Abstract
Advances in systems biology and bioinformatics have highlighted that no cell population is truly uniform and that stochastic behavior is an inherent property of many biological systems. As a result, bulk measurements can be misleading even when particular care has been taken to isolate a single cell type, and measurements averaged over multiple cell populations in a tissue can be as misleading as the average height at an elementary school. There is a growing need for experimental techniques that can provide a combination of single cell resolution, large cell populations, and the ability to track cells over multiple time points. In this article, a microwell array cytometry platform was developed to meet this need and investigate the heterogeneity and stochasticity of cell behavior on a single cell basis. The platform consisted of a microfabricated device with high-density arrays of cell-sized micro-wells and custom software for automated image processing and data analysis. As a model experimental system, we used primary hepatocytes labeled with fluorescent probes sensitive to mitochondrial membrane potential and free radical generation. The cells were exposed to oxidative stress and the responses were dynamically monitored for each cell. The resulting data was then analyzed using bioinformatics techniques such as hierarchical and k-means clustering to visualize the data and identify interesting features. The results showed that clustering of the dynamic data not only enhanced comparisons between the treatment groups but also revealed a number of distinct response patterns within each treatment group. Heat-maps with hierarchical clustering also provided a data-rich complement to survival curves in a dose response experiment. The microwell array cytometry platform was shown to be powerful, easy to use, and able to provide a detailed picture of the heterogeneity present in cell responses to oxidative stress. We believe that our microwell array cytometry platform will have general utility for a wide range of questions related to cell population heterogeneity, biological stochasticity, and cell behavior under stress conditions.
Keywords: cytometry, microfabrication, microwells, hepatocytes, mitochondria, membrane potential, free radicals
Introduction
Measurements averaged over the population of cells in a tissue can be as misleading as the average height in an elementary school, but such measurements can still be misleading even when particular care has been taken to isolate a single cell type. Recent advances in systems biology and bioinformatics have highlighted that no cell population is truly uniform and that stochasticity is an inherent property of many biological systems.1–4 Population heterogeneity has long been observed in primary cells such as hepatocytes, which are well known to specialize within the tissue based on anatomical and environmental cues.5 More surprising is that significant cell-to-cell variation can be present even in presumably uniform populations like clonal cell lines, and is a natural consequence of positive feedback in the mechanisms controlling gene expression.6,7 There is also growing evidence that this population heterogeneity has a meaningful biological role, such as the behavior of mammalian macrophages in response to pathogen exposure.8
In many cases, the variation observed at the cellular level is closely linked to the stochastic nature of underlying biological systems. Studies of ion channels, signaling pathway dynamics, transcription factor networks, and differentiation have also shown that the behavior of a biological system is often much more stochastic and discrete than would be assumed from population averages. At the level of individual molecules and even cells, biological processes tend to have probabilistic rather than continuous response curves and undergo rapid shifts between metastable states.9–11 What appears as a smooth transition at the bulk level may be anything but smooth at the level of each participant. These issues can pose a significant challenge for traditional experimental approaches that rely on bulk assays, because the average values obtained may not be representative of any given cell in the population. In many cases, high-throughput, single-cell approaches will greatly increase the accuracy, content, and statistical power of the collected biological data.
Recently, there has been much recent interest in high-throughput tools that can probe biological systems at the single-cell and subcellular levels.12–14 Flow cytometry has been the tool of choice for fluorescence-based assays at the single cell level. Even though it is a well established technique and can provide single-time-point measurements of multiple parameters, including cell size and granularity, it is limited by the fact that cells must be kept in suspension and that there is no way to track individual cells across multiple time points. With digital cameras, rapid increases in computational power, and the development of better analysis techniques, several forms of image cytometry have become viable alternatives.13,15 Unlike flow cytometry, image cytometry is generally used with attached cells and can provide dynamic data for individual cells. Unfortunately, advanced image processing and data analysis techniques are often needed to extract useful results, making the systems much harder to use and automate than flow cytometry. The information content is potentially higher, but this comes at a price in complexity.
One way to reduce this complexity is by better controlling the spatial arrangement of the cell population being studied. Much of the difficulty in image cytometry comes from interpreting random seeding patterns and identifying cell boundaries. These issues can both be addressed by seeding the cells in a regular pattern and isolating them from their neighbors. Microfabrication technologies such soft lithography have led to the development of several widely-used cell patterning techniques based on the deposition of adhesion molecules and/or adhesion inhibitors in precisely controlled arrangements.16,17 Microfluidics, dielectrophoretic traps, and laser capture microscopy have also been used to control cell patterning in novel ways.13 A recent addition to the cell patterning arsenal is the physical entrapment of individual cells in high density microwell arrays.18–24 Combining such an array with integrated alignment and identification features greatly simplifies the image processing needed for effective image cytometry and allows thousands of cells to be reliably tracked across multiple time points, even when the devices undergo significant manipulation between measurements.
