Abstract
Living cells are remarkably complex. To unravel this complexity, living-cell assays have been developed that allow delivery of experimental stimuli and measurement of the resulting cellular responses. High-throughput adaptations of these assays, known as living-cell microarrays, which are based on microtiter plates, high-density spotting, microfabrication, and microfluidics technologies, are being developed for two general applications: (a) to screen large-scale chemical and genomic libraries and (b) to systematically investigate the local cellular microenvironment. These emerging experimental platforms offer exciting opportunities to rapidly identify genetic determinants of disease, to discover modulators of cellular function, and to probe the complex and dynamic relationships between cells and their local environment.
Keywords: high-throughput screening, cell microarrays, reverse transfection, microfabrication, microfluidics, single-cell analysis
“The purpose of a culture on a plate is not the observation of the morphology of the cells, but the study of the dynamic changes undergone by the cells during their life outside of the organism, and the nature of their secretions.”
Carrel & Burrows, 1911 (1)
1. INTRODUCTION
The living cell is a dynamic entity of seemingly infinite complexity. Despite the apparent monotony of being equipped with identical genetic codes, cells within the human body develop into diverse well-differentiated entities exhibiting unique patterns of gene expression, which orchestrate adaptive tissue-specific responses to dynamic environmental cues. Living-cell assays are developed as experimental tools for characterizing relationships between cell states, environmental stimuli (e.g., soluble factors, extracellular matrix, neighboring cells), and resulting cell responses. A secondary goal of these assays is to provide insight into the molecular mechanisms underlying these relationships. There are three general requirements for performing living-cell assays: (a) the ability to maintain cells alive outside of the organism, (b) the ability to control delivery of stimuli or assay inputs, and (c) the ability to measure cellular responses or assay outputs.
The ability to culture cells in vitro dates back to the early 1900s, when Harrison, Carrel, Burrows, and their contemporaries first tamed the techniques (1). In heroic efforts, they overcame challenges of infection in a preantibiotic era and developed methods for performing long-term cell culture that continue to serve as the foundation of modern techniques. Efforts to automate and parallelize the process led to the development of new culture vessels, such as continuous perfusion bioreactors. Today, cell culture methodologies are standardized, and there are a variety of culture-compatible flasks, dishes, and multiwell plates from which to choose. Meanwhile, methods for controlling stimuli and measuring responses remain areas of rapid development and active research. This review describes experimental platforms specifically designed for delivering stimuli to and measuring responses from living cells in high-throughput formats. These platforms, which are relevant to both industry and academia, are emerging as important tools for drug discovery, toxicology, functional genomics, and investigation of cellular microenvironments. Moreover, they are changing how living-cell experiments are performed, and they are broadly impacting the way we organize our understanding of the cell, its molecular machinery, and its behavior within the context of complex tissues.
Historically, biologists organized their understanding of the cell into pathways, limited sets of molecules and genes arranged in roughly linear sequences that described the propagation of signals from the cell surface through the cytoplasm into the nucleus and back. This conceptualization was attractive because it allowed questions to be posed in an experimentally tractable fashion. One investigator could specialize in a specific pathway; acquire the necessary reagents, primers, antibodies, cell lines, and knockout mice; and proceed to explore undescribed molecular details. However, the drawback of such a pathway-oriented approach was that cross talk and feedback between pathways often went unappreciated. Even today, our understanding of dynamic interactions between pathways and the emergent properties of the networks they collectively create are only beginning to be explored.
Recently, a movement termed systems biology emerged to address this issue through quantitative measurement and modeling of interconnected cell signaling networks. At the same time, large genomic libraries were being developed to functionally characterize newly sequenced genomes through -omics-based studies. DNA microarrays were enabling unbiased expression profiling, cDNA libraries were allowing overexpression of genes, and RNAi libraries were making genome-wide knockdown experiments practical. In the drug development arena, synthetic and natural biochemical libraries were being assembled for high-throughput screening of putative therapeutic agents. In areas of basic cell biology, scientists and engineers were joining to explore tissue complexity by building microscale tools and characterizing spatial and temporal properties of cellular responses. Together, the confluence of these events fueled the development of new high-throughput experimental platforms for studying stimulus-response relationships and spatiotemporal dynamics in living cells.
This review describes high-throughput experimental platforms termed living-cell microarrays. We group the platforms into two categories, those used for screening large chemical and genomic libraries and those used to dissect the cellular microenvironment. In the first category, we discuss living-cell microarrays based on microtiter plates and spotting technologies that have been optimized for screening applications. In the second, we discuss microfabrication and microfluidic living-cell arrays optimized to control various aspects of the local cell environment.
2. ATTRIBUTES OF A LIVE-CELL ASSAY
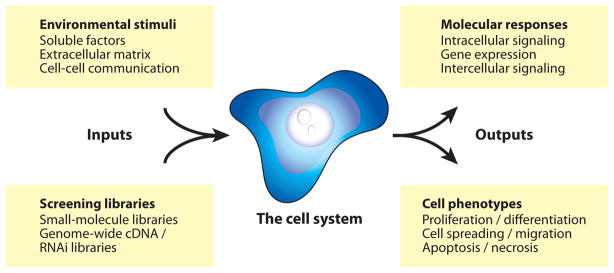
Living-cell assays can be divided into three fundamental components: (a) the inputs, (b) the outputs, and (c) the cells under investigation, as illustrated in Figure 1. High-throughput cell assay platforms are designed to systematically vary one or more of these components between elements of the array while holding the remaining assay variables constant. Therefore, before embarking on a discussion of the living-cell microarray technologies themselves, it is first useful to review the assay variables they aim to control.
Figure 1.
Attributes of a cell assay.
2.1. Inputs: Controlling the Microenvironment
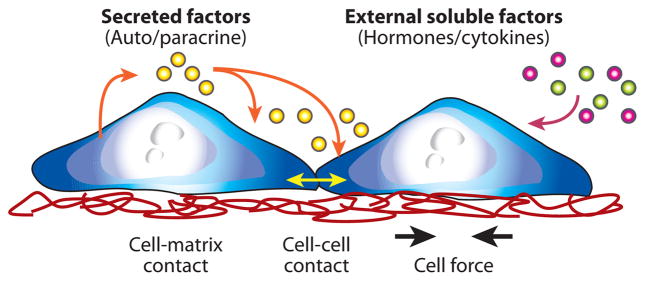
The inputs of a cell assay span all aspects of the cellular microenvironment—soluble, insoluble, and cellular—as illustrated in Figure 2. The most commonly controlled variables in conventional cell assays are those in the soluble environment. Soluble inputs consist of standardized culture medium, metabolic substrates, vitamins, antibiotics, and other unspecified additives derived from animal serum. They also include experimental stimuli, such as cytokines, growth factors, hormones, and putative therapeutics, which are added at various doses, durations, and combinations to study their effects on cell responses (2). Another important group of assay inputs are insoluble factors, such as extracellular matrix (ECM). The ECM provides mechanical and biochemical cues that modulate the shape, migration patterns, and overall phenotype of associated cells (3). The local arrangement of neighboring cells also plays an important role in modulating cell responses to stimuli. Intercellular communication between cells can occur via secreted factors, which communicate signals at a distance in a paracrine fashion (4) or via contact-mediated signals, which can be transferred through gap junctions (5), transduced by cadherins (6) or transmitted by mechanical forces (7). Together, these diverse environmental inputs are integrated by the cell to dynamically determine its behavior.
Figure 2.
The cell microenvironment.
To a first approximation, the cellular environment might be assumed to be simple. The soluble environment might appear static and homogeneous, the extracellular matrix might seem spatially uniform, and cells might be taken to be uniform in orientation and density. However, upon closer inspection, the local vicinity of adherent cells is found to be highly variable and remarkably complex. Concentrations of factors are continuously evolving in time and space owing to uptake and secretion of metabolites and proteins by cells and their neighbors (8). Extracellular matrix is continually synthesized, deposited, and degraded to modulate cell migration, tissue organization, and repair. Cells are continuously varying the direction, degree, and arrangement of their contacts with each other, creating dynamic functional units that coordinate adaptive responses to external stimuli (9, 10). It is on this length scale of <1 mm that environmental changes in culture best simulate those of dense tissues, and it is the desire to understand these additional layers of complexity that motivates the engineering of novel experimental culture systems with increasing degrees of microscale control.
