Abstract
Understanding the nature and mechanism of congenital defects of the different organ systems in humans has heavily relied on the analysis of the corresponding mutant phenotypes in rodent models. Optical coherence tomography (OCT) has recently emerged as a powerful tool to study early embryonic development. This non-invasive optical methodology does not require labeling and allows visualization of embryonic tissues with single cell resolution. Here, we will discuss how OCT can be applied for structural imaging of early mouse and rat embryos in static culture, cardiodynamic and blood flow analysis, and in utero embryonic imaging at later stages of gestation, demonstrating how OCT can be used to assess structural and functional birth defects in mammalian models.
Keywords: optical coherence tomography (OCT), mouse, rat, embryo, live embryonic imaging
Introduction
Thousands of different birth defects have been reported which affect the structure or function of every part of the human body. Studies in mouse models are invaluable for defining genetic and environmental factors affecting the anatomical and physiological development of different organ systems in humans. For this purpose, hundreds of mouse mutants linked to human birth defects and diseases have been reported [1] and several large-scale, international, genome-wide screens for new and advanced models of human disease have been initiated [2,3,4]. However, the success of these efforts depends on the ability to analyze phenotypic outcomes, raising an urgent need for better phenotyping tools.
Recently, great progress has been made in developing imaging methods for developmental biology. High-frequency ultrasound allows visualizing whole embryos in utero. The resolution of this method even for high-frequency systems is limited to about 30–100 μm, yet it has proven successful in screening for specific defects [5,6]. Micro-MRI is another technique used for structural 3D embryo imaging, and proof-of-concept studies have shown that it can be used to screen for different phenotypes [7,8,9]. However, the spatial resolution of micro-MRI is also about 25–100 μm and this method is particularly sensitive to motion due to long integration times, limiting its applicability for live embryo imaging. Higher resolution optical microscopy methods such as confocal or multiphoton imaging can be used to image early stage post-implantation embryos in culture (E5.5–10.5) with sub-micron spatial resolution, but the imaging depth is limited to a few hundreds of micrometers [10,11,12]. Because of the limited field-of-view and the need for vital, fluorescent cell or tissue markers (such as fluorescent proteins), these methods are most appropriate to answer well-defined questions in a limited number of mutants rather than as a screening tool. Optical projection tomography (OPT) is a very exciting, innovative method for high-resolution 3-D imaging of mouse embryos and has been used successfully to define phenotypic variations within mutants in several studies. The main advantage of this method is the ability to image whole mount immunostaining in embryos, but imaging through such thick 3-D volumes is enhanced by clearing agents that are not compatible with live embryonic imaging [13,14].
Optical coherence tomography has a unique niche for live mouse embryonic imaging and phenotyping. OCT has an order of magnitude higher spatial resolution (2–10 μm) than ultrasound or Micro-MRI, while imaging an order of magnitude deeper (1–3 mm) than that of confocal/multiphoton microscopy [19]. The high spatial resolution and true 4-D capability, without the need for applied contrast agents are advantageous for imaging mouse embryos. Here, we will discuss OCT applications for live embryonic imaging and functional analysis at different stages of mammalian development and how this method can be used for identifying mutant phenotypes.
Optical Coherence Tomography
Optical Coherence Tomography (OCT) was introduced as a method for generating 3-D tomograms of the eye in 1991 by Fujimoto’s group [15]. The non-ionizing light of the OCT is considered to be safe and is FDA approved for ophthalmologic use in humans [16]. OCT systems are now routinely found in ophthalmology practices worldwide and these systems are becoming increasingly more popular with researchers and clinicians in ophthalmology as well as cardiology, dermatology and oncology [17,18]. Modern OCT systems can achieve a spatial resolution of about 2 to 10 μm and an imaging depth of about 1 to 3 mm in tissues [19]; thus, it is ideal for live “microscopic histology”.
