather than acting through a catalytic mechanism. The oxygen-dependent activity of the Ofd1N-REG dioxygenase determines the binding partner for Ofd1CTD.
A non-catalytic mechanism analogous to that discovered here may exist in the HIF system to provide an additional layer of regulatory control. Recently, genetic analyses in C. elegans showed that the prolyl hydroxylase EGL-9 can repress HIF-1 transcriptional activity through a VHL-independent pathway. In these experiments, catalytically deficient EGL-9 failed to destabilize HIF-1, but still repressed HIF-1 transcriptional activity (Shao et al., 2009). Other studies suggest that mammalian PHD prolyl hydroxylases may regulate HIF-1α in a VHL-independent mechanism (Ozer et al., 2005; To and Huang, 2005). Whether these VHL-independent mechanisms, act by regulating HIF DNA binding is unknown.
Previously, we demonstrated that Ofd1 accelerates Sre1N degradation by the proteasome in the presence of oxygen (Hughes and Espenshade, 2008; Lee et al., 2009). Here, we identify additional components required for Sre1N degradation: the E2 ubiquitin-conjugating enzyme Rhp6 and the E3 ligase Ubr1. These two proteins also cooperate to target the nuclear envelope protein Cut8 for degradation in fission yeast (Takeda and Yanagida, 2005). Ofd1 acts upstream of Ubr1 in the same pathway, because deletion of ofd1+ in sre1N-MP ubr1Δ cells had no effect on Sre1N protein level (Fig 2B and 2C). Whether Ofd1 acts directly on Sre1N to accelerate degradation or whether, for example, Ofd1 affects Ubr1 activity is unknown. Ubr1 functions in the N-end rule degradation pathway in which N-terminal residues determine protein stability (Bartel et al., 1990; Meinnel et al., 2006). Indeed, the Sre1 N-terminus contains a tertiary destabilizing residue (Gln) that may affect protein stability. Alternatively, Sre1N may contain an internal degron, as is the case for the Ubr1 substrate Cup9 in S. cerevisiae (Byrd et al., 1998; Turner et al., 2000). Future experiments will address whether Sre1N is a substrate for Ubr1 and the sequence requirements for degradation.
Ofd1 Now has two oxygen-dependent functions in the Sre1 pathway: control of Sre1N DNA binding and Sre1N degradation. Both functions require Nro1 and the Ofd1N-REG dioxygenase for oxygen-dependent regulation. How the Ofd1N-REG dioxygenase relays information regarding oxygen availability to Ofd1CTD is currently unknown. Crystal structure data for the S. cerevisiae Ofd1 homolog, Tpa1, indicate that Ofd1 is likely a prolyl hydroxylase (Henri et al., 2010; Kim et al., 2010). It remains to be determined whether Ofd1 hydroxylates itself or another protein, and what role hydroxylation plays in regulation of Sre1N DNA binding. Recently, the mammalian ortholog of Ofd1, named Ogfod1, was shown to be involved in the recovery of cells from stress and to decrease viability of cells under ischemic conditions (Saito et al., 2010; Wehner et al., 2010). Whether Ogfod1 also regulates hypoxic gene expression is unknown, but our results provide a mechanistic understanding of how such a pathway could be controlled. Current studies focus on these unanswered questions.
Experimental Procedures
Materials and standard methods (antibody preparation, plasmids, measurement of protein turnover, co-immunoprecipitation, recombinant protein expression and purification) are described in the Supplemental Experimental Procedures.
Yeast Strains and Culture
Wild-type haploid S. pombe KGY425 (h-, his3-D1, leu1-32, ura4-D18 and ade6-M210) was obtained from American Type Culture Collection (Burke and Gould, 1994). S. pombe strains used in this study are described in Supplemental Table S1 and were generated from haploid yeast by mating or homologous recombination using established techniques and oligonucleotides listed in Supplemental Table S2 (Bahler et al., 1998; Hughes and Espenshade, 2008). Yeast strains were grown to exponential phase at 30°C in rich medium containing yeast extract plus supplements (YES)(225 μg/ml each of histidine, leucine, adenine, lysine and uracil) or in Edinburgh minimal medium as indicated (Hughes et al., 2005). Anaerobic growth conditions were maintained using an In Vivo 400 hypoxia workstation (Biotrace Inc.) as described (Hughes et al., 2005; Todd et al., 2006).