2.2. Outputs: Measuring Cell Responses
Cell responses can be measured and quantified in many ways. They can be monitored with molecular specificity using Western blotting or enzyme-linked immunosorbant assays (ELISAs), which are commonly quantified in microtiter plates using bulk fluorescence, luminescence, or absorbance as a readout. Unfortunately, these methods require cell lysis and therefore cannot be used to follow cell responses across space and time. Instead, dynamic changes must be inferred by assembling average responses measured at individual time points from different cell populations. Microscopy can be used to gain spatial resolution either at fixed time points using fixed and stained cells or by nondestructively monitoring dynamic responses of living cells. In this manner, morphological features can be quantified and analyzed in real time to study phenotypes such as proliferation, migration, or differentiation.
The development of live-cell imaging tools, such as the genetically encoded green fluorescent protein (GFP) and its variants, represented a major advance in the ability to study intracellular molecular events across space and time (11–14). A wide range of fluorescent proteins have been engineered into plasmids and viral vectors and delivered to cells by transfection and infection, respectively, to visualize processes, including protein-protein interactions (15), activation of kinases (16), nuclear translocation (17), and the transcriptional activity of an engineered promoter (18). Enhanced variants, such as enhanced green fluorescent protein (EGFP), have been optimized for increased intensity (19), and a wide range of spectral variants (20) have broadened the utility of this molecular tool. Although GFP is commonly used for its spatial resolution, dynamic measurements initially presented challenges because the stable protein decays at a slow and often unpredictable rate. Therefore, to improve dynamic monitoring and minimize background fluorescence from leaky expression, a destabilized GFP variant was developed by adding a peptide sequence rich in proline, glutamic acid, serine, and threonine to the C terminus of EGFP that leads to proteasome degradation and a characteristic half-life (21). More recently, a gradual fluorescence emission shift was also introduced as a molecular timer to study dynamic events (22).
Live-cell responses are ideally measured by image-based methods collectively known as high-content or high-throughput microscopy. In this approach, cells are imaged by a microscope with a programmable motorized stage, and images are subsequently quantified by automated image analysis software. This method has been applied to a wide range of systems biology and drug discovery studies as recently reviewed (23–26). Specialized routines have now been developed based on free and commercial software packages, and recently a modular open-source Matlab-based tool kit called CellProfiler was released for analyzing and quantifying results from cell microarray experiments (27, 28). Such software typically performs segmentation, object identification, feature extraction, and quantification. These measurements are relatively straightforward to make when applied to images of fixed cells with stained nuclei (29); however, quantification of live cells based on time-lapse imaging requires additional steps to correct for culture medium background fluorescence, to track objects and account for cell movement, and to identify individual cells without using a nuclear stain for reference. If these challenges are overcome, the resulting measurements offer a unique window into the rich spatial and temporal complexity of the cell and its local microenvironment. These high dimensionality quantitative data sets can then be analyzed by data-driven methods, such as clustering, principle component analysis, and partial least-squares regression (30) or used for parameter fitting in physicochemical cell-signaling models (31).
2.3. The Cell System: Differentiation, Genetics, and Population Heterogeneity
The choice of cell type is an important decision in any live-cell assay. Cells can be chosen from a range of commercial cell lines, harvested as primary cells from animals, or isolated from surgically resected human tissues or tumors. They can also be isolated from genetically modified animals, such as knockout mice, engineered to stably overexpress or knock down a gene of interest, or designed to express fluorescent fusion proteins or transcriptional reporters to monitor molecular events across space and time. Once a cell type is chosen, it is also useful to monitor or control its initial state. One need only glance at a phase-contrast image of cells to appreciate the wide range of individual cell shapes and the varying degrees of contact with neighboring cells. Adding further complexity is the fact that cell division is highly asynchronous, such that cells can be found at every stage of the cell cycle at any moment in time, unless specific synchronization strategies are employed (32). As a result of these varied initial states, cells can exhibit dramatic differences in their response to stimuli (33). It is therefore important to consider both the cell type and the initial state when performing a cell assay.
One must also choose whether to study cells as a population or as single cells. Conventional cell assays measure average responses of large cell populations. An implicit assumption of this approach is that the average response is representative of a typical cell within the population. In many cases, however, this is an oversimplification. One of the most striking examples is encountered when examining cells that exhibit all-or-none responses. Because activation occurs stochastically, such cells become activated asynchronously, and the average population response increases gradually, whereas at the single-cell level, responses are abrupt but exhibit a distribution of activation times. Importantly, if one examines the distribution of single cells midway through the response, no individual cell will exhibit the response level predicted by the average population. Instead, there will be only fully activated and nonactivated cells (34). Accurately characterizing single-cell responses as a function of space and time can play an important role in understanding the variations and emergent properties of cell populations. To this end, efforts to profile cell responses at the single-cell level are growing increasingly common (35, 36).
3. MICROTITER PLATE TECHNOLOGIES
The industry standard for high-throughput living-cell assays is the microtiter plate (96- 384-and 1536-well), a miniaturized and parallelized version of a conventional tissue culture dish. Initial challenges related to evaporation, increased surface-area-to-volume ratios, small-volume fluid transfers, plate handling, and large-scale imaging have been largely overcome; today, a wide range of laboratory automation equipment is available, including plate-handling robotics, high-precision fluid dispensers, nanoliter-scale transfer pipettes, multiwell plate scanners, and automated microscopes (summarized in a recent review in Reference 37). The microtiter plate platform was initially developed for drug discovery and toxicology; however, recently, it has become accessible to academic labs for basic science investigations through the creation of high-throughput screening (HTS) core facilities with access to a growing number of chemical and genomic libraries.
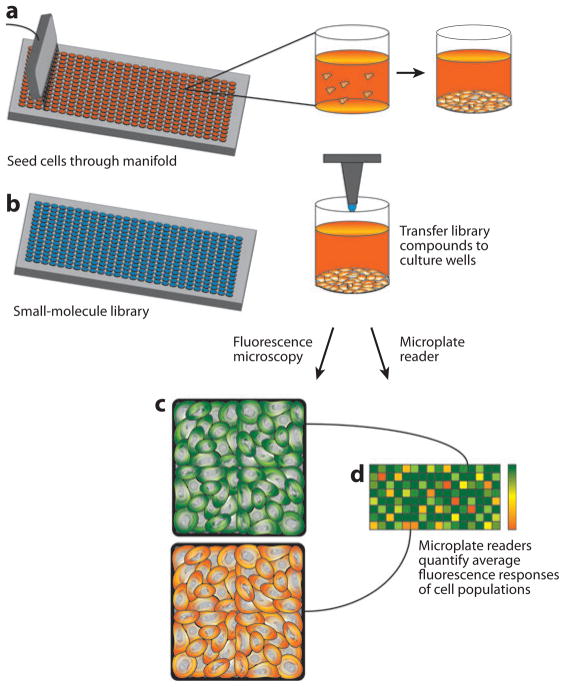
The typical workflow for a high-throughput cell-based chemical screening assay is shown in Figure 3. Cells are first delivered from suspension through fluidic manifolds into the wells of a microtiter plate using automated fluidic dispensers. They are allowed to attach and spread, and nanoliter volumes of concentrated experimental compounds are transferred from wells of the library storage plate into the cell-seeded wells of microtiter plates. Cells are incubated with the compounds for a specified duration, additional stimuli or assay-specific reagents are added and removed as needed, and the experiment is quantified using either a fluorescence or luminescence plate reader or using high-throughput microscopy with image-based quantification. Measurements based on imaging, although more information dense, are inherently slower because they require time for stage movement, autofocusing, and long exposures to detect low-intensity signals, such as an expressed intracellular fluorescent protein. Image-based measurements also require significant storage resources. For example, at 1 MB per image and multiple images per well, it has been estimated that a 200,000 compound screen would require 1 TB of data storage space. Such image-based approaches were recently reviewed in the context of chemical screening (38).
Figure 3.
High-throughput screening. (a) Cells are seeded through a manifold into wells of a high-density microtiter plate and are allowed to sediment and attach. (b) Concentrated compounds from the small-molecule library are transferred from their storage in library microtiter plates to the wells of the cell-seeded microtiter plates. Cells in each well are then exposed to a well-calibrated stimulus, (c) responses are captured by fluorescence microscopy, quantified by automated image analysis, and (d) displayed as a heatmap to identify potential hits from the molecular library.
Once an assay has been developed, repeatability is parameterized by the Z factor, a quality measure for an assay, defined as
| (1) |
where σp,n and μp,n represent the standard deviation and mean, respectively, of positive and negative control samples (p and n subscripts). Typically, pilot studies are performed to estimate the Z factor for a given assay, allowing for further optimization prior to initiating a large-scale high-throughput screen. A Z factor of 1.0 indicates an ideal assay with large dynamic range and small standard deviations; 0.5–1.0 is considered excellent, 0–0.5 is marginal, and less than 0 indicates that positive and negative controls are overlapping (39). Once an assay has been selected and optimized, a screen is performed, and so-called hits are identified that are commonly repeated for verification and then investigated by lower-throughput assays for validation and mechanistic exploration. Methods for developing a good high-throughput small-molecule screen and reporting the resulting data were recently reviewed (40, 41).