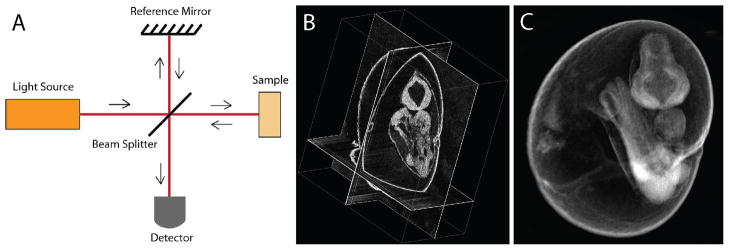
Briefly, OCT systems are based on interferometry, utilizing a coherent, broadband light source. Similarly to the ultrasound imaging, the in-depth resolution of the OCT is achieved by analysis of an echo time delay and an amplitude of the signal backscattered from different sample depth. Because the speed of light is much higher than the speed of sound, the time delay of the backscattered light cannot be measured directly, as in the ultrasound approach. To overcome this limitation, OCT utilizes light interferometry (Figure 1A). The laser light is split into a sample arm that is scanned through tissue and a reference path that does not pass through tissue. Images are generated from the interference signals that are created when the light from both arms is recombined, providing contrast where there are differences in the optical properties of tissues, such as between tissue layers, fluid-tissue interfaces or from different cellular densities. OCT relies on natural tissue contrast and does not require application of any contrast agents. A single unit of OCT data is an in-depth intensity profile (called A-scan). By lateral scanning of the beam, 2-D cross-sectional images (B-scans) and 3-D volumes of OCT data are acquired. C-scan in OCT refers to a cross-sectional image perpendicular to the scanning beam. A more complete description of how OCT imaging works and ways to improve contrast, speed and resolution can be found in [19].
Figure 1.
Live embryonic imaging with optical coherence tomography (OCT). (A) Simplified diagram of OCT principle. (B) A cross-sectional view through the 9.5 dpc mouse embryo cultured on the imaging stage, (C) 3-D rendering view of the same OCT data set. The data set consists of 512×512 A-scans.
Structural OCT imaging of early embryonic development
OCT has been successfully applied for live structural embryonic imaging in different non-mammalian model systems, such as Drosophila [20], zebrafish [21], Xenopus laevis [22,23], quail [24,25], and chick [26,27,28,29] with a primary focus on cardiovascular analysis. Jenkins et al. applied OCT for imaging of extracted mouse embryonic hearts at E12.5 and E13.5, which demonstrated the potential for this technique to visualize structural and functional consequences of genetic manipulations [30]. The same group reported 3-dimensional OCT images of E13.5 beating, embryonic mouse hearts that were excised and externally paced [31]. Likewise, Luo et al. imaged beating E10.5 hearts in embryos that were maintained outside the uterus, but with dramatically slower than normal heart rates [32]. By combining OCT imaging with robust embryo culture methods [11,33], our group has been able to use OCT for structural and hemodynamic analysis of the cardiovascular system in live mouse embryos [34,35,36]. Although not in a perfectly natural environment, early embryos (E7.5–10.5) cultured on the imaging stage for over 24 hours show similar developmental milestones as those in utero including vessel remodeling and heart looping, providing a window into otherwise inaccessible information [10]. Figure 1(B,C) shows an example of a live mouse embryo imaged with OCT at E9.5. Structural OCT is very effective as a method to produce 3-D reconstructions of whole live cultured mouse embryos from E7.5 to about E10.5 as well as for dynamic 2-D imaging with single cell resolution [34,35,36].
Live OCT imaging protocols have also been extended to the rat at embryonic stages E10.5 – E11.5, equivalent to E8.5 – 9.5 mouse embryos [19]. Structural 3-D imaging of live rat embryos, dynamic heart imaging, and Doppler OCT analysis of the beating heart was demonstrated [19]. The rat model is preferred for a variety of physiological studies, and recent derivation of rat embryonic stem (ES) cells by Buehr et al. and Li et al. [37,38] opened a door for a wide range of genetic alterations to create rat models with more relevance to human disorders, including better models of congenital defects. OCT imaging is a useful tool for live dynamic embryonic analysis of rat mutants.
Functional OCT imaging
In addition to structural imaging, OCT can be used for Doppler analysis to obtain velocity measurements from moving structures, with the same spatial and temporal resolution [39]. Doppler OCT relies on detection of a phase shift between adjacent in-depth OCT scans at each point, which is caused by movement of the light scatterers. Blood flow velocity at each pixel can be reconstructed according to the formula (11):
| (1) |
where Δϕ - is a Doppler shift-induced phase shift calculated between successive A-scans, n - is a refractive index, <k> - is the average wave number, τ - is time between the successive A-scans, and β - is an angle between the flow direction and the laser beam. The angle β can be calculated from structural 2-D and 3-D data sets.