Using this approach, high throughput single cell fluorescence dynamic data can be generated with high reproducibility and minimal user intervention during the image analysis phase. Integrating this system with bioinformatics techniques in the data analysis phase allows for the rich characterization of cell behavior at the single cell level. This robust micro-well array cytometry platform facilitated investigation of a number of biological problems that would be difficult to tackle with more traditional techniques. Our initial efforts have focused on generating dynamic single cell fluorescence images of primary hepatocytes under oxidative stress using a combination of mitochondrial and free radical probes. The dynamic single cell fluorescence images were then analyzed using bioinformatics techniques such as hierarchical and k-means clustering to visualize the data and extract interesting features.
Methods and Materials
Device fabrication
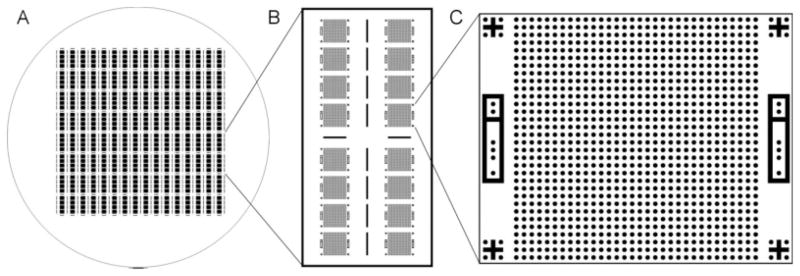
The devices used in this study were fabricated by PDMS replica molding. A 4 inch silicon wafer was first spin-coated with 25 μm SU-8 negative photoresist and patterned by photolithography to generate a reusable master. The most recent design included 512 microwell arrays arranged in groups of four and surrounded by cutting guides. Each microwell array consisted of a 32 by 32 grid of 25-μm diameter cylindrical wells along with alignment and identification features to aid later analysis (Figure 1). For efficient imaging, the layout was designed such that each array filled the 4× visual field of our microscopy system. Replicas were fabricated by mixing PDMS prepolymer with initiator and pouring the mixture onto the master in a 4 mm layer, briefly degassing in a vacuum chamber, and curing at 75°C overnight. The cured PDMS sheet was then peeled from the mold and cut as needed into individual devices.
Figure 1. A reusable mold for casting the PDMS devices is fabricated on a 4 inch silicon wafer using photolithography.
As seen in Panel A, each wafer contains 512 microwell arrays arranged in groups of four and separated by cutting guides. Each PDMS sheet is typically divided into 32 devices measuring 1.6 × 0.8 cm for easy mounting on mini glass slides. As shown in Panel B, each device contains 16 individual arrays with an empty region at the center for pipetting. In some experiments, half sized devices with eight arrays were used to allow for faster imaging. Panel C shows the layout of a typical array, including a regular 32 × 32 grid of 25 μm wells designed to hold single cells, alignment features at each corner, and unique binary identification features on each side.
Assembly and coating
Individual devices containing eight arrays each were cut from the cast PDMS and reversibly mounted well-side-up on 2.5 × 1.0 cm miniature slides for easier handling. The devices were sterilized by either autoclaving after assembly or rinsing in 100% ethanol before assembly. The outer surfaces of the arrays were blocked by applying a 200 μL droplet of 2% Bovine Serum Albumin (BSA, Sigma), incubating for 10 min at 25°C, and rinsing well with PBS. Because the PDMS is hydrophobic, the BSA solution does not enter the microwells and serves to reduce cell adhesion outside of the wells. A 1.0 mg/mL stock solution of fibronectin (Sigma) was diluted 1:40 in phosphate buffered saline (PBS) immediately before use. Each device was then covered with a 200 μL droplet of the fibronectin solution and placed under vacuum for 30 min to promote microwell filling. The devices were then incubated for another 30 min at 25°C, rinsed with media, and kept wet until seeding.
Hepatocyte seeding
Primary rat hepatocytes were freshly isolated from female Lewis rats and resuspended at 1 × 106 cells/mL in C+H hepatocyte culture medium.25 With proper preparation, the resulting cell suspension contains less than 1% nonparenchymal cells. The isolated hepatocytes are typically 15–20 μm in diameter, while the nonparenchymal cells are typically less than 10 μm in diameter. Particular care was taken with the pipetting to gently break up clumps of cells and achieve a single cell suspension. Droplets of the suspension were then placed on each device and periodically recirculated by gentle pipetting to improve filling efficiency and reduce cell attachment outside of the microwells. After 5–10 min, excess cells were removed by rinsing with fresh media, and the devices were briefly examined under the microscope. If loading was poor after the first round of seeding, it was repeated a second time. Once fully loaded, each device was transferred to a dish of fresh medium and gently agitated to remove any cells remaining on the surface of the device.
