Abstract
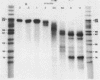
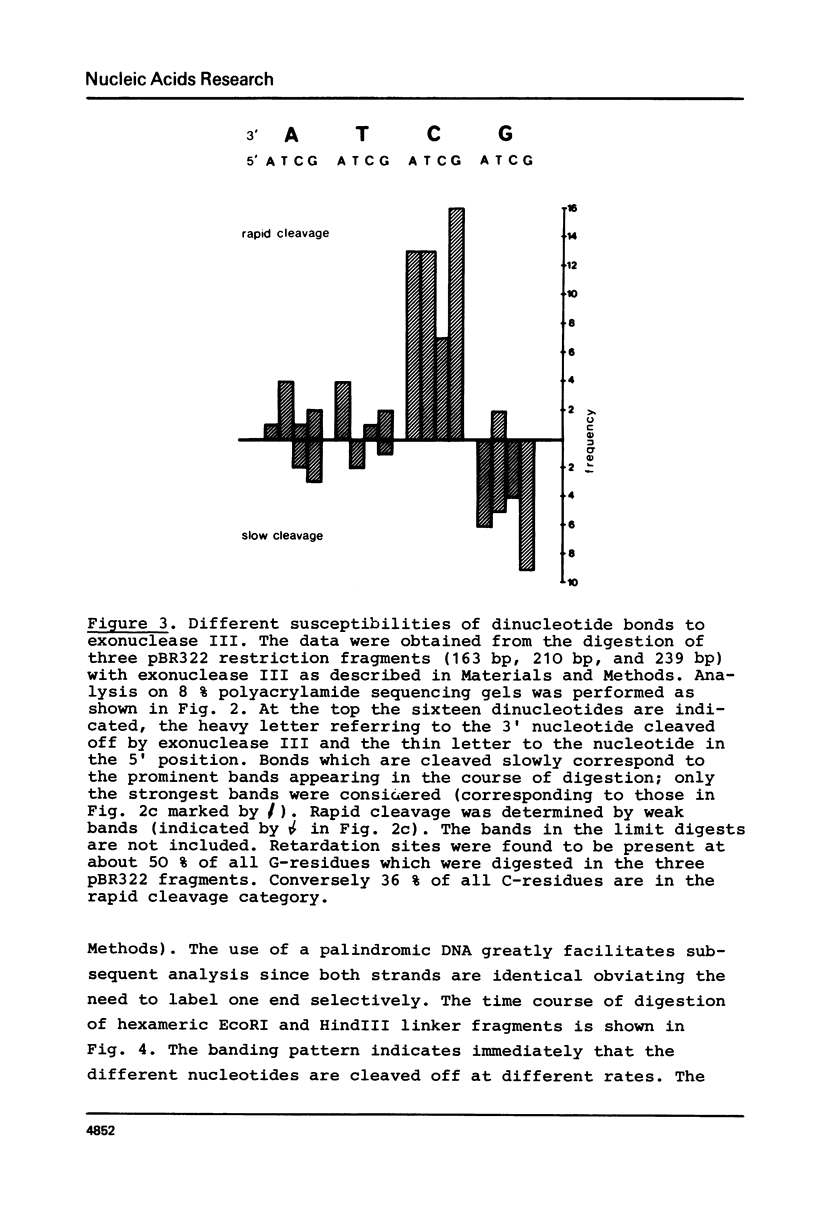
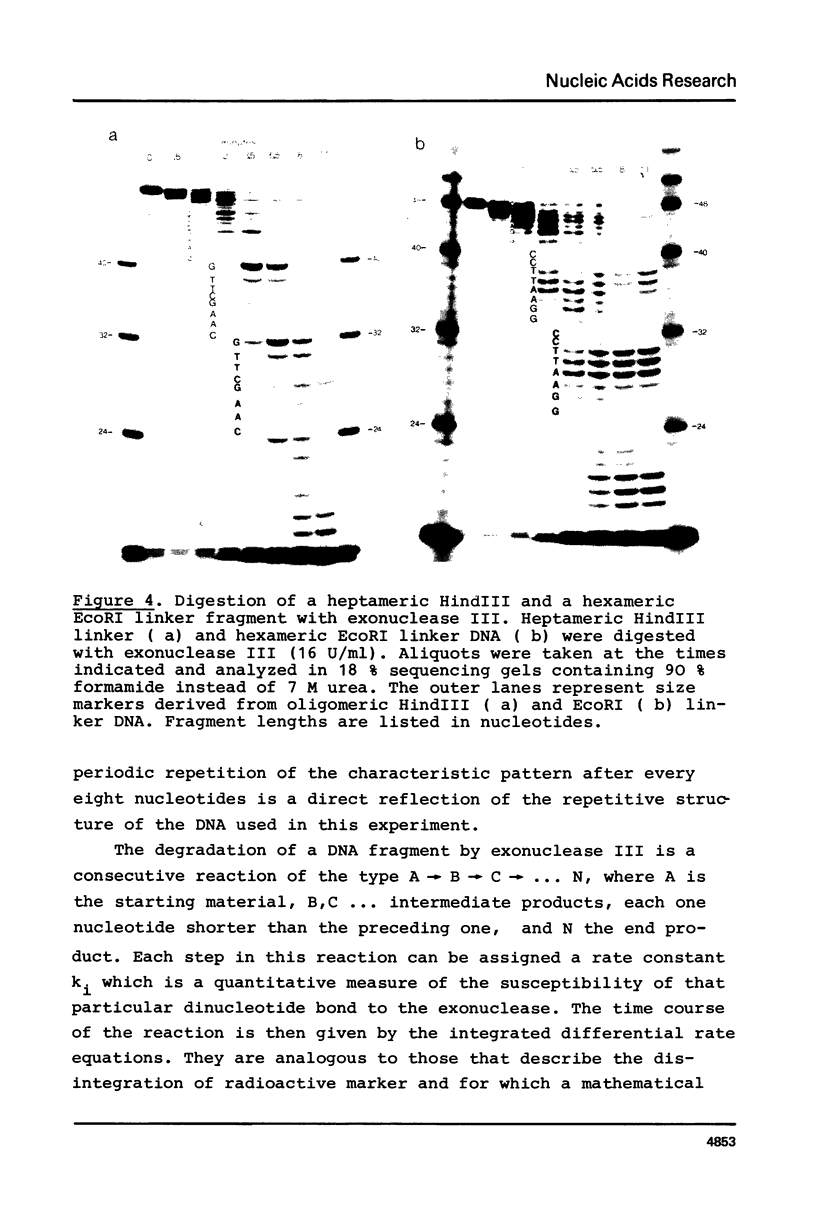
The influence of the nucleotide sequence on the digestion of deoxyribonuclease from E. coli, has been investigated. It was found that the rate at which mononucleotides are released varies in a sequence dependent fashion. C-residues are cleaved off rapidly and G-residues slowly while A and T are released at an intermediate rate. Quantitative analyses of digestion experiments with synthetic DNA fragments made it possible to determine rate constants for the cleavage of several dinucleotide bonds by exonuclease III. These values were found to differ by up to a factor of 3. Summation of the differences can lead to appreciable variation in the overall rate of digestion of a DNA strand. The nucleotide specificity of exonuclease III leads to a transient appearance of a series of discrete DNA fragments intermediate in digestion and a stable set of fragments in limit digests, i.e. at the point when all DNA has become single-stranded. This property of exonuclease III needs to be taken into account for the application of the enzyme in the analysis of nucleoprotein complexes.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chao M. V., Gralla J., Martinson H. G. DNA sequence directs placement of histone cores on restriction fragments during nucleosome formation. Biochemistry. 1979 Mar 20;18(6):1068–1074. doi: 10.1021/bi00573a021. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Frank R., Müller D., Wolff C. Identification and suppression of secondary structures formed from deoxy-oligonucleotides during electrophoresis in denaturing polyacrylamide-gels. Nucleic Acids Res. 1981 Oct 10;9(19):4967–4979. doi: 10.1093/nar/9.19.