Doppler OCT is an effective way to characterize blood flow dynamics in early embryos analysis. It can be applied to reconstruct spatially and temporally resolved Doppler shift velocity profiles from yolk sac vessels and embryonic vessels when the blood flow is well established [35], as well as at early stages of circulation while blood flow is being established, based on velocity measurements from individual circulating blood cells [34]. This is highly important as it allows visualizing earliest blood circulation defects in mouse mutants with cardiovascular abnormalities, which is currently not possible with other modalities.
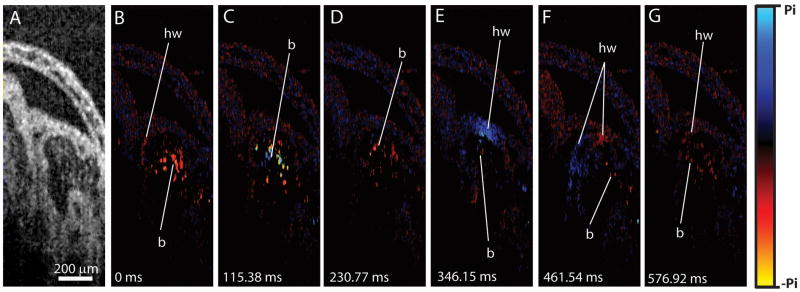
Doppler OCT imaging in rat embryos has also been performed [40]. Figure 2 shows an example of a structural OCT image through the E10.5 rat embryonic heart and a series of color-coded Doppler images of the same area taken from a time lapse and representing different phases of the heartbeat cycle. The Doppler shift signal is generated by the velocity component, which is parallel to the OCT laser beam. The blue shift corresponds to movement toward the detector, while the red shift corresponds to movement away from the detector. As one can see from the figure, the Doppler shift was detected from the circulating blood cells inside the heart (labeled as b) as well as from the moving heart wall (labeled as hw).
Figure 2.
Doppler OCT velocity imaging in the live rat embryonic heart at 10.5 dpc. (A) Structural image acquired from the primitive ventricle of the beating embryonic heart. (B–G) Corresponding representative Doppler color-coded maps from the same area taken out of a time lapse. Doppler imaging was performed at about 26 fps at 512 A-lines per frame. Adopted from [40].
In many cases, hemodynamic analysis in the heart is complicated by the Doppler shift phase wrapping (at high flow velocities, the Doppler OCT shift exceeds 2π, which makes velocity calculations ambiguous) and the difficulty of mapping the exact flow direction, which is required for the velocity calculation. Several groups are developing and optimizing algorithms to overcome these limitations in avian models [26,27]. Potentially, these methods can be applied for quantitative Doppler OCT hemodynamic analysis in mammalian embryonic hearts.
Traditionally, the vascular structure is reconstructed based on Doppler OCT analysis of the blood flow. The major drawback of Doppler OCT for blood flow analysis and reconstruction of vascular structure is its insensitivity to the transverse component of blood flow, which prevents Doppler OCT visualization of vessels perpendicular to the scanning laser beam. Another disadvantage of the Doppler OCT is its dependence on phase stability of the system. Alternatively, Speckle Variance (SV) OCT analysis can be used for 3-D reconstruction of the vasculature in cultured embryos [41]. SV OCT analysis relies on statistical properties of time-varying speckle pattern in OCT images, as the decorrelation of speckles from moving scatterers is faster than that of static scatterers. Because the most dynamic scatterers in the embryo are circulating blood cells, this algorithm allows to visualize 3-D circulatory network. SV OCT imaging provides more accurate 3-D visualization of the vasculature than the Doppler OCT method, since Doppler OCT relies on the axial component of the blood flow and SV OCT it is not sensitive to the flow direction.
In utero embryonic imaging
OCT imaging of cultured embryos described above reveals unique information about early mammalian development; however, the major limitation of this method is that embryos grown in culture can only be maintained for 24–48 hours. Furthermore, embryos obtained beyond the E10.5 stage do not survive long in culture due to the need for maternal support.