Dyes and treatments
Hepatocyte mitochondrial membrane potential and mitochondrial superoxide generation was monitored after treatment with the free radical generator, menadione. The fluorescent dye rhodamine 123 (Rh123) was used to qualitatively assess mitochondrial membrane potential (ΔΨ) in the cells.26,27 A 1,000× stock Rh123 solution was prepared by dissolving 10 mg lyophilized Rh123 in 1 mL DMSO and used at a final concentration of 10 μg/mL. Mitochondrial superoxide generation was assessed using dihydroethidium (DHE), which is initially nonfluorescent but is oxidized in the presence of superoxide to form the fluorescent dye ethidium.28 A 1,000× stock DHE solution was prepared by dissolving 3.15 mg lyophilized DHE in 1 mL DMSO and used at a final concentration of 10 μM. A 25 mM stock menadione solution was prepared by dissolving 4.3 mg menadione in 1 mL ethanol and used at a final concentration of 0–100 μM. All stock solutions were protected from light and stored at −20°C in small aliquots.
Seeded microwell devices were first incubated for 15 min at 37°C in C+H culture media with Rh123 at 10 μg/mL, rinsed to remove excess dye, and incubated at 37°C in fresh media for an additional 30 min. Immediately before imaging, DHE and menadione stock solutions were added to warm C+H culture medium in the appropriate amounts, and the seeded devices were transferred into the prepared solutions. Array imaging was started within 15 min of initial exposure to the dye and treatment.
Fluorescence microscopy
Brightfield and fluorescence images were captured on a Zeiss 200 Axiovert microscope with an AxioCam MRm digital camera, typically using a 2.5× objective and 1.6× optovar for full images of a single array. Rh123 fluorescence was measured in the green channel using a Zeiss #38 filter set, and DHE fluorescence was measured in the red channel using a Zeiss #31 filter set. Exposure times were selected to maximize the dynamic range of the resulting images. A brightfield image was taken along with each set of fluorescence images for alignment, quality control, and display purposes.
Image processing and data extraction
Custom image analysis software was developed with the Insight Segmentation and Registration Toolkit (ITK) and used for general image processing tasks, automated alignment of array images, and calculation of pixel intensity statistics for each well.29 Alignment was performed in three stages. Brightfield images of the arrays were first normalized, thresholded, and flood filled to generate simplified black and white images. A rough alignment was then performed between these images and a centered template image of the entire array. From these results, the approximate locations of the alignment features were calculated and used as the seed positions for fine alignment of each feature. In both cases, alignment was performed using a mean squares distance metric and a one-plus-one evolutionary optimizer. The expected coordinates of each well were then calculated based on the identified positions of the four alignment features.
A number of processing steps were applied to the fluorescence images before computing fluorescence statistics for each well. The images were first corrected for stray background illumination and spatial variation in lamp intensity using black and white reference images.30 In some cases, morphological image filters were used to create a background image and eliminate local variation arising from device orientation and nonuniformities in the device or surrounding media. The resulting pixel intensities were finally renormalized to a standard range using either a fixed multiplier or by setting black and white pixel percentages. Pixel intensity statistics including the mean, median, mode, and standard deviation were calculated for the pixels belonging to each well and exported as a spreadsheet file. Further analysis and plotting was performed using the R statistical package.31
Quality control
A simple filtering algorithm was used to automatically identify empty and poorly seeded wells and tag them as such so that they could be excluded from later analysis. Empty wells were identified by settings fluorescence intensity thresholds. Obstructed wells were identified by first examining brightfield images of the annular region immediately surrounding each well. Using the thresholded images generated for alignment purposes, the number of black pixels in each annular region was counted. Properly seeded wells typically contained no more than a few such pixels, and wells were considered obstructed if the percent of black pixels in the region exceeded 5%. This value was chosen as a strict threshold based on histograms of annular pixel counts and manual validation with a subset of the arrays. A similar procedure was carried out with each fluorescence channel to identify additional wells with improper seeding. Wells identified as empty or obstructed were excluded from later plotting and data analysis.
Clustering of fluorescence time courses
Several unsupervised clustering techniques such as k-means and agglomerative hierarchical clustering were applied to Rh123 and DHE fluorescence time courses to help visualize the data and identify distinct patterns of cell behavior. Clustering was performed in R using routines from the Bioconductor software package.32 k-means clustering was performed multiple times for both raw and normalized time course data. The number of clusters was optimized by examining the within cluster sum of squares and the Gap statistic.33 Cluster assignments were also validated by running the algorithm multiple times with random starting points. Hierarchical clustering was performed using the Euclidean distance and either Ward’s method or complete linkage.