The strengths of the microtiter plate platform are its standardized dimensions and its compatibility with existing laboratory automation equipment. The platform is ideal for screening chemical libraries that have been assembled and stored in multiwell formats, and at present, there is no better format for studying arrays of living-cell assays in which each element is to be exposed to a different soluble stimulus. However, although microtiter plate technologies represent the industry standard for high-throughput screening, they also place several important limitations on the attributes of a living-cell assay that can be controlled. For example, time-varying stimuli, such as transient chemical exposures used in pulse-chase experiments, are difficult to control because they require complete removal of the soluble stimulus, followed by multiple washing steps. Similarly, it is difficult to deliver spatially heterogeneous stimuli, such as gradients of chemotactic factors, and to control the delivery of one stimulus with respect to another in time or space. Initial cell position, shape, and local density are also difficult to control, which limits investigation of intercellular communication and the emergent properties of cell communities. Microtiter plate experiments are also not well suited for studying phenomena that occur on short timescales because stimulation and measurement are performed using different equipment. These limitations, combined with the need for additional scaling, have motivated the development of several next-generation living-cell arrays.
4. HIGH-DENSITY SPOTTING TECHNOLOGY
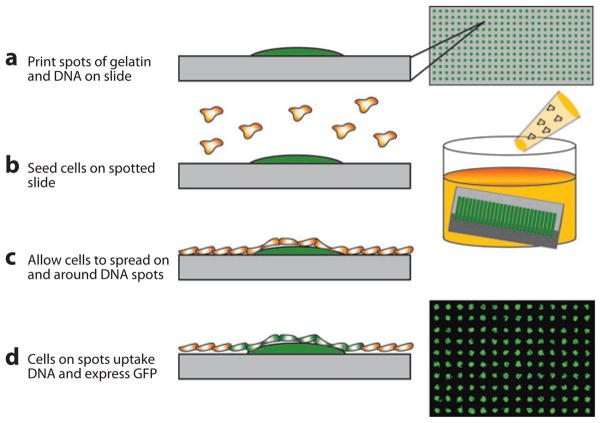
In an effort to improve upon the miniaturization and parallelization provided by multiwell plates, investigators took inspiration from printing strategies used to create DNA and protein microarrays and applied them to living-cell assays. This platform, which is illustrated in Figure 4, was termed cell microarrays (42). This technology has been extensively reviewed (43–45) and involves culturing cells in confluent monolayers over high-density arrays of nanoliter droplets preprinted on glass slides. Compared with microtiter plates, the approach offers reduced assay volume, increased assay density and throughput, compatibility with long-term storage, and eliminates the need for high-throughput fluid transfer equipment once the spotted array is created.
Figure 4.
Spotted cell microarray technology. (a) DNA is mixed with gelatin and spotted as a high-density array on a glass slide, and it is dried and stored until needed for an experiment. (b) To perform an experiment, the array is rehydrated, and cells are seeded uniformly across the array. (c) Cells attach and spread across the array, and (d) those cells residing above DNA spots take up the underlying nucleotides and become transfected in a spatially localized fashion (42).
4.1. Cell Microarray Construction and Experimentation
Cell microarrays are created by mixing genomic material, such as plasmid DNA, with gelatin solutions or by directly spotting solutions of virus (42, 46, 47). These solutions are transferred from their storage in 96- or 384-well plates to a glass microscope slide using contact printing, after which the resulting high-density array of nanoliter spots can be dried and stored for months. To perform a cell microarray experiment, the spotted array is rehydrated and a single cell type is seeded uniformly across the entire array to create a confluent monolayer of cells. Remarkably, only those cells seeded above the printed spots are influenced by the underlying genomic material, and to a first approximation, there is negligible cross talk between spots. As a result, each element of the array is treated as an independent experiment, making high-density studies of large genomic and biochemical libraries practical. Typical spot sizes and spacings are hundreds of micrometers, allowing thousands of spots to be printed per slide. Importantly, results from cell microarray experiments, in contrast to microtiter plate assays, must always be quantified using automated microscopy and image-based analysis because the experiments are all performed on the same substrate in the same culture solution.
4.2. Genomic Microarrays
Gain- and loss-of-function experiments have revolutionized functional characterization of the genome. Although these experiments were initially performed one gene at a time, the recent construction of plasmid and viral vector libraries has enabled dramatic scaling of the approach. Cell microarrays represent an ideal experimental platform for miniaturizing and parallelizing genome-wide gain- and loss-of-function studies on living cells. To make this possible, a method called reverse transfection was developed, which allowed cells to be added to prespotted vectors rather than adding the vectors to preseeded cells as in conventional forward transfection. In this way, a different gene can be overexpressed in each element of the array, and large genomes can be screened in a small number of experiments. In a similar fashion, RNA interference (RNAi) technologies such as siRNA and shRNA, which promote gene-specific knockdown after transfection or infection, can also be studied in a massively parallel fashion. For background on RNAi methods, see Reference 48. The high density of the cell microarray platform coupled with a growing number of DNA and RNAi libraries is enabling remarkably efficient functional characterization of the genome (45, 49).
Since the first description of cell microarrays, they have been applied to a broad range of applications in basic and applied biology. For example, overexpression studies were performed to study cAMP (50), to monitor serum responses using a GFP reporter (51), and to study intracellular localization of a library of genetic fusion proteins (52). RNAi microarrays were first described and validated by targeting genes involved in cytokinesis and proteolysis (53). Since then, reverse-transfected gene-silencing cell microarrays have been used for a range of applications, including the identification of host factors required for intracellular bacterial infection (54), identification of genes associated with mycobacterial infection in Drosophila (55), and identification of genes associated with cell proliferation and morphology in Drosophila (56). It was also used to study Caenorhabditis elegans, a live intact organism susceptible to ingested RNAi, to identify genes required for protection from genomic mutations (57) and gene inactivations that modulated fat metabolism (58). Applications of cell microarrays in genomic studies have been the topic of several recent reviews (59, 60).
4.3. Small-Molecule Arrays
The application of cell microarrays to small-molecule screening presents unique challenges compared with genomic libraries. First, small molecules have significantly lower molecular weight and therefore readily diffuse. This was addressed by embedding small molecules into biodegradable poly-(D),(L)-lactide/glycolide (PLGA) flat-disc particles of nanoliter volume (61). Optimal conditions were identified to avoid cell toxicity and retain compatibility with existing libraries, which are commonly dissolved in dimethylsulfoxide (DMSO). Controlled-release particles were then coated with fibronectin to facilitate cell adhesion onto the array. In one demonstration of this approach, molecules were studied for their effect on cell viability and proliferation. Importantly, although spot sizes were only 200 μm in diameter, cytotoxic effects were measurable several hundred micrometers from the spot, suggesting that the entrapped molecules were released to diffuse. Furthermore, the diffusivity and release kinetics will likely vary between molecules of a library. Together, these points highlight the need to design arrays with sufficient replicate spots and interspot spacing to not only provide a statistically significant number of cells for analysis but also to characterize interspot cross talk. The desire to achieve high-density experimentation must therefore be balanced by the need to maintain approximate experiment independence.
4.4. Microenvironment Arrays
Spotting technologies are powerful ways of studying the complexity of extracellular matrix. In one study, investigators spotted different combinations of extracellular matrix molecules (62) and studied the functional behavior of cells seeded on the array. In this way, they were able to rank ECM combinations based on their ability to induce albumin production in primary hepatocytes, and they were able to examine the influence of the ECM combinations on the differentiation of embryonic stem cells.
This approach was recently expanded on by printing additional aspects of the microenvironment and studying the effects on cell fate determination of a bipotential neural stem cell population (63). In these studies, two sets of four identical spots were placed at different locations to partially compensate for potential cross talk between cells on different microenvironments. Each spot was 400 μm separated by 650 μm of space, such that a few hundred cells were contained in each spot, and nonidentical neighboring microenvironments were separated by a distance of 1.45 mm. In summary, spotted arrays allow screening of cellular responses to various genes, soluble small molecules, and insoluble components of ECM.
5. MICROFABRICATED CELL ARRAYS
The cellular microenvironment consists of a strikingly diverse collection of soluble factors, extracellular matrix cues, and cellular networks (64, 65). These have proven difficult to experimentally control using conventional techniques, in large part due to the wide range of length- and timescales involved. Microfabrication, a technology originally developed for creating integrated circuits in the microelectronics industry, offers precise reproducible spatial control and can be adapted to achieve temporal control as well. As a result, microfabrication is ideal for controlling cellular microenvironments in living-cell assays. Moreover, because the methods are inherently scalable and lend themselves to screening geometrical parameters, they are ideally suited for constructing high-throughput living-cell arrays. This section briefly reviews the general methodologies used to create microfabricated cell-based devices and provides specific examples of microfabricated arrays that utilize chemical, mechanical, and electromagnetic cell patterning.