Beginning at E12.5 through the remainder of embryogenesis, mouse embryos can be visualized in utero with OCT [42]. Prior to E12.5, each embryo in the uterus is surrounded by a thick layer of decidua, which scatters the signal extensively making embryos inaccessible to optical imaging. As the embryos grow and the placenta forms, the decidua thins and degenerates by E12.5, enabling embryo imaging through the uterine wall. For longitudinal imaging, the female is anesthetized, and the uterine horn is externalized through an abdominal incision for access. After imaging, the incision is closed with surgical sutures. Although semi-invasive, this method allows repeated imaging of the same living embryos to characterize temporal changes in organ development at unprecedented spatial resolution. High resolution OCT imaging of different embryonic organs, such as brain, limb, and eye have also been demonstrated [42]. Figure 3 shows examples of 3-D reconstructions of mouse embryos in the uterus at three embryonic stages. The head and the forelimb are visible in all the reconstructions, and there is a clear morphological and size difference between the stages shown in the Figure. Changes in craniofacial details over time are remarkable. This approach can be useful in analyzing embryos from genetic screens to avoid histological sectioning of multiple litters and to study the effects of pharmacological and toxicological agents on embryo development. Limited imaging depth is the major drawback of this method, however, optical clearing methods might potentially prove useful for enhancing the OCT signal from internal structures [43].
Figure 3. 3D OCT imaging of mouse embryos in utero at different developmental stages.
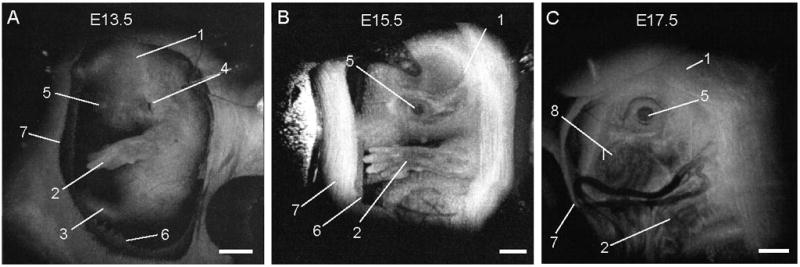
Each data set for the reconstructions was acquired as a series of 512 × 512 A-scans over a 10mm × 10mm × 2.2 mm volume. 1, head; 2, forelimb; 3, hindlimb; 4, pinna of ear; 5, eye; 6, yolk sac; 7, uterine wall; 8, follicles of vibrissae. Scale bars correspond to 1mm. Adopted from [42].
Future applications
Live OCT imaging is an exciting innovative approach to study mammalian development. While some experience is needed to maintain live embryo culture on the imaging stage and to perform survival surgeries for longitudinal in utero analysis, no other methods are currently available to visualize live mouse embryos at a similar spatial and temporal resolution. Currently, functional annotation of the mammalian genome will be carried out on large numbers of mice obtained from the International Knockout Mouse Consortium by the International Mouse Phenotyping Consortium [44,45]. Approximately one-third of all single gene mutations result in early, post-implantation embryonic lethality; abnormal development of the cardiovascular system results in the death of nearly two thirds of these [46]. Because there is no need for additional contrast agents on living embryos and because entire litters can be imaged within minutes, OCT imaging is suitable for secondary screening of embryonic lethal mutations to define the morphological or functional basis of the phenotype. Furthermore, the ability to scan live, mid-gestation stages with high resolution in utero will enable chronic imaging of phenotype progression in tissues such as the eye, limb, and brain with much better spatial resolution than that offered by ultrasound or MRI analysis, and it may be adapted to other organ systems as well. High throughput phenotype analysis of mice will be required for functional studies, as well as pre-clinical, toxicological and systems biology approaches [47]. Current studies are underway to define protocols for routine phenotype analysis using OCT imaging. With increasing commercial availability of turn-key machines and the growing popularity of OCT imaging for a variety of applications, OCT systems are likely to become common instruments in many rodent phenotyping core labs.