Results
Array loading efficiency
Loading efficiencies were determined for 64 microwell arrays on four separate devices. For each well, the presence of a cell was determined by gating on total Rh123 fluorescence. No additional dye was needed because essentially all cells stained to a level well above background. In experiments with a significant population of nonstaining cells, an additional dye such as DAPI could be added for this purpose. Poorly seeded and debris obstructed wells were identified using brightfield images of the arrays as described in the methods. For the set of 64 arrays tested, the percentage of properly filled wells was 70.4 ± 14.5% (mean ± s.d.). The percentage of debris obstructed wells was 5.0 ± 2.2% (mean ± s.d.). Only properly seeded wells were included in later analysis.
Reproducibility of fluorescence values
To test the reproducibility of the intensity values obtained with the system, repeated images were taken of the same array positioned at various points within the visual field. Each image was processed independently, empty and poorly seeded wells were filtered out, and the resulting time courses were plotted for each well. With each image taken, a slight global decrease in well intensity values was observed across the entire array, most likely due to photobleaching. This systematic global variation could largely be eliminated by normalizing the well intensities in a given array image by the median well intensity for that image. After this correction, the well intensities were remarkably consistent from image to image. The coefficient of variation (CV%) across the time course was calculated individually for each well. For Rh123 fluorescence, the CV% (mean ± SD) was 1.1 ± 0.5% with very few outliers. Similar results were achieved for other vital dyes not discussed here.
Single fluorescence time courses
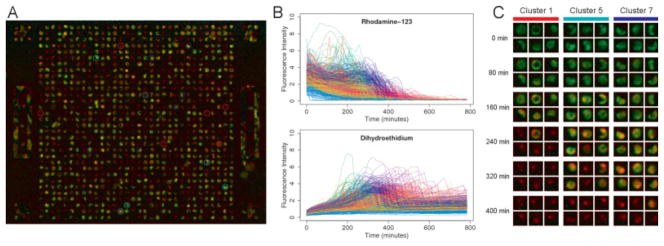
Combining the microwell devices with an automated microscopy system and custom image analysis software allows single cell fluorescence time courses to be obtained with minimal user intervention. Time courses of Rh123 and DHE fluorescence were generated for cells treated with varying amounts of the free radical generator menadione. Shown in Figure 2 is a sample of these time courses for cells treated with 25 μM menadione and colored based on the estimated time of cell death.
Figure 2. Using the microwell array cytometry system, single cell fluorescence time courses can be generated with minimal user intervention.
Panel A is a false color fluorescence image of primary hepatocytes stained with rhodamine 123 (green channel) and dihydroethidium (red channel) and exposed to 25 μM menadione for 4 h. Panel B shows the single cell time courses of Rh123 and DHE fluorescence generated from this and the other images in the time series, with images collected every 16 min for approximately 13 h. The color of each time course corresponds to the survival time determined for that cell. Panel C is an abbreviated time series highlighting individual cells in the array and the diversity of fluorescence patterns observed. k-means clustering was used to identify clusters of cells with similar time courses, and the highlighted cells in each column were randomly selected from these clusters.
The resulting time courses tend to follow characteristic patterns reflecting the mechanism and staining pattern of each dye, but the timing and magnitude of the individual time course features vary significantly from cell to cell. Because the cells were loaded with Rh123 and rinsed before any imaging occurs, the Rh123 time courses begin with a roughly exponential decrease in intensity due to photobleaching and dye leakage. There is then a spike in fluorescence as the mitochondria begin to lose polarization, releasing the dye into the cytoplasm, and reducing the amount of self quenching. Shortly thereafter, there is a complete loss of fluorescence representing plasma membrane depolarization and cell death.
The DHE time courses also follow a characteristic and expected pattern with a steadily increasing baseline intensity due in part to photoactivation and nonspecific conversion of the dye. Superimposed on this baseline is a period of rapid intensity change corresponding to mitochondrial superoxide generation as nonfluorescent DHE is converted to fluorescent ethidium in the presence of superoxide radicals. The timing of this peak roughly corresponds to the spike in Rh123 fluorescence and is followed by a drop in fluorescence back to the baseline as the mitochondria lost their integrity and released trapped dye.
It is important to note that even within this single treatment condition, the cell population displayed a wide range of response patterns that would be difficult to characterize without an unbiased and high throughput experimental system. With manual image analysis, cells at the extremes of this range would likely be missed or excluded as outliers, when in fact the behavior of these rare cells may be more important than the “average” cells when trying to understanding the biological processes involved.
k-means clustering of single cell time courses
Having individual time courses for hundreds to thousands of cells opens up the possibility of using bioinformatics techniques such as k-means and hierarchical clustering to better characterize the data and explore the behavior of cell populations with distinct behaviors. With bulk measurement or low cell count experiments, the typical diversity of cell responses is treated as a source of unwanted biological noise and one is limited to generating a single average response.