Northern and Western Blotting
Total RNA isolation and northern blot analysis were performed as described previously (Hughes et al., 2005). PCR fragments for northern probe synthesis were generated using gene-specific primer pairs (Supplemental Table S2)(Todd et al., 2006). Whole-cell yeast extract preparation and western blot analysis using indicated antibodies and horseradish peroxidase-conjugated anti-rabbit IgG were performed as described previously (Hughes et al., 2005)(Lee et al., 2009).
Chromatin Immunoprecipitation
Log-phase cells (1 × 108 cells/immunoprecipitation) were used for chromatin immunoprecipitation as described in detail in Supplemental Experiment Procedures. Bound DNA was recovered by phenol/chloroform extraction. Isolated DNA was quantified by real-time PCR using gene-specific primers to amplify a 100 bp fragment 200-500 bp upstream of the start codon. Fraction bound was calculated using the formula 2(Ctinput− Ctpulldown) for each treatment. Percent input was calculated by dividing the average starting quantity for each sample by the corresponding starting quantity of input DNA
β-Galactosidase Assays
sre1N-MP ubr1Δ 4xSRE-lacZ and sre1N-MP ubr1Δ ofd1Δ 4xSRE-lacZ cells were grown overnight in YES rich medium at 30°C to exponential phase. For DMOG treatment, cells were then collected and resuspended at 4 × 106 cells/ml in YES without or with DMOG for 6 hours. For low oxygen treatment, cells were resuspended in YES or deoxygenated YES for 3 hours. To assay lacZ expression, β-galactosidase activity was determined in permeabilized cells as previously described (Burke et al., 2000). Activity is expressed as Miller units {1,000 × (A420)/[(tmin)(Vml)(A600)]}.
Cell Extract Preparation
sre1N ofd1Δ nro1Δ ubr1Δ cells were grown in rich medium at 30°C to exponential phase and cells (2.8 × 1010) were pelleted and resuspend in high salt buffer (50 mM Hepes pH 7.5, 1.5 mM MgCl2, 0.42 M NaCl, 1 mM EDTA, 1 mM EGTA, 2.5% glycerol plus protease inhibitors). Cells were lysed using an EmulisiFlex-C3 and insoluble material was removed by centrifugation at 100,000 g at 4°C for 30 minutes. Cell extract concentration was determined using Coomassie plus protein assay reagent (Thermo Scientific).
Electrophoretic Mobility Shift Assays
DNA binding reactions were performed in gel shift buffer (50 mM HEPES-KOH, pH 7.5, 100 mM KCl, 5 mM MgCl2, 40 μg/ml bovine serum albumin, 20% [v/v] glycerol, 1 mM dithiothreitol) as described previously (Todd et al., 2006). Reaction mixtures containing 40 μg of cell extract, indicated amount of Ofd1CTD and Nro1 protein, 1 NM of radiolabeled DNA probe, 0.5 mg/ml poly(dI-dC)(GE Amersham) and BSA to achieve a total of 80 μg protein/reaction. Reaction mixtures were incubated at 25°C for 40 min in siliconized tubes. DNA-protein complexes were resolved by 4% polyacrylamide gel electrophoresis and detected by autoradiography. 32P-radiolabeled DNA probes were prepared by annealing two complementary oligonucleotides as previously described (Sehgal et al., 2007).