5.1. Fabrication Technology
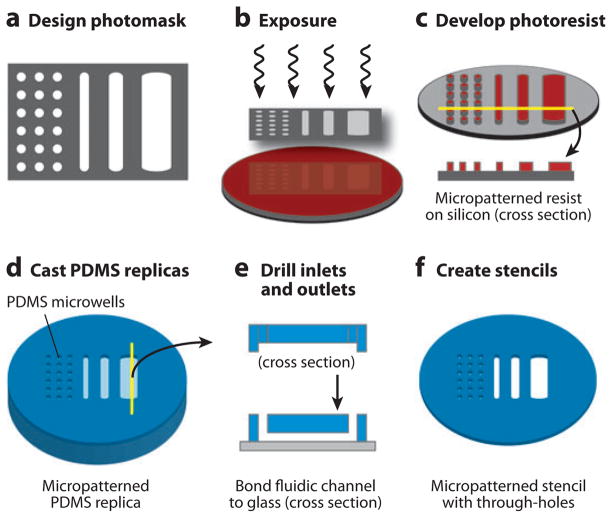
The most commonly used method for constructing microfabricated cell-based devices is soft lithography (66). In this approach, a photomask is drawn using a computer-aided design tool and printed on a high-resolution printer with minimum feature sizes as small as 10 μm. Photolithography is then performed by spin-coating a polished silicon wafer with a thin film of photosensitive material, such as SU-8 epoxy, exposing the film through the printed photomask and developing it to create permanent raised microstructured patterns on the silicon substrate. Using this method, microstructure heights are determined by thin-film thickness (typically 1 μm–1000 μm), and the remaining dimensions are determined by the photomask. Once the silicon master mold is fabricated, elastomeric replicas can be repeatedly formed by mixing polydimethylsiloxane (PDMS) prepolymer (resin plus a curing agent), casting or spin-coating it on the microstructured master, thermally curing the polymer, and peeling the resulting flexible and transparent films. Because the uncured prepolymer flows and conforms to the underlying microstructures, patterns are transferred with high fidelity from silicon masters to the polymer replicas. The resulting microstructured PDMS films can then be used directly as stamps, open microwells, or three-walled microchannels. When PDMS stencils with full thickness through-holes are required, the prepolymer can be spin-coated onto the microstructures and a rigid nonadhesive flat plate used to apply downward force during curing. Finally, permanent closed-channel microfluidic networks can be constructed by irreversibly bonding the three-walled PDMS microchannel networks onto glass slides using an oxygen plasma surface treatment. These methods are summarized pictorially in Figure 5. Examples of microfabricated cell arrays based on mechanical, chemical, and electromagnetic patterning are discussed below and are shown in Figure 6.
Figure 5.
Microfabrication and soft lithography. (a) Photomasks are drawn using a computer-aided design tool and are printed by a high-resolution printer on mylar transparency films. (b) Photoresist-coated silicon wafers are exposed through the photomask and (c) developed to translate the photomask design to the silicon surface. (d) The silicon then serves as a master mold for casting transparent polydimethylsiloxane (PDMS) polymer replicas, which can be (e) drilled to create inlets and outlets and bonded to a glass substrate to create closed microfluidic devices or (f) used to create stencils with micropatterned through-holes.
Figure 6.
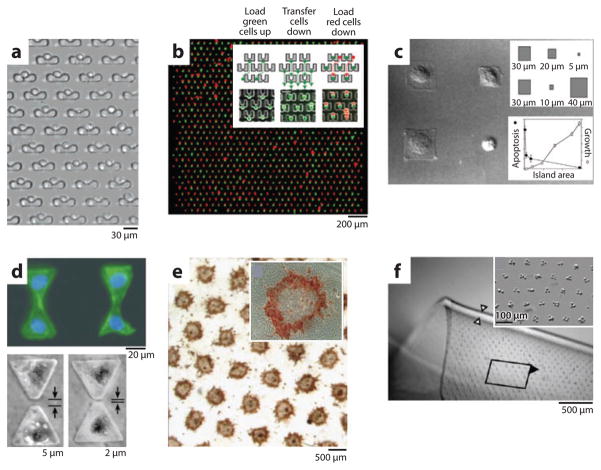
Examples of microfabricated cell arrays. (a) An array of mechanical cell traps captures single cells and allows dynamic exposure to compounds in solution (72). (b) An array of mechanical traps captures individual cells from two different populations (one green, one red) and places them in contact for investigation of cell fusion. Inset illustrates cell pairing method (73). (c) Chemical surface patterning is used to control cell location and spread area. Cell area (inset top) was found to correlate with cell phenotype (apotosis and growth) (inset bottom) (116). (d) Surface patterning is used to create pairs of cells in contact or with controlled gaps to dissect contact-dependent and -independent behavior (75). (e) The hepatocyte-fibroblast contact area is systematically varied while holding the cell ratio constant by patterning hepatocyte islands of different diameters and densities surrounded by fibroblasts. Albumin synthesis (red) was preferentially induced in hepatocytes at island edges where they were in contact with adjacent fibroblasts. Inset shows a single island (80). (f) Fibroblasts are patterned by dielectrophoresis and embedded in a flexible hydrogel film. Inset shows close-up of the embedded cell array elements (89).
5.2. Mechanical Patterning
Microfabricated cell arrays are often created directly, using the PDMS microstructures (stencils, fluidic channels, or microwell traps) as mechanical barriers to capture cells. Microwell geometries have been optimized for single cells, allowing large numbers of individual cells to be stimulated in a massively parallel fashion (67). Furthermore, because the cells are regularly spaced and limited to the area of the predefined well, they can be readily quantified by microscopy and automated image analysis software. This simple yet powerful technique has been used with numerous cell lines, primary cells, and stem cells in a variety of applications. For example, in one study, the platform was used to relate the initial state of a cell, with its survival following cryopreservation (67a). In this work, primary hepatocytes were isolated, labeled with a fluorescent dye that correlated with the mitochondrial membrane potential, loaded into the wells of the single-cell array, and measured with single-cell resolution. The entire array was then cryopreserved, thawed, and because cells remained in their respective wells, survival was assessed and correlated with initial mitochondrial membrane potential. Single-cell studies such as these are critical for understanding treatments in which only a fraction of cells survive, as they offer the possibility of using initial cell state to uncover why a seemingly homogeneous population of cells gives rise to two distinct responses to a single stimulus (33).
Microwell cell arrays have also been used to allow selection of cells based on functional phenotypes. For example, large 234,000-element single-cell arrays were used to identify antigen-specific B cells, which were then retrieved for subsequent analysis by reverse transcriptase polymerase chain reaction (RT-PCR) (68). In another study, single hematopoietic stem cells (HSCs) were sparsely seeded in an array of large 300 μm × 400 μm wells, such that there was initially only one cell seeded per well. Their proliferation into clonal populations was then followed using time-lapse video microscopy, and the timing of proliferations was correlated with their subsequent ability to reconstitute the marrow after retrieval and animal transplantation (69). The drawback of using large wells to create clonal populations is that it relies on probability to achieve single-cell seeding. This method was improved upon in a more recent study by a group aiming to grow clonal islands of mouse embryonic stem cells (mESCs). They created a single-cell array with widely spaced microwell elements, loaded the wells with mESCs, and inverted them prior to cell attachment, such that cells sedimented to an underlying adhesive culture substrate, a method termed Bio Flip Chip (BFC). This technique not only seeded a well-spaced single-cell array, but it did so without placing mechanical or chemical barriers on the culture surface that would impede subsequent proliferation. In this way, they created high-density arrays of clonal stem cell colonies. By controlling the spacing between islands, they were then able to decouple secreted and contact-dependent growth signals and gain insight into their relative contributions to mESC colony formation (70).
Mechanical barriers have also been used to capture single cells from flowing cell suspensions in closed fluidic devices. In this approach, each barrier was designed with a small fluidic leak that allowed incoming single cells into the traps but prevented multiple cells from being captured. Once loaded with cells, the arrays were used to characterize the functional heterogeneity of intracellular enzyme activity across a seemingly homogeneous population of individual cells (71). Using this device, researchers demonstrated that the single cells can sediment, attach, and multiply in the array for long-term experimentation (72). More recently, an advanced version of this approach was described that captures two different cell types flowing in opposite directions to reliably create pairs of two different cell types for studying cell fusion (73).