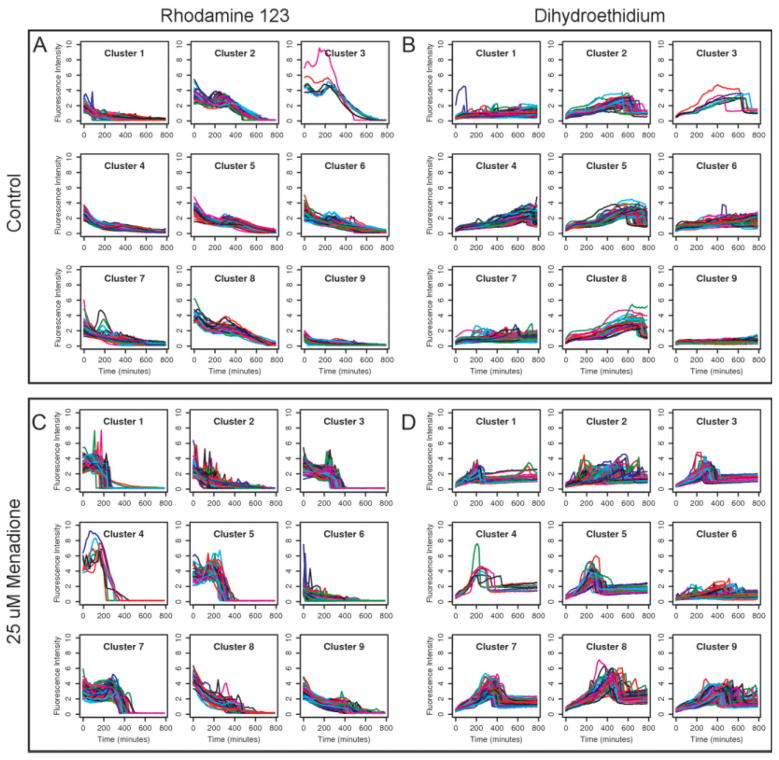
k-means clustering was performed on Rh123 and DHE time courses for a range of cluster counts and evaluated using several figures of merit. For each channel, all of the time courses were rescaled by a fixed factor such that the average Euclidean distance between two time courses would be equal to one. This was done so that the two channels would have equal weight when combined in a single vector for clustering. Shown in Figure 3 are the results for nine clusters under two different treatment conditions. Clustering was performed separately for each treatment condition, but within a treatment condition, the cluster assignments are shared for the two channels and numbered according to the median time of cell death within the cluster.
Figure 3. k-means clustering was performed on Rh123 and DHE time course data under two experimental conditions.
Because clustering was performed on combined data, the cluster assignments in the two channels correspond to each other. Control cells with no menadione treatment are shown in Panels A and B. Cells treated with 25 μM menadione are shown in Panels C and D. Several distinct response patterns can be identified from the clustered data. The response patterns differ in shape and timing between the two conditions, but also within each condition, reflecting the wide diversity of behaviors possible in a cell population. For each fluorescence channel, the general shape of each time course is constrained by the mechanism of the dye used, but there is considerable variation in the magnitude and timing of the responses.
Visualizing single cell dose response experiments using hierarchical clustering
Dose response curves are a common and well established way to present data from pharmacological tests with a binary endpoint such as cell death. Such curves can be generated from high throughput single cell experiments, but this does not take full advantage of the collected data. Reducing a single cell fluorescence time courses to a simple endpoint such as cell death does not account for the initial state of the cell and its behavior leading up to that endpoint. Here, we took a different approach for analyzing the retrieved data to yield a more detailed picture of hepatocyte behavior in response to menadione treatment.
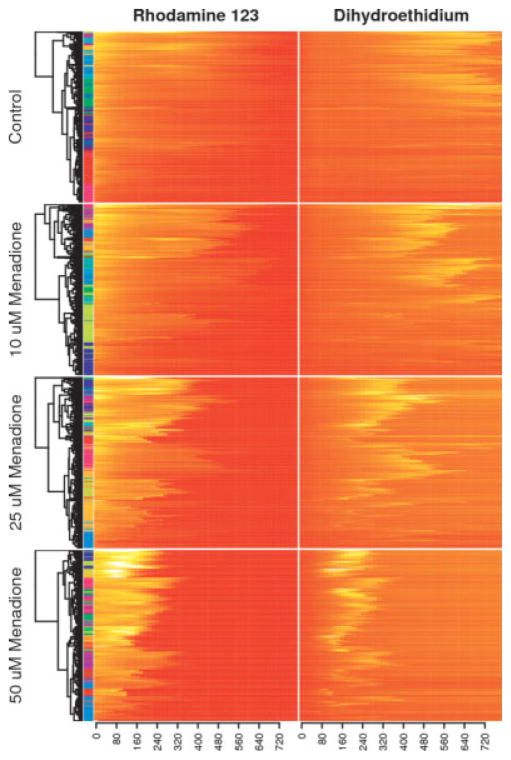
Hierarchical clustering was performed on the normalized and combined Rh123 and DHE time courses using the Euclidean distance and complete linkage. The dendrograms were then reordered such that branches with the highest mean intensity appear at the top. The results are shown in Figure 4. The colored bars between the heat maps and the dendrograms indicate the cluster assignment for each cell using the k-means algorithm. As the menadione dosage increases, the rate of free radical generation also increases, and cell death tends to occur earlier. Even within a single treatment condition, however, there is a great diversity of cell survival times. For the 25 μM treatment condition, survival times range anywhere from 150 to 600 min. Another striking feature is the temporal correlation between the fluorescence channels, with the contours of the paired heat maps following each other very closely.