Supplementary Material
Acknowledgments
We thank Espenshade and Amzel Lab members for experimental suggestions and for reviewing the paper. We thank Wayne Lai and Ling-Chu Hung in the Department of Pathology at UT-Southwestern Medical Center at Dallas for help in generating monoclonal antibody to Sre1. We thank Takashi Toda (London Research Institute) and Dieter Wolf (Sanford-Burnham Medical Research Institute) for kindly providing yeast strains. This study was supported by a grant from the National Institutes of Health to PJE (HL-077588). C-YSL and BTH are recipients of American Heart Association Predoctoral Fellowships, and PJE is an Established Investigator of the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; New York: 2000. [Google Scholar]
- Burke JD, Gould KL. Molecular cloning and characterization of the Schizosaccharomyces pombe his3 gene for use as a selectable marker. Mol Gen Genet. 1994;242:169–176. doi: 10.1007/BF00391010. [DOI] [PubMed] [Google Scholar]
- Byrd C, Turner GC, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev. 2007;17:71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Crack JC, Thomson AJ, LeBrun NE. Bacterial sensors of oxygen. Curr Opin Microbiol. 2009;12:145–151. doi: 10.1016/j.mib.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Helmann JD. OxyR: a molecular code for redox sensing. Sci STKE. 2002;2002:pe46. doi: 10.1126/stke.2002.157.pe46. [DOI] [PubMed] [Google Scholar]
- Henri J, Rispal D, Bayart E, van Tilbeurgh H, Seraphin B, Graille M. Structural and functional insights into Saccharomyces cerevisiae Tpa1, a putative prolylhydroxylase influencing translation termination and transcription. J Biol Chem. 2010;285:30767–30778. doi: 10.1074/jbc.M110.106864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Semenza GL. Regulation of hypoxia-inducible factor 1 by prolyl and asparaginyl hydroxylases. Biochem Biophys Res Commun. 2005;338:610–616. doi: 10.1016/j.bbrc.2005.08.193. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27:1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- Kim HS, Kim HL, Kim KH, Kim do J, Lee SJ, Yoon JY, Yoon HJ, Lee HY, Park SB, Kim SJ, et al. Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex. Nucleic Acids Res. 2010;38:2099–2110. doi: 10.1093/nar/gkp1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Stewart EV, Hughes BT, Espenshade PJ. Oxygen-dependent binding of Nro1 to the prolyl hydroxylase Ofd1 regulates SREBP degradation in yeast. EMBO J. 2009;28:135–143. doi: 10.1038/emboj.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T, Serero A, Giglione C. Impact of the N-terminal amino acid on targeted protein degradation. Biol Chem. 2006;387:839–851. doi: 10.1515/BC.2006.107. [DOI] [PubMed] [Google Scholar]
- Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- Ozer A, Wu LC, Bruick RK. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF) Proc Natl Acad Sci USA. 2005;102:7481–7486. doi: 10.1073/pnas.0502716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Adachi N, Koyama H, Matsushita M. OGFOD1, a member of the 2-oxoglutarate and iron dependent dioxygenase family, functions in ischemic signaling. FEBS Lett. 2010;584:3340–3347. doi: 10.1016/j.febslet.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Schofield CJ, Ratcliffe PJ. Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun. 2005;338:617–626. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Hughes BT, Espenshade PJ. Oxygen-dependent, alternative promoter controls translation of tco1+ in fission yeast. Nucleic Acids Res. 2008;36:2024–2031. doi: 10.1093/nar/gkn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Lee CY, Espenshade PJ. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 2007;3:1389–1396. doi: 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Life with oxygen. Science. 2007;318:62–64. doi: 10.1126/science.1147949. [DOI] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Powell-Coffman JA. Two distinct roles for EGL-9 in the regulation of HIF-1-mediated gene expression in Caenorhabditis elegans. Genetics. 2009;183:821–829. doi: 10.1534/genetics.109.107284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Yanagida M. Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell. 2005;122:393–405. doi: 10.1016/j.cell.2005.05.023. [DOI] [PubMed] [Google Scholar]
- To KK, Huang LE. Suppression of hypoxia-inducible factor 1alpha (HIF-1alpha) transcriptional activity by the HIF prolyl hydroxylase EGLN1. J Biol Chem. 2005;280:38102–38107. doi: 10.1074/jbc.M504342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol. 2006;26:2817–2831. doi: 10.1128/MCB.26.7.2817-2831.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- Wehner KA, Schutz S, Sarnow P. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Mol Cell Biol. 2010;30:2006–2016. doi: 10.1128/MCB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TL, Lee CY, Amzel LM, Espenshade PJ, Bianchet MA. The hypoxic regulator of sterol synthesis Nro1 is a nuclear import adaptor. Structure. 2011;19:503–14. doi: 10.1016/j.str.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.