5.3. Chemical Patterning
Microfabrication can be combined with surface-modification strategies to indirectly pattern cells by controlling the spatial localization of biochemical cues that affect the adherence of cells. In this method, microstructured films are coated with biomolecules and used to stamp or transfer them to the culture surface wherever raised microscale features are present. Depending on whether the printed molecules are more adhesive (e.g., fibronectin or collagen) or less adhesive (e.g., polyethylene glycol or albumin) than the surrounding culture substrate, the cells will preferentially localize onto or avoid the stamped regions, respectively. The transfer of surface molecules can be achieved by physisorption, but more commonly, covalent bonding strategies based on self-assembled monolayer chemistry are used to achieve high-density surface modification. These methods were recently reviewed in detail (74). In this way, cells can be patterned individually or as colonies of controlled sizes and shapes to systematically investigate the functional significance of cell shape, cell density, and intercellular contact, as demonstrated by the following examples.
One of the earliest applications of this approach used covalent attachment of fibronectin to decouple the effects of cell adhesive area from cell shape. Patterns were created to maintain a constant total adhesive area while the cell’s spatial distribution was varied to promote increasingly larger spread cell areas. When phenotypes were assessed, results revealed that small islands of fibronectin were associated with apoptosis, and large islands were associated with proliferation. Moreover, this biological effect appeared to be due to cell shape, because increased proliferation correlated with increasing spread area, even when the total adhesive area was maintained constant. Control of cell shape was then extended to pairs of cells, which were arrayed either in contact or separated by a small 20-μm gap to systematically dissect the effects of contact-dependent and contact-independent cell communication (75). Evidence from these studies again supported a role for mechanical force and cell spreading as a proliferative signal and further emphasized the importance of the local cellular environment in modulating cell phenotypes. Similar methods were later used to study the cell shape and tension in mesenchymal stem cells (MSCs) (76). Results from this work suggested that MSCs on small adhesive islands tended toward adipocyte differentiation, whereas cells spread on larger islands tended toward an osteoblastic lineage. This was later extended to multicellular MSC colonies, revealing that the pattern of differentiation correlated with gradients in mechanical stress, such that areas of low stress were systematically associated with adipogenesis, and areas of high stress were associated with osteogenesis (77).
Cell patterning has also played an important role in studying intercellular communication between different cell types. For example, it had long been appreciated that interactions between primary rat hepatocytes and nonparenchymal cells, such as endothelial cells or fibroblasts, improved liver-specific functions in vitro. However, it was not known whether this was the result of the ratio of the different cell types or of the amount of contact area between them. Therefore, photolithographic surface patterning was used to create micropatterned collagen islands that controlled the placement of primary hepatocytes and nonparenchymal fibroblasts, allowing the effects of cell ratio and cell contact area to be systematically decoupled. Results from these studies provided compelling evidence that the coculture phenomenon was primarily determined by the amount of heterotypic interactions rather than the overall cell ratio (78–80), again demonstrating that the spatial arrangement of cells is an important determinant of phenotype. Since these early studies, additional microfabrication techniques have been employed to investigate the underlying molecular details of the coculture effect; however, it remains a complex spatiotemporal phenomenon that appears to depend strongly on cadherin signaling (81, 82). Knowledge gained from these basic studies was recently employed to optimize liver-specific function of primary hepatocytes using microfabricated stencils that are compatible with conventional multiwell dishes and pipetting equipment. In doing so, they created a highly practical and functional cell culture model that exhibited characteristic patterns of gene expression, phase I/II metabolism, canalicular transport, secretion of liver-specific products, and susceptibility to hepatotoxins (83), making it a promising platform for high-throughput metabolic and toxicologic screening studies.
5.4. Electromagnetic Patterning
Electromagnetic forces can be used to manipulate cells based on their charge (electrophoresis) or polarizability (dielectrophoresis), as recently reviewed (84). These forces exhibit particularly advantageous scaling laws and provide powerful tools for localizing cells in space and constructing living-cell arrays. Typically, electrodes are fabricated by depositing a thin film of metal and photolithographically patterning it with a wet chemical etch. A more detailed discussion of the underlying physics and the rationale for various electrode designs was recently reviewed in Reference 85. Briefly, by varying electrode designs, investigators have created single-cell and multicellular arrays for long- and short-term applications. Electrodes can be activated transiently to trap cells in suspension and release them individually on demand, as demonstrated by a closed-volume fluidic device that created a linear microfluidic cell-sorting cytometer array. By balancing viscous, gravitational, and dielectrophoretic forces, a linear array of cells was trapped, functionally characterized, and released as single cells on demand (86). This method, which relied on extruded electrodes, was later adapted to planar electrodes, generalized for use in physiologic salt solutions, and expanded to create two-dimensional cell-sorting arrays with a combination of floor and ceiling electrodes (87). Dieletrophoretic traps have also been used to position cells for long-term experimentation by trapping them in space and allowing them to sediment, attach, and spread on the underlying surface. This was used to create a regular array of clonal stem cell colonies that would be extraordinarily tedious to create by conventional techniques. Another long-term arraying strategy involved using dielectrophoresis to control cell position in three dimensions and subsequently encasing the cells in a photopolymerizable hydrogel. This cell-laden flexible film could then be manually manipulated for tissue engineering applications (88, 89). To demonstrate this approach, dielectrophoresis was used to arrange clusters of chondracytes and decouple the effects of cell density from local cell organization (88).
6. MICROFLUIDIC LIVING-CELL ARRAYS
The cell arrays described thus far utilize open-volume culture formats with free liquid-air interfaces. As a result, it is difficult to experimentally control the soluble environment in space and time. For example, providing a transient stimulus would require manually removing fluid, performing multiple washes, and providing new medium to each well. In contrast, closed-volume culture systems, such as perfusion bioreactors, allow stimuli to be replaced in seconds and thus enable real-time spatial and temporal control of soluble cellular microenvironments. Microfluidics is a technology aimed at creating microscale closed-volume networks of channels that has emerged as a promising technology for miniaturizing and parallelizing fluid-addressable cultures and creating continuous-flow living-cell microarrays.
6.1. Design and Construction
Microscale devices are unique in that they are often dominated by different physical phenomena than their macroscale counterparts (90, 91). For example, on the microscale, flow is dominated by viscous forces and is therefore laminar. In addition, solute transport, which ultimately determines the kinetics of solution mixing, is dominated by diffusion. Therefore, fluid flow rates, pressures, and solute concentrations can be predicted from microchannel design, offering unique opportunities to precisely control the spatiotemporal attributes of the soluble cell microenvironment.
As with other microfabricated cell-based devices, the most commonly used material for constructing cell-based microfluidics is elastomeric PDMS. Device transparency, mechanical elasticity, flexibility of design, speed of fabrication, and the ability to chemically modify channel surfaces are among the many advantages offered by this material. Some concerns have been raised about evaporative losses that might concentrate culture medium, but currently their impact on live-cell assays remains uncertain (92–94). Most microfluidic devices are controlled using external pumps and tubing; however, recent advances have enabled design of integrated valve and pumping strategies (95). In addition, efforts have been made to adapt microfluidic devices to achieve compatibility with conventional microtiter plate laboratory automation equipment (96, 97).
The functionality of microfluidic devices is closely linked to the types of integrated components that can be fabricated. In this regard, the development of integrated microvalves enabled dramatic improvements in device design, functionality, and complexity (95, 98). By bonding two fluidic networks together, separated by a thin PDMS membrane, large numbers of monolithic pressure-controlled microvalves could be designed at arbitrary locations and constructed in parallel (98). This approach offered tremendous advantages over previously described valve designs because there is no leakage and the fabrication is remarkably simple. Since its introduction, the complexity of valve-controlled microfluidic circuits has progressed dramatically, leading to a host of new applications.
6.2. Cell-Based Microfluidics
Microfluidic cell-based assays are still in their infancy, but have been used to perform a number of unique studies with living cells that would be extraordinarily difficult using conventional techniques. For example, methods were developed to create and dynamically control stable soluble gradients of chemotactic factors to study the molecular mechanisms underlying recruitment of neutrophils to areas of infection, inflammation, and injury (99, 100). Soluble factors were selectively delivered to subcellular cell structures, allowing investigation of intracellular signal propagation following localized extracellular stimulation (101, 102). Local concentrations of oxygen were controlled to allow development of physiologic oxygen gradients in the direction of flow, thereby recreating physiological patterns that led to hepatic zonation in vivo (103). In another study, spatial gradients of oxygen tension were controlled to investigate oxygen-induced migration and olfactory sensing in C. elegans (104, 105). Microfluidic devices have been used to control the location and diffusion paths of secreted molecules to probe intercellular communication in neuroregeneration (106). Many of these device concepts remain at the proof-of-principle stage; however, they will be most powerful when realized on a large scale, at which point they can serve as tools for controlling the microenvironment in high-throughput living-cell arrays.