Figure 4. Hierarchical clustering with the Euclidean distance and complete linkage was performed on combined Rh123 and DHE time course data in a dose response experiment, with dendrogram nodes reordered according to average fluorescence intensities.
Each heatmap row corresponds to the behavior of a single cell from one of the arrays, and the color bar on the left side indicates cluster assignments using the k-means algorithm. Presenting dose response data as a set of clustered heatmaps is an information-rich alternative to classic survival curves. Although it is straightforward to represent the collected data as a set of survival curves, the heatmap approach better highlights the diversity of cell responses to each stimulus and allows one to appreciate the temporal relationships between multiple fluorescence channels.
Heat maps are particularly well suited to visualizing the data from high throughput single cell dose response experiments, particularly in combination with hierarchical clustering. Placing time courses with similar profiles close together made it easier to identify major trends in the data, both within each treatment condition and between them and highlighted the temporal relationships between multiple fluorescence channels. As an alternative to clustering, the time courses were sorted according to the time at which a particular end point is reached, but this tended to obscure potentially interesting trends in the data.
Discussion
The microwell array cytometry system presented in this article is a novel, high-throughput, high-content platform for the study of cell behavior, cell population heterogeneity, and biological stochasticity. Most importantly, the system is able to generate dynamic single cell fluorescence data for multiple vital probes and correlate pretreatment phenotypes with post-treatment outcomes at the single cell level. The system also provides sufficiently large cell populations for meaningful analysis, easy loading and staining protocols, automated data handling, and reproducibility of the measured fluorescence values. In addition to presenting the cytometry system, a number of powerful bioinformatics techniques were adapted and applied to the study of single cell fluorescence time courses, demonstrating the advantages of this approach over classic bulk assays and analysis techniques.
The need for a microwell array cytometry platform arose in direct response to the limitations of bulk assays and currently available cytometry technologies. Bulk assays can only deal with population averages, which represents a loss of potentially useful information and can mislead the user in a number of significant ways.12 Single cell methods such as flow and image cytometry avoid this problem by providing the entire distribution of values seen in the population, but each has its own limitations. The major limitation of flow cytometry is its inability to track individual cells across multiple time points. One can track the overall behavior of the population across time, but not the individual behavior of each cell. With array cytometry, each cell is associated with a single microwell in a rugged PDMS device and remains attached and in place through significant manipulation of the device. The inclusion of alignment and identification features around each array makes it even easier to track cells across time.
Image cytometry is limited by the complexity of identifying and segmenting fluorescence images of cells, particularly with random seeding and high cell densities. Although analysis techniques are improving, it is not trivial to extract meaningful single cell data from such images.15 With array cytometry, image processing is greatly simplified by seeding each cell in its own microwell, which physically separates the cells and places them in known positions relative to easily identified alignment features. This technological approach makes it easier to automate the data analysis and use the system in a high-throughput fashion. Because of its low cost and relative simplicity, microwell array cytometry has the potential to become as widely used and versatile as flow cytometry.
Bioinformatics techniques such as k-means and hierarchical clustering are a powerful set of tools that can be used to organize, analyze, and visualize the voluminous data generated by high throughput techniques such as image cytometry and microwell array cytometry. As demonstrated in this article, the clustering of single cell fluorescence time courses helps identify groups of cells with distinct behaviors, makes it easier to compare the results of different treatment conditions, and highlights the diversity of responses to treatment within a presumably uniform cell population. The use of heatmaps with hierarchical clustering provides a data-rich complement to survival curves for dose response experiments where additional time course data is available.
Although they are superficially similar, fluorescence time course data from a microwell array cytometry experiment has a number of features that distinguish it from the gene expression data generated by cDNA and oligonucleotide microarrays. Most importantly, the physical properties and behavior of each fluorescent dye impose a great deal of structure on the resulting time courses. For a particular dye, much of the observed variation between cells relates to timing and magnitude rather than the overall shape of the curve. This poses a different set of challenges than are addressed by the time course analysis often done for gene expression data, which emphasizes the grouping of genes with similar time course shapes, but not magnitudes.34 Another important difference is that the number of features examined is relatively small when compared with the number of replicates, the opposite of most gene expression studies.35 These differences will require the development and refinement of bioinformatics techniques suited to this type of data and represent a great opportunity for future bioinformatics research.