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C. P., Sommer R., Polke C., Beck E., Schaller H. Structure of the orgin of DNA replication of bacteriophage fd. Proc Natl Acad Sci U S A. 1978 Jan;75(1):50–53. doi: 10.1073/pnas.75.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. H., Wu R. New rapid methods for DNA sequencing based in exonuclease III digestion followed by repair synthesis. Nucleic Acids Res. 1982 Mar 25;10(6):2065–2084. doi: 10.1093/nar/10.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981 Feb 11;9(3):683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Zachau H. G. Characterization of distinct segments in mouse satellite DNA by restriction nucleases. Eur J Biochem. 1977 Mar 1;73(2):383–392. doi: 10.1111/j.1432-1033.1977.tb11329.x. [DOI] [PubMed] [Google Scholar]
- Hörz W., Zachau H. G. Deoxyribonuclease II as a probe for chromatin structure. I. Location of cleavage sites. J Mol Biol. 1980 Dec 15;144(3):305–327. doi: 10.1016/0022-2836(80)90093-5. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Omori A., Zachau H. G. Non-random arrangement of nucleosomes in satellite I containing chromatin of rat liver. Nucleic Acids Res. 1980 Nov 25;8(22):5377–5390. doi: 10.1093/nar/8.22.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Masamune Y., Fleischman R. A., Richardson C. C. Enzymatic removal and replacement of nucleotides at single strand breaks in deoxyribonucleic acid. J Biol Chem. 1971 Apr 25;246(8):2680–2691. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Relation of nucleosomes to DNA sequences. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):103–108. doi: 10.1101/sqb.1978.042.01.011. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Variable center to center distance of nucleosomes in chromatin. J Mol Biol. 1982 Jan 25;154(3):515–523. doi: 10.1016/s0022-2836(82)80010-7. [DOI] [PubMed] [Google Scholar]