6.3. Examples of Microfluidic Cell Arrays
Several linear microfluidic cell arrays have already been created to systematically vary one parameter across a row of elements containing small adherent cell populations (107). For example, in one device, diffusive mixing was performed in an upstream microfluidic network to generate several doses of a cytokine stimulus. This was delivered to downstream preseeded NFκB-GFP transcriptional reporter cells while time-lapse fluorescence microscopy was used to measure the resulting gene-expression dynamics. In another study, microfluidic channels with different resistances were used to deliver a logarithmic distribution of different flow rates to an array of embryonic stem cells. The various soluble environments led to dramatic differences in cell morphology and colony growth kinetics (108). Microfluidics have also been used to vary the temporal aspects of the soluble environment. A novel design strategy termed flow-encoded switching was used to control delivery of many different dynamic stimulus patterns to downstream GFP reporter cells using a comparatively small number of inputs. Patterns included variable pulse widths, pulse train lengths, and pulse frequencies (109). Importantly, highly dynamic parallel stimulus regimens such as these would be impossible to control using conventional pipetting and multiple rinses.
Several two-dimensional microfluidic living-cell arrays have also been created. One study used mechanical barriers with controlled leaks to trap populations of cells. Once seeded in rows, cells were stimulated with different concentrations of serum (110) in the orthogonal direction. To improve control of cell seeding and avoid cross talk during exposure to soluble stimuli, a valve-controlled array was created that allowed independent isolation of rows and columns for cell seeding and stimulation. Using this approach, dynamic gene expression profiling was performed by seeding a library of stable monoclonal GFP reporter cell lines in rows and stimulating them with a panel of inflammatory stimuli, inhibitors, and modulators in columns (111). This proof-of-principle experiment measured dynamic responses of eight cell lines to eight stimuli with three replicates across 24 h, yielding ~5000 single-time-point measurements per automated experiment. Valve-controlled two-dimensional fluidic arrays have also been built for highly automated culture of stem cells (112) and for high-throughput cell-toxicity studies (113). Finally, these methods were recently adapted to create tissue-like arrays that can be perfused with physiologically relevant flow rates, adding a third dimension to microfluidic living-cell arrays (114). Together, these early microfluidic cell arrays (examples of which are shown in Figure 7) demonstrate the potential of controlling spatiotemporal aspects of the soluble environment while performing live-cell monitoring.
Figure 7.
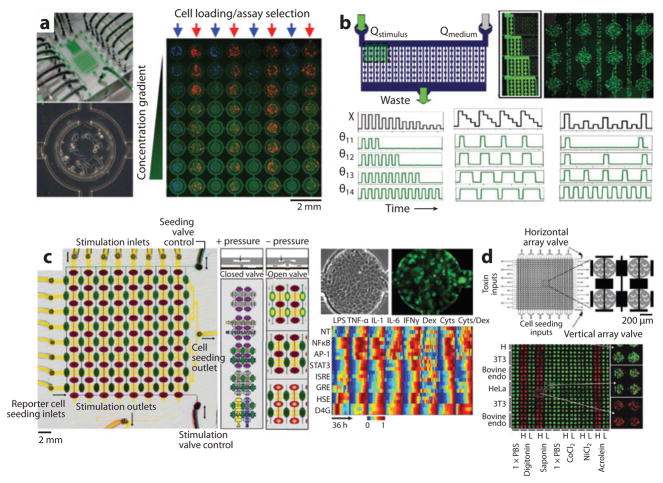
Examples of microfluidic cell arrays. (a) A microfluidic array delivers a concentration gradient of fluorescent dextran in columns to independently seeded rows of fluorescently labeled cells. The figure illustrates the full device (top left), the 64-element cell culture array (right), and a single element (lower left) (110). (b) A microfluidic device uses flow-encoded switching to control parallel delivery of different stimulus timing regimens to preseeded GFP reporter cells. The figure illustrates device design (top left), dye filled channels at different time points (top middle), channels seeded with fluorescent GFP reporter cells (top right), example excitation sequences (black) and the resulting dynamic exposures of 4 channels (green) varying pulse duration, width, and frequency (109). (c) A microfluidic valve-controlled array enables dynamic gene expression profiling. The figure illustrates device design, valve design, reporter cell seeding, stimulation strategy (left), phase and fluorescence images of GFP reporter cells in an array element (top right), and a heat map quantifying dynamic GFP responses across the array (bottom right) (111). (d) A microfluidic array for toxin screening. The figure illustrates device design (top left), close-up of valve-controlled array elements (top right), and fluorescence images of cell viability (green, live: red, dead) across the array of cell lines and putative toxins (bottom) (113).
7. CONCLUSION
Historically, cell assays focused on investigation of single pathways, and there was minimal need for miniaturization or parallelization. Stimuli were homogeneous and static, and responses were population averages measured at single time points. Today, the need for genome-wide screening, characterization of large chemical libraries, and decoupling of complex microenvironmental inputs has prompted development of novel scalable experimental platforms. This review describes high-throughput live-cell assay systems beginning with microtiter plates, the current industry standard, and then we introduce next-generation living-cell microarrays based on high-density spotting, microfabrication, and microfluidics. These emerging platforms promise to improve the density, scale, and efficiency of existing assays, and in doing so, offer unique and exciting opportunities to rapidly identify genetic determinants of disease, discover modulators of cellular function, and create a window into the complex and dynamic relationships between cells and their local environments.
SUMMARY POINTS.
Living-cell microarrays are high-throughput experimental systems that allow parallel control of cell stimuli while measuring cellular responses.
Microtiter plates are the current industry standard for high-throughput screening but require large laboratory automation equipment and lack spatiotemporal control of the cell microenvironment.
Reverse transfection allows cell arrays to be created on a single glass slide, such that each array element is exposed to a different stimulus. This approach has been particularly useful for screening cDNA and RNAi libraries.
Spotted arrays can also be used to study small-molecule libraries or to screen the effects of extracellular matrix combinations.
Microfabricated arrays based on mechanical (microwells), chemical (surface patterning), or electromagnetic (dielectrophoresis) patterning are ideal for creating single-cell and multicell colonies of controlled shape. In doing so, they provide powerful tools for studying intercellular communication.
Microfluidics are ideal for creating high-throughput closed-volume perfusion bioreactors. In doing so, they complement other living-cell array technologies by offering precise spatiotemporal control of the soluble cellular microenvironment.