In vivo, primary cells such as hepatocytes are known to have significant heterogeneity in function and gene expression due to metabolic zonation and other factors.5 Future work with the array cytometry system will explore how the initial state of a hepatocyte affects its response to stress. Distinct responses to oxidative stress are seen in vivo between hepatocytes in different zones of the liver, and some of this is likely intrinsic to the hepatocytes rather than the structure of the environment. Hepatocytes will be stained or tagged based on zone of origin and other properties, and differences in stress response will then be tracked using the array system.
There are also many planned enhancements to both the physical and software aspects of the system. On the software side, this includes computing additional statistics for each well such as cell size, granularity, and fluorescence colocalization. On the hardware side, the array will be combined with an existing combinatorial microfluidic device, greatly expanding the number of markers and conditions that can be tested at one time.36 More complex array designs are being developed that allow for controlled cell–cell interaction between adjacent wells. This will take the form of clustered wells with small connecting trenches that allow for cell-cell contact but preserve the current seeding efficiency. Efforts are also underway to automate the collection of higher resolution images of interesting wells based on position and phenotype data from low resolution scout images.
Conclusion
As our understanding of biology grows, single-cell and subcellular analysis techniques will become increasingly important. Already it has become clear that the forced population averaging of bulk experimental techniques provides a limited view of the rich variation and stochasticity present in biological systems. Studying large populations does have its advantages, however, and high throughput experimental techniques are beginning to recover those advantages without sacrificing the richness of single cell analysis. Further advances will require the development of improved algorithms for automated data collection, physical device designs to support this automation, and new approaches in bioinformatics to analyze the results. It is expected that the system described here is a step in this direction.
Contributor Information
Kenneth L. Roach, Center for Engineering in Medicine, BioMEMS Resource Center, Massachusetts General Hospital, Harvard Medical School, Shriners Hospital for Children, Boston, MA. Harvard-MIT Division of Health Sciences and Technology, Cambridge, MA
Kevin R. King, Center for Engineering in Medicine, BioMEMS Resource Center, Massachusetts General Hospital, Harvard Medical School, Shriners Hospital for Children, Boston, MA. Harvard-MIT Division of Health Sciences and Technology, Cambridge, MA
Basak E. Uygun, Center for Engineering in Medicine, BioMEMS Resource Center, Massachusetts General Hospital, Harvard Medical School, Shriners Hospital for Children, Boston, MA
Isaac S. Kohane, Harvard-MIT Division of Health Sciences and Technology, Cambridge, MA. Informatics Program, Children’s Hospital Boston and Harvard Medical School, Boston, MA
Martin L. Yarmush, Center for Engineering in Medicine, BioMEMS Resource Center, Massachusetts General Hospital, Harvard Medical School, Shriners Hospital for Children, Boston, MA. Harvard-MIT Division of Health Sciences and Technology, Cambridge, MA
Mehmet Toner, Center for Engineering in Medicine, BioMEMS Resource Center, Massachusetts General Hospital, Harvard Medical School, Shriners Hospital for Children, Boston, MA. Harvard-MIT Division of Health Sciences and Technology, Cambridge, MA.
