Abstract
Large chromosomal deletions are among the most common molecular abnormalities in cancer, yet the identification of relevant genes has proven difficult. The 5q− syndrome, a subtype of myelodysplastic syndrome (MDS), is a chromosomal deletion syndrome characterized by anemia and thrombocytosis. Although we have previously shown that hemizygous loss of RPS14 recapitulates the failed erythroid differentiation seen in 5q− syndrome, it does not affect thrombocytosis. Here we show that a microRNA located in the common deletion region of 5q− syndrome, miR-145, affects megakaryocyte and erythroid differentiation. We find that miR-145 functions through repression of Fli-1, a megakaryocyte and erythroid regulatory transcription factor. Patients with del(5q) MDS have decreased expression of miR-145 and increased expression of Fli-1. Overexpression of miR-145 or inhibition of Fli-1 decreases the production of megakaryocytic cells relative to erythroid cells, whereas inhibition of miR-145 or overexpression of Fli-1 has a reciprocal effect. Moreover, combined loss of miR-145 and RPS14 cooperates to alter erythroid-megakaryocytic differentiation in a manner similar to the 5q− syndrome. Taken together, these findings demonstrate that coordinate deletion of a miRNA and a protein-coding gene contributes to the phenotype of a human malignancy, the 5q− syndrome.
Introduction
The 5q− syndrome is a subtype of myelodysplastic syndrome (MDS) characterized by a macrocytic anemia, a normal or elevated platelet count, and hypolobated micromegakaryocytes.1 Patients have heterozygous deletions of chromosome 5q, and no genetic lesions have been identified on the intact allele that would cause homozygous inactivation of a gene, raising the hypothesis that haploinsufficiency for one or more genes is sufficient to cause the clinical phenotype of the 5q− syndrome. A common deleted region (CDR) for the 5q− syndrome has been mapped that spans approximately 1.5 megabases, encompassing 40 protein-coding genes.2 In a screen of each of the genes within this CDR, we found that decreased expression of RPS14, encoding a member of the 40S ribosomal subunit, causes the erythroid phenotype of the 5q− syndrome.3 Similarly, inherited mutations that inactivate one allele of other ribosomal proteins cause Diamond-Blackfan anemia (DBA), another disease in which patients have a severe macrocytic anemia.4
Whereas macrocytic anemia is a defining feature of the 5q− syndrome and DBA, both of which are associated with haploinsufficiency for ribosomal genes, thrombocytosis is characteristic of the 5q− syndrome but not DBA.5 We hypothesized that additional genes must contribute to the molecular basis of the 5q− syndrome given that RPS14 deficiency probably does not explain the full clinical phenotype of the 5q− syndrome, that deletions are universally large, and that no mutations have been found that inactivate RPS14 or any other single gene within the 5q− syndrome CDR.
In our genetic screen of the 5q− syndrome CDR, we evaluated all protein coding genes in the CDR for effects on differentiation, but no genes other than RPS14 altered the ratio of megakaryocytic to erythroid cells, either alone or in combination with RPS14.3 Two miRNAs within the CDR, miR-143 and miR-145, are down-regulated in a variety of human cancer types.2,6–9 Reports have shown that these 2 miRNAs play an important role in the differentiation of both embryonic stem cells and smooth muscle.10–12 Notably, a recent study has described a role for miR-145 (in combination with miR-146a) in contributing to thrombocytosis in murine hematopoietic progenitors via non–cell-autonomous paracrine regulation of IL-6 signaling.13 We therefore examined whether these miRNAs might alter hematopoietic differentiation and contribute to the phenotype of the 5q− syndrome.
Methods
Cell culture
Human erythroleukemia (HEL) cells were acquired from ATCC and were cultured under standard conditions. Cryopreserved human bone marrow CD34+ cells were obtained from Poietics. Erythroid and megakaryocytic differentiation was induced in vitro using a 2-phase liquid culture system as previously described.3 Cells were cultured in serum-free expansion medium (StemCell Technologies) supplemented with 100 U/mL penicillin/streptomycin, 2mM glutamine, 40 μg/mL lipids (Sigma-Aldrich), 100 ng/mL SCF, 10 ng/mL IL-3, 50 ng/mL thrombopoietin, and 10 ng/mL IL-6. Erythropoietin was added on day 7 at a concentration of 3 U/mL. Cells were harvested for flow cytometry after 10 days of liquid culture.
Flow cytometry
Lineage-specific differentiation was evaluated by flow cytometry. Human hematopoietic cells were incubated for 15 minutes on ice with phycoerythrin, phycoerythrin-Cy5, or FITC-conjugated antibodies against glycophorin-A (BD Biosciences PharMingen), CD71 (BD Biosciences Pharmingen), and CD41 (BD Biosciences Pharmingen) to assess