Acknowledgments
The authors’ work in this area is supported by funds from NIH grants R01 AI063795 and P41 EB002503 and by a postdoctoral fellowship, #8495, from the Shriners Burns Hospital.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Carrel A, Burrows MT. Cultivation of tissues in vitro and its technique. J Exp Med. 1911;14:244–47. doi: 10.1084/jem.13.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingber DE. Mechanical control of tissue morphogenesis during embryological development. Int J Dev Biol. 2006;50:255–66. doi: 10.1387/ijdb.052044di. [DOI] [PubMed] [Google Scholar]
- 4.Werner S, Krieg T, Smola H. Keratinocyte-fibroblast interactions in wound healing. J Investig Dermatol. 2007;127:998–1008. doi: 10.1038/sj.jid.5700786. [DOI] [PubMed] [Google Scholar]
- 5.Saez JC, Berthoud VM, Branes MC, Martinez AD, Beyer EC. Plasma membrane channels formed by connexins: their regulation and functions. Physiol Rev. 2003;83:1359–400. doi: 10.1152/physrev.00007.2003. [DOI] [PubMed] [Google Scholar]
- 6.Wheelock MJ, Johnson KR. Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol. 2003;19:207–35. doi: 10.1146/annurev.cellbio.19.011102.111135. [DOI] [PubMed] [Google Scholar]
- 7.Chen CS, Tan J, Tien J. Mechanotransduction at cell-matrix and cell-cell contacts. Annu Rev Biomed Eng. 2004;6:275–302. doi: 10.1146/annurev.bioeng.6.040803.140040. [DOI] [PubMed] [Google Scholar]
- 8.Janes KA, Gaudet S, Albeck JG, Nielsen UB, Lauffenburger DA, Sorger PK. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124:1225–39. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 9.Lecuit T. Adhesion remodeling underlying tissue morphogenesis. Trends Cell Biol. 2005;15:34–42. doi: 10.1016/j.tcb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Montell DJ. Morphogenetic cell movements: diversity from modular mechanical properties. Science. 2008;322:1502–5. doi: 10.1126/science.1164073. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Shyy JY, Chien S. Fluorescence proteins, live-cell imaging, and mechanobiology: seeing is believing. Annu Rev Biomed Eng. 2008;10:1–38. doi: 10.1146/annurev.bioeng.010308.161731. [DOI] [PubMed] [Google Scholar]
- 12.van Roessel P, Brand AH. Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins. Nat Cell Biol. 2002;4:E15–20. doi: 10.1038/ncb0102-e15. [DOI] [PubMed] [Google Scholar]
- 13.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 14.Tsien RY. Building and breeding molecules to spy on cells and tumors. FEBS Lett. 2005;579:927–32. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 15.Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–11. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 16.Ng T, Squire A, Hansra G, Bornancin F, Prevostel C, et al. Imaging protein kinase Calpha activation in cells. Science. 1999;283:2085–89. doi: 10.1126/science.283.5410.2085. [DOI] [PubMed] [Google Scholar]
- 17.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–8. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 18.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–5. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 19.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–64. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 20.Patterson G, Day RN, Piston D. Fluorescent protein spectra. J Cell Sci. 2001;114:837–38. doi: 10.1242/jcs.114.5.837. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Zhao X, Fang Y, Jiang X, Duong T, et al. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–75. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 22.Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, et al. “Fluorescent timer”: protein that changes color with time. Science. 2000;290:1585–88. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 23.Bailey SN, Wu RZ, Sabatini DM. Applications of transfected cell microarrays in high-throughput drug discovery. Drug Discov Today. 2002;7:S113–18. doi: 10.1016/s1359-6446(02)02386-3. [DOI] [PubMed] [Google Scholar]
- 24.Wu RZ, Bailey SN, Sabatini DM. Cell-biological applications of transfected-cell microarrays. Trends Cell Biol. 2002;12:485–88. doi: 10.1016/s0962-8924(02)02354-1. [DOI] [PubMed] [Google Scholar]
- 25.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006;7:690–96. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 26.Starkuviene V, Pepperkok R. The potential of high-content high-throughput microscopy in drug discovery. Br J Pharmacol. 2007;152:62–71. doi: 10.1038/sj.bjp.0707346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. High-throughput image analysis software designed for cell microarray analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 29.Conrad C, Erfle H, Warnat P, Daigle N, Lorch T, et al. Automatic identification of subcellular phenotypes on human cell arrays. Genome Res. 2004;14:1130–36. doi: 10.1101/gr.2383804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–28. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- 31.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195–203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 32.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–98. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 33.St-Pierre F, Endy D. Determination of cell fate selection during phage lambda infection. Proc Natl Acad Sci USA. 2008;105:20705–10. doi: 10.1073/pnas.0808831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Carlo D, Lee LP. Dynamic single-cell analysis for quantitative biology. Anal Chem. 2006;78:7918–25. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 35.Loo LH, Wu LF, Altschuler SJ. Image-based multivariate profiling of drug responses from single cells. Nat Methods. 2007;4:445–53. doi: 10.1038/nmeth1032. Single-cell multidimensional dynamic characterization in multiwell plates. [DOI] [PubMed] [Google Scholar]
- 36.Neumann B, Held M, Liebel U, Erfle H, Rogers P, et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3:385–90. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 37.Blow N. Lab automation: tales along the road to automation. Nat Methods. 2008;5:109–12. doi: 10.1038/nmeth0108-109. [DOI] [PubMed] [Google Scholar]
- 38.Carpenter AE. Image-based chemical screening. Nat Chem Biol. 2007;3:461–65. doi: 10.1038/nchembio.2007.15. [DOI] [PubMed] [Google Scholar]
- 39.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 40.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–79. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 41.Inglese J, Shamu CE, Guy RK. Reporting data from high-throughput screening of small-molecule libraries. Nat Chem Biol. 2007;3:438–41. doi: 10.1038/nchembio0807-438. [DOI] [PubMed] [Google Scholar]
- 42.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–10. doi: 10.1038/35075114. Original description of cell microarrays based on the reverse transfection method. [DOI] [PubMed] [Google Scholar]
- 43.Palmer E, Freeman T. Cell-based microarrays: current progress, future prospects. Pharmacogenomics. 2005;6:527–34. doi: 10.2217/14622416.6.5.527. [DOI] [PubMed] [Google Scholar]
- 44.Angres B. Cell microarrays. Expert Rev Mol Diagn. 2005;5:769–79. doi: 10.1586/14737159.5.5.769. [DOI] [PubMed] [Google Scholar]
- 45.Wheeler DB, Carpenter AE, Sabatini DM. Cell microarrays and RNA interference chip away at gene function. Nat Genet. 2005;37(Suppl):S25–30. doi: 10.1038/ng1560. [DOI] [PubMed] [Google Scholar]
- 46.Erfle H, Neumann B, Liebel U, Rogers P, Held M, et al. Reverse transfection on cell arrays for high content screening microscopy. Nat Protoc. 2007;2:392–99. doi: 10.1038/nprot.2006.483. [DOI] [PubMed] [Google Scholar]
- 47.Bailey SN, Ali SM, Carpenter AE, Higgins CO, Sabatini DM. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nat Methods. 2006;3:117–22. doi: 10.1038/nmeth848. [DOI] [PubMed] [Google Scholar]
- 48.Hannon GJ. RNA interference. Nature. 2002;418:244–51. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 49.Carpenter AE, Sabatini DM. Systematic genome-wide screens of gene function. Nat Rev Genet. 2004;5:11–22. doi: 10.1038/nrg1248. [DOI] [PubMed] [Google Scholar]
- 50.Redmond TM, Ren X, Kubish G, Atkins S, Low S, Uhler MD. Microarray transfection analysis of transcriptional regulation by cAMP-dependent protein kinase. Mol Cell Proteomics. 2004;3:770–79. doi: 10.1074/mcp.M400018-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Webb BL, Diaz B, Martin GS, Lai F. A reporter system for reverse transfection cell arrays. J Biomol Screen. 2003;8:620–23. doi: 10.1177/1087057103259324. [DOI] [PubMed] [Google Scholar]
- 52.Palmer E, Freeman T. Investigation into the use of C- and N-terminal GFP fusion proteins for subcellular localization studies using reverse transfection microarrays. Comp Funct Genomics. 2004;5:342–53. doi: 10.1002/cfg.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva JM, Mizuno H, Brady A, Lucito R, Hannon GJ. RNA interference microarrays: high-throughput loss-of-function genetics in mammalian cells. Proc Natl Acad Sci USA. 2004;101:6548–52. doi: 10.1073/pnas.0400165101. First demonstration of RNAi cell microarray. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248–51. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 55.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–53. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 56.Wheeler DB, Bailey SN, Guertin DA, Carpenter AE, Higgins CO, Sabatini DM. RNAi living-cell microarrays for loss-of-function screens in Drosophila melanogaster cells. Nat Methods. 2004;1:127–32. doi: 10.1038/nmeth711. [DOI] [PubMed] [Google Scholar]
- 57.Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, et al. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17:443–48. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–72. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 59.Kassner PD. Discovery of novel targets with high throughput RNA interference screening. Comb Chem High Throughput Screen. 2008;11:175–84. doi: 10.2174/138620708783877744. [DOI] [PubMed] [Google Scholar]
- 60.Vanhecke D, Janitz M. High-throughput gene silencing using cell arrays. Oncogene. 2004;23:8353–58. doi: 10.1038/sj.onc.1208027. [DOI] [PubMed] [Google Scholar]