Literature Cited
- 1.Levsky JM, Singer RH. Gene expression and the myth of the average cell. Trends Cell Biol. 2003;13:4–6. doi: 10.1016/s0962-8924(02)00002-8. [DOI] [PubMed] [Google Scholar]
- 2.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 3.Longo D, Hasty J. Dynamics of single-cell gene expression. Mol Syst Biol. 2006;2:64. doi: 10.1038/msb4100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artyomov MN, Das J, Kardar M, Chakraborty AK. Purely stochastic binary decisions in cell signaling models without underlying deterministic bistabilities. Proc Natl Acad Sci USA. 2007;104:18958–18963. doi: 10.1073/pnas.0706110104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gebhardt R. Metabolic zonation of the liver: regulation and implications for liver function. Pharmacol Ther. 1992;53:275–354. doi: 10.1016/0163-7258(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 6.Rao CV, Wolf DM, Arkin AP. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- 7.Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramsey S, Ozinsky A, Clark A, Smith KD, de Atauri P, Thorsson V, Orrell D, Bolouri H. Transcriptional noise and cellular heterogeneity in mammalian macrophages. Philos Trans R Soc Lond B Biol Sci. 2006;361:495–506. doi: 10.1098/rstb.2005.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 10.Blake WJ, MKA, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 11.Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 12.Di Carlo D, Lee LP. Dynamic single-cell analysis for quantitative biology. Anal Chem. 2006;78:7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 13.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 14.Sims CE, Allbritton NL. Analysis of single mammalian cells on-chip. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 15.Gordon A, Colman-Lerner A, Chin TE, Benjamin KR, Yu RC, Brent R. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat Methods. 2007;4:175–181. doi: 10.1038/nmeth1008. [DOI] [PubMed] [Google Scholar]
- 16.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 17.Falconnet D, Csucs G, Grandin HM, Textor M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials. 2006;27:3044–3063. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 18.Rettig JR, Folch A. Large-scale single-cell trapping and imaging using microwell arrays. Anal Chem. 2005;77:5628–5634. doi: 10.1021/ac0505977. [DOI] [PubMed] [Google Scholar]
- 19.Revzin A, Sekine K, Sin A, Tompkins RG, Toner M. Development of a microfabricated cytometry platform for characterization and sorting of individual leukocytes. Lab Chip. 2005;5:30–37. doi: 10.1039/b405557h. [DOI] [PubMed] [Google Scholar]
- 20.Ochsner M, Dusseiller MR, Grandin HM, Luna-Morris S, Textor M, Vogel V, Smith ML. Micro-well arrays for 3D shape control and high resolution analysis of single cells. Lab Chip. 2007;7:1074–1077. doi: 10.1039/b704449f. [DOI] [PubMed] [Google Scholar]
- 21.Chin VI, Taupin P, Sanga S, Scheel J, Gage FH, Bhatia SN. Microfabricated platform for studying stem cell fates. Biotechnol Bioeng. 2004;88:399–415. doi: 10.1002/bit.20254. [DOI] [PubMed] [Google Scholar]
- 22.Biran I, Walt DR. Optical imaging fiber-based single live cell arrays: a high-density cell assay platform. Anal Chem. 2002;74:3046–3054. doi: 10.1021/ac020009e. [DOI] [PubMed] [Google Scholar]
- 23.Deutsch M, Deutsch A, Shirihai O, Hurevich I, Afrimzon E, Shafran Y, Zurgil N. A novel miniature cell retainer for correlative high-content analysis of individual untethered non-adherent cells. Lab Chip. 2006;6:995–1000. doi: 10.1039/b603961h. [DOI] [PubMed] [Google Scholar]
- 24.Ozawa T, Kinoshita K, Kadowaki S, Tajiri K, Kondo S, Honda R, Ikemoto M, Piao L, Morisato A, Fukurotani K, Kishi H, Muraguchi A. MAC-CCD system: a novel lymphocyte micro-well-array chip system equipped with CCD scanner to generate human monoclonal antibodies against influenza virus. Lab Chip. 2009;9:158–163. doi: 10.1039/b810438g. [DOI] [PubMed] [Google Scholar]
- 25.Dunn JC, Tompkins RG, Yarmush ML. Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol Prog. 1991;7:237–245. doi: 10.1021/bp00009a007. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro HM. Membrane potential estimation by flow cytometry. Methods. 2000;21:271–279. doi: 10.1006/meth.2000.1007. [DOI] [PubMed] [Google Scholar]
- 27.Duchen MR, Surin A, Jacobson J. Imaging mitochondrial function in intact cells. Methods Enzymol. 2003;361:353–389. doi: 10.1016/s0076-6879(03)61019-0. [DOI] [PubMed] [Google Scholar]
- 28.Rothe G, Valet G. Flow cytometric analysis of respiratory burst activity in phagocytes with hydroethidine and 2′,7′-dichloro-fluorescin. J Leukoc Biol. 1990;47:440–448. [PubMed] [Google Scholar]
- 29.Yoo TS, Ackerman MJ, Lorensen WE, Schroeder W, Chalana V, Aylward S, Metaxas D, Whitaker R. Engineering and algorithm design for an image processing API: a technical report on ITK--the Insight Toolkit. Stud Health Technol Inform. 2002;85:586–592. [PubMed] [Google Scholar]
- 30.Souchier C, Brisson C, Batteux B, Robert-Nicoud M, Bryon PA. Data reproducibility in fluorescence image analysis. Methods Cell Sci. 2003;25:195–200. doi: 10.1007/s11022-004-2383-4. [DOI] [PubMed] [Google Scholar]
- 31.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 32.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc B. 2001;63:411–423. [Google Scholar]
- 34.Luan Y, Li H. Clustering of time-course gene expression data using a mixed-effects model with B-splines. Bioinformatics. 2003;19:474–482. doi: 10.1093/bioinformatics/btg014. [DOI] [PubMed] [Google Scholar]
- 35.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King KR, Wang S, Irimia D, Jayaraman A, Toner M, Yarmush ML. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip. 2007;7:77–85. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]