Acknowledgments
The studies described in this manuscript were supported in part by grants from the National Institutes of Health (R01HL095586) and the American Heart Association (10SDG3830006).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moon AM. Mouse models for investigating the developmental basis of human birth defects. Pediatr Res. 2006;59:749–755. doi: 10.1203/01.pdr.0000218420.00525.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown SD, Wurst W, Kuhn R, Hancock JM. The functional annotation of mammalian genomes: the challenge of phenotyping. Annu Rev Genet. 2009;43:305–333. doi: 10.1146/annurev-genet-102108-134143. [DOI] [PubMed] [Google Scholar]
- 3.Kim IY, Shin JH, Seong JK. Mouse phenogenomics, toolbox for functional annotation of human genome. BMB Rep. 2010;43:79–90. doi: 10.5483/bmbrep.2010.43.2.079. [DOI] [PubMed] [Google Scholar]
- 4.Morgan H, Beck T, Blake A, Gates H, Adams N, et al. EuroPhenome: a repository for high-throughput mouse phenotyping data. Nucleic Acids Res. 2010;38:D577–585. doi: 10.1093/nar/gkp1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster FS, Zhang M, Duckett AS, Cucevic V, Pavlin CJ. In vivo imaging of embryonic development in the mouse eye by ultrasound biomicroscopy. Invest Ophthalmol Vis Sci. 2003;44:2361–2366. doi: 10.1167/iovs.02-0911. [DOI] [PubMed] [Google Scholar]
- 6.Phoon CKL, Turnbull DH. Ultrasound biomicroscopy-Doppler in mouse cardiovascular development. Physiol Genomics. 2003;14:3–15. doi: 10.1152/physiolgenomics.00008.2003. [DOI] [PubMed] [Google Scholar]
- 7.Pallares P, Fernandez-Valle ME, Gonzalez-Bulnes A. In vivo virtual histology of mouse embryogenesis by ultrasound biomicroscopy and magnetic resonance imaging. Reproduction, Fertility and Development. 2009;21:283–292. doi: 10.1071/rd08124. [DOI] [PubMed] [Google Scholar]
- 8.Nieman BJ, Bock NA, Bishop J, Chen XJ, Sled JG, et al. Magnetic resonance imaging for detection and analysis of mouse phenotypes. NMR Biomed. 2005;18:447–468. doi: 10.1002/nbm.981. [DOI] [PubMed] [Google Scholar]
- 9.Hogers B, Gross D, Lehmann V, Zick K, De Groot HJ, et al. Magnetic resonance microscopy of mouse embryos in utero. Anat Rec. 2000;260:373–377. doi: 10.1002/1097-0185(20001201)260:4<373::AID-AR60>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 10.Jones EAV, Crotty D, Kulesa PM, Waters CW, Baron MH, et al. Dynamic in vivo imaging of postimplantation mammalian embryos using whole embryo culture. Genesis. 2002;34:228–235. doi: 10.1002/gene.10162. [DOI] [PubMed] [Google Scholar]
- 11.Lucitti JL, Jones EAV, Huang C, Chen J, Fraser SE, et al. Vascular remodeling of the mouse yolk sac requires hemodynamic force. Development. 2007;134:3317–3326. doi: 10.1242/dev.02883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nowotschin S, Hadjantonakis AK. Use of KikGR a photoconvertible green-tored fluorescent protein for cell labeling and lineage analysis in ES cells and mouse embryos. BMC Dev Biol. 2009;9:49. doi: 10.1186/1471-213X-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walls JR, Coultas L, Rossant J, Henkelman RM. Three-dimensional analysis of vascular development in the mouse embryo. PLoS One. 2008;3:e2853. doi: 10.1371/journal.pone.0002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colas JF, Sharpe J. Live optical projection tomography. Organogenesis. 2009;5:129–134. doi: 10.4161/org.5.4.10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, et al. Optical coherence tomography. Science. 1991;254:1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Lee L. Clinical applications and new developments of optical coherence tomography: an evidence-based review. Clin Exp Optom. 2007;90:317–335. doi: 10.1111/j.1444-0938.2007.00151.x. [DOI] [PubMed] [Google Scholar]
- 17.Marschall S, Sander B, Mogensen M, Jorgensen TM, Andersen PE. Optical coherence tomography-current technology and applications in clinical and biomedical research. Anal Bioanal Chem. 2011;400:2699–2720. doi: 10.1007/s00216-011-5008-1. [DOI] [PubMed] [Google Scholar]