- 61.Bailey SN, Sabatini DM, Stockwell BR. Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. Proc Natl Acad Sci USA. 2004;101:16144–49. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–25. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 63.Soen Y, Mori A, Palmer TD, Brown PO. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol. 2006;2:37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–79. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 65.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116:769–78. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 66.Xia Y, Whitesides GM. Soft lithography. Angew Chem Int Ed Engl. 1998;37:550–75. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. Early paper describing the soft lithography approach. [DOI] [PubMed] [Google Scholar]
- 67.Rettig JR, Folch A. Large-scale single-cell trapping and imaging using microwell arrays. Anal Chem. 2005;77:5628–34. doi: 10.1021/ac0505977. [DOI] [PubMed] [Google Scholar]
- 67a.Roach KL, King KR, Uygun K, Hand SC, Kohane IS, Yarmush ML, et al. High throughput single cell arrays as a novel tool in biopreservation. Cryobiology. 2009 doi: 10.1016/j.cryobiol.2009.03.001. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tajiri K, Kishi H, Tokimitsu Y, Kondo S, Ozawa T, et al. Cell-microarray analysis of antigen-specific B-cells: single cell analysis of antigen receptor expression and specificity. Cytometry A. 2007;71:961–67. doi: 10.1002/cyto.a.20471. [DOI] [PubMed] [Google Scholar]
- 69.Dykstra B, Ramunas J, Kent D, McCaffrey L, Szumsky E, et al. High-resolution video monitoring of hematopoietic stem cells cultured in single-cell arrays identifies new features of self-renewal. Proc Natl Acad Sci USA. 2006;103:8185–90. doi: 10.1073/pnas.0602548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosenthal A, Macdonald A, Voldman J. Cell patterning chip for controlling the stem cell microenvironment. Biomaterials. 2007;28:3208–16. doi: 10.1016/j.biomaterials.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Carlo D, Aghdam N, Lee LP. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal Chem. 2006;78:4925–30. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- 72.Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6:1445–49. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 73.Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. Microfluidic control of cell pairing and fusion. Nat Methods. 2009;6(2):147–52. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Folch A, Toner M. Microengineering of cellular interactions. Annu Rev Biomed Eng. 2000;2:227–56. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 75.Nelson CM, Chen CS. Cell-cell signaling by direct contact increases cell proliferation via a PI3K-dependent signal. FEBS Lett. 2002;514:238–42. doi: 10.1016/s0014-5793(02)02370-0. [DOI] [PubMed] [Google Scholar]
- 76.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–95. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 77.Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921–27. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhatia SN, Yarmush ML, Toner M. Controlling cell interactions by micropatterning in co-cultures: hepatocytes and 3T3 fibroblasts. J Biomed Mater Res. 1997;34:189–99. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 79.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Probing heterotypic cell interactions: hepatocyte function in microfabricated co-cultures. J Biomater Sci Polym Ed. 1998;9:1137–60. doi: 10.1163/156856298x00695. [DOI] [PubMed] [Google Scholar]
- 80.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883–900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 81.Khetani SR, Szulgit G, Del Rio JA, Barlow C, Bhatia SN. Exploring interactions between rat hepatocytes and nonparenchymal cells using gene expression profiling. Hepatology. 2004;40:545–54. doi: 10.1002/hep.20351. [DOI] [PubMed] [Google Scholar]
- 82.Khetani SR, Chen AA, Ranscht B, Bhatia SN. T-cadherin modulates hepatocyte functions in vitro. FASEB J. 2008;22:3768–75. doi: 10.1096/fj.07-105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–26. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 84.Voldman J. Electrical forces for microscale cell manipulation. Annu Rev Biomed Eng. 2006;8:425–54. doi: 10.1146/annurev.bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- 85.Voldman J. Electrical forces for microscale cell manipulation. Annu Rev Biomed Eng. 2006;8:425–54. doi: 10.1146/annurev.bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- 86.Voldman J, Gray ML, Toner M, Schmidt MA. A microfabrication-based dynamic array cytometer. Anal Chem. 2002;74:3984–90. doi: 10.1021/ac0256235. Dielectrophoretic addressable array for trapping, characterizing, and sorting single cells. [DOI] [PubMed] [Google Scholar]
- 87.Taff BM, Voldman J. A scalable addressable positive-dielectrophoretic cell-sorting array. Anal Chem. 2005;77:7976–83. doi: 10.1021/ac0513616. [DOI] [PubMed] [Google Scholar]
- 88.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Probing the role of multicellular organization in three-dimensional microenvironments. Nat Methods. 2006;3:369–75. doi: 10.1038/nmeth873. Dielectrophoretic cell patterning and hydrogel encapsulation are combined to create three-dimensional cell arrays. [DOI] [PubMed] [Google Scholar]
- 89.Albrecht DR, Tsang VL, Sah RL, Bhatia SN. Photo- and electropatterning of hydrogel-encapsulated living cell arrays. Lab Chip. 2005;5:111–18. doi: 10.1039/b406953f. [DOI] [PubMed] [Google Scholar]
- 90.Squires TMQ, Quake SR. Microfluidics: Fluid physics at the nanoliter scale. Rev Mod Phys. 2005;77:977–1025. [Google Scholar]
- 91.Beebe DJ, Mensing GA, Walker GM. Physics and applications of microfluidics in biology. Annu Rev Biomed Eng. 2002;4:261–86. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 92.Heo YS, Cabrera LM, Song JW, Futai N, Tung YC, et al. Characterization and resolution of evaporation-mediated osmolality shifts that constrain microfluidic cell culture in poly(dimethylsiloxane) devices. Anal Chem. 2007;79:1126–34. doi: 10.1021/ac061990v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Berthier E, Warrick J, Yu H, Beebe DJ. Managing evaporation for more robust microscale assays. Part 1 Volume loss in high throughput assays. Lab Chip. 2008;8:852–59. doi: 10.1039/b717422e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Berthier E, Warrick J, Yu H, Beebe DJ. Managing evaporation for more robust microscale assays. Part 2 Characterization of convection and diffusion for cell biology. Lab Chip. 2008;8:860–64. doi: 10.1039/b717423c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–16. doi: 10.1126/science.288.5463.113. First description of integrated PDMS microvalves. [DOI] [PubMed] [Google Scholar]
- 96.Yu H, Alexander CM, Beebe DJ. A plate reader-compatible microchannel array for cell biology assays. Lab Chip. 2007;7:388–91. doi: 10.1039/b612358a. [DOI] [PubMed] [Google Scholar]
- 97.Warrick J, Meyvantsson I, Ju J, Beebe DJ. High-throughput microfluidics: improved sample treatment and washing over standard wells. Lab Chip. 2007;7:316–21. doi: 10.1039/b613350a. [DOI] [PubMed] [Google Scholar]
- 98.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–84. doi: 10.1126/science.1076996. Demonstration of PDMS microvalve scalability. [DOI] [PubMed] [Google Scholar]
- 99.Li Jeon N, Baskaran H, Dertinger SK, Whitesides GM, Van de Water L, Toner M. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat Biotechnol. 2002;20:826–30. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 100.Irimia D, Liu SY, Tharp WG, Samadani A, Toner M, Poznansky MC. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab Chip. 2006;6:191–98. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sawano A, Takayama S, Matsuda M, Miyawaki A. Lateral propagation of EGF signaling after local stimulation is dependent on receptor density. Dev Cell. 2002;3:245–57. doi: 10.1016/s1534-5807(02)00224-1. [DOI] [PubMed] [Google Scholar]
- 102.Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature. 2005;434:1134–38. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Allen JW, Khetani SR, Bhatia SN. In vitro zonation and toxicity in a hepatocyte bioreactor. Toxicol Sci. 2005;84:110–19. doi: 10.1093/toxsci/kfi052. [DOI] [PubMed] [Google Scholar]
- 104.Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–22. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 105.Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–84. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- 106.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thompson DM, King KR, Wieder KJ, Toner M, Yarmush ML, Jayaraman A. Dynamic gene expression profiling using a microfabricated living cell array. Anal Chem. 2004;76:4098–103. doi: 10.1021/ac0354241. [DOI] [PubMed] [Google Scholar]
- 108.Kim L, Vahey MD, Lee HY, Voldman J. Microfluidic arrays for logarithmically perfused embryonic stem cell culture. Lab Chip. 2006;6:394–406. doi: 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- 109.King KR, Wang S, Jayaraman A, Yarmush ML, Toner M. Microfluidic flow-encoded switching for parallel control of dynamic cellular microenvironments. Lab Chip. 2008;8:107–16. doi: 10.1039/b716962k. [DOI] [PubMed] [Google Scholar]
- 110.Lee PJ, Hung PJ, Rao VM, Lee LP. Nanoliter scale microbioreactor array for quantitative cell biology. Biotechnol Bioeng. 2006;94:5–14. doi: 10.1002/bit.20745. [DOI] [PubMed] [Google Scholar]
- 111.King KR, Wang S, Irimia D, Jayaraman A, Toner M, Yarmush ML. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip. 2007;7:77–85. doi: 10.1039/b612516f. High-throughput microfluidic living cell array for dynamic gene expression profiling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Versatile, fully automated, microfluidic cell culture system. Anal Chem. 2007;79:8557–63. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 113.Wang Z, Kim MC, Marquez M, Thorsen T. High-density microfluidic arrays for cell cytotoxicity analysis. Lab Chip. 2007;7:740–45. doi: 10.1039/b618734j. [DOI] [PubMed] [Google Scholar]
- 114.Lee PJ, Gaige TA, Ghorashian N, Hung PJ. Microfluidic tissue model for live cell screening. Biotechnol Prog. 2007;23(4):946–51. doi: 10.1021/bp070053l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–73. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 116.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–28. doi: 10.1126/science.276.5317.1425. Early single-cell patterning paper dissecting the role of cell shape and degree of adhesion on phenotype. [DOI] [PubMed] [Google Scholar]