- RICHARDSON C. C., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. I. PURIFICATION OF THE ENZYME AND CHARACTERIZATION OF THE PHOSPHATASE ACTIVITY. J Biol Chem. 1964 Jan;239:242–250. [PubMed] [Google Scholar]
- RICHARDSON C. C., LEHMAN I. R., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. II. CHARACTERIZATION OF THE EXONUCLEASE ACTIVITY. J Biol Chem. 1964 Jan;239:251–258. [PubMed] [Google Scholar]
- Riley D., Weintraub H. Nucleosomal DNA is digested to repeats of 10 bases by exonuclease III. Cell. 1978 Feb;13(2):281–293. doi: 10.1016/0092-8674(78)90197-6. [DOI] [PubMed] [Google Scholar]
- Rogers S. G., Weiss B. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 1980;65(1):201–211. doi: 10.1016/s0076-6879(80)65028-9. [DOI] [PubMed] [Google Scholar]
- Schreier P. H., Davies R. W., Kotewicz M. L. Protection against exonuclease III digestion. A new way to investigate protein-DNA interactions. FEBS Lett. 1980 Jan 1;109(1):159–163. doi: 10.1016/0014-5793(80)81333-0. [DOI] [PubMed] [Google Scholar]
- Shalloway D., Kleinberger T., Livingston D. M. Mapping of SV40 DNA replication origin region binding sites for the SV40 T antigen by protection against exonuclease III digestion. Cell. 1980 Jun;20(2):411–422. doi: 10.1016/0092-8674(80)90627-3. [DOI] [PubMed] [Google Scholar]
- Smith A. J. The use of exonuclease III for preparing single stranded DNA for use as a template in the chain terminator sequencing method. Nucleic Acids Res. 1979 Mar;6(3):831–848. doi: 10.1093/nar/6.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck R. E., Zachau H. G. A long-range and two short-range periodicities are superimposed in the 1.706-g/cm3 satellite DNA from calf thymus. Eur J Biochem. 1978 Aug 15;89(1):267–279. doi: 10.1111/j.1432-1033.1978.tb20923.x. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Olivera B. M. Processivity of DNA exonucleases. J Biol Chem. 1978 Jan 25;253(2):424–429. [PubMed] [Google Scholar]