- 18.Fercher AF. Optical coherence tomography - development, principles, applications. Z Med Phys. 2010;20:251–276. doi: 10.1016/j.zemedi.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 19*.Drexler W, Fujimoto JG, editors. Optical coherence tomography: technology and applications. Berlin: Springer; 2008. This is a deep comprehensive review of OCT principle and applications in biomedicine. [Google Scholar]
- 20.Choma MA, Izatt SD, Wessells RJ, Bodmer R, Izatt JA. In vivo imaging of the adult drosophila melanogaster heart with real-time optical coherence tomography. Circulation. 2006;114:e35–36. doi: 10.1161/CIRCULATIONAHA.105.593541. [DOI] [PubMed] [Google Scholar]
- 21.Kagemann L, Ishikawa H, Zou J, Charukamnoetkanok P, Wollstein G, et al. Repeated, noninvasive, high resolution spectral domain optical coherence tomography imaging of zebrafish embryos. Mol Vis. 2008;14:2157–2170. [PMC free article] [PubMed] [Google Scholar]
- 22.Mariampillai A, Standish BA, Munce NR, Randall C, Liu G, et al. Doppler optical cardiogram gated 2D color flow imaging at 1000 fps and 4D in vivo visualization of embryonic heart at 45 fps on a swept source OCT system. Optics Express. 2007;15:1627–1638. doi: 10.1364/oe.15.001627. [DOI] [PubMed] [Google Scholar]
- 23.Boppart SA, Tearney GJ, Bouma BE, Southern JF, Brezinski ME, et al. Noninvasive assessment of the developing Xenopus cardiovascular system using optical coherence tomography. Proceedings of the National Academy of Sciences. 1997;94:4256–4261. doi: 10.1073/pnas.94.9.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins MW, Chughtai OQ, Basavanhally AN, Watanabe M, Rollins AM. In vivo gated 4D imaging of the embryonic heart using optical coherence tomography. Journal of Biomedical Optics. 2007;12:030505. doi: 10.1117/1.2747208. [DOI] [PubMed] [Google Scholar]
- 25.Garita B, Jenkins MW, Han M, Zhou C, Vanauker M, et al. Blood flow dynamics of one cardiac cycle and relationship to mechanotransduction and trabeculation during heart looping. Am J Physiol Heart Circ Physiol. 2011;300:H879–891. doi: 10.1152/ajpheart.00433.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins MW, Adler DC, Gargesha M, Huber R, Rothenberg F, et al. Ultrahigh-speed optical coherence tomography imaging and visualization of the embryonic avian heart using a buffered Fourier Domain Mode Locked laser. Optics Express. 2007;15:6251–6267. doi: 10.1364/oe.15.006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis A, Izatt J, Rothenberg F. Quantitative measurement of blood flow dynamics in embryonic vasculature using spectral Doppler velocimetry. Anat Rec (Hoboken) 2009;292:311–319. doi: 10.1002/ar.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manner J, Thrane L, Norozi K, Yelbuz TM. In vivo imaging of the cyclic changes in cross-sectional shape of the ventricular segment of pulsating embryonic chick hearts at stages 14 to 17: A contribution to the understanding of the ontogenesis of cardiac pumping function. Developmental Dynamics. 2009;238:3273–3284. doi: 10.1002/dvdy.22159. [DOI] [PubMed] [Google Scholar]
- 29.Rugonyi S, Shaut C, Liu A, Thornburg K, Wang RK. Changes in wall motion and blood flow in the outflow tract of chick embryonic hearts observed with optical coherence tomography after outflow tract banding and vitelline-vein ligation. Phys Med Biol. 2008;53:5077–5091. doi: 10.1088/0031-9155/53/18/015. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins MW, Patel P, Deng HY, Montano MM, Watanabe M, et al. Phenotyping transgenic embryonic murine hearts using optical coherence tomography. Applied Optics. 2007;46:1776–1781. doi: 10.1364/ao.46.001776. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins MW, Rothenberg F, Roy D, Nikolski VP, Hu Z, et al. 4D embryonic cardiography using gated optical coherence tomography. Optics Express. 2006;14:736–748. doi: 10.1364/opex.14.000736. [DOI] [PubMed] [Google Scholar]
- 32.Luo W, Marks DL, Ralston TS, Boppart SA. Three-dimensional optical coherence tomography of the embryonic murine cardiovascular system. Journal of Biomedical Optics. 2006;11:021014. doi: 10.1117/1.2193465. [DOI] [PubMed] [Google Scholar]
- 33.Jones EAV, Baron MH, Fraser SE, Dickinson ME. Measuring hemodynamic changes during mammalian development. Am J Physiol Heart Circ Physiol. 2004;287:H1561–1569. doi: 10.1152/ajpheart.00081.2004. [DOI] [PubMed] [Google Scholar]
- 34**.Larina IV, Ivers S, Syed S, Dickinson ME, Larin KV. Hemodynamic measurements from individual blood cells in early mammalian embryos with Doppler swept source OCT. Optics Letters. 2009;34:986–988. doi: 10.1364/ol.34.000986. The manuscript demonstrates that OCT can be used to reconstruct spatially and temporally resolved blood flow profiles in deep embryonic vessels based on velocity measurements from individual blood cells. This is especially important for hemodynamic analysis at early stages of circulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Larina IV, Sudheendran N, Ghosn M, Jiang J, Cable A, et al. Live imaging of blood flow in mammalian embryos using Doppler swept source optical coherence tomography. Journal of Biomedical Optics. 2008;13:0605061–0605063. doi: 10.1117/1.3046716. The first study demonstrating live structural imaging and hemodynamic analysis in mouse embryos cultured on the imaging stage with OCT. [DOI] [PubMed] [Google Scholar]
- 36.Larin KV, Larina IV, Liebling M, Dickinson ME. Live Imaging of Early Developmental Processes in Mammalian Embryos with Optical Coherence Tomography. Journal of Innovative Optical Health Sciences. 2009;2:253–259. doi: 10.1142/S1793545809000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buehr M, Meek S, Blair K, Yang J, Ure J, et al. Capture of Authentic Embryonic Stem Cells from Rat Blastocysts. Cell. 2008;135:1287. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z, Milner T, Srinivas S, Wang X, Malekafzali A, et al. Noninvasive imaging of in vivo blood flow velocity using optical Doppler tomography. Optics Letters. 1997;22:1119–1121. doi: 10.1364/ol.22.001119. [DOI] [PubMed] [Google Scholar]
- 40*.Larina IV, Furushima K, Dickinson ME, Behringer RR, Larin KV. Live imaging of rat embryos with Doppler swept-source optical coherence tomography. J Biomed Opt. 2009;14:050506. doi: 10.1117/1.3241044. The first study demonstrating live structural OCT imaging and Doppler OCT cardiodynamic analysis in cultured rat embryos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Sudheendran N, Syed S, Dickinson ME, Larina IV, Larin KV. Speckle variance OCT imaging of the vasculature in live mammalian embryos. Laser Physics Letters. 2011;8:247–252. This study demonstrates visualization of the mouse embryonic vasculature using speckle variance OCT approach. [Google Scholar]
- 42**.Syed SH, Larin KV, Dickinson ME, Larina IV. Optical coherence tomography for high-resolution imaging of mouse development in utero. J Biomed Opt. 2011;16:046004. doi: 10.1117/1.3560300. The manuscript introduces a methodology for live OCT imaging of different organs of developing mouse embryos through the uterine wall. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larina IV, Carbajal EF, Tuchin VV, Dickinson ME, Larin KV. Enhanced OCT imaging of embryonic tissue with optical clearing. Laser Physics Letters. 2008;5:476–480. [Google Scholar]
- 44.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, et al. The knockout mouse project. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Justice MJ, Siracusa LD, Stewart AF. Technical approaches for mouse models of human disease. Dis Model Mech. 2011;4:305–310. doi: 10.1242/dmm.000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hentges KE, Nakamura H, Furuta Y, Yu Y, Thompson DM, et al. Novel lethal mouse mutants produced in balancer chromosome screens. Gene Expr Patterns. 2006;6:653–665. doi: 10.1016/j.modgep.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 47.Justice MJ. Removing the cloak of invisibility: phenotyping the mouse. Dis Model Mech. 2008;1:109–112. doi: 10.1242/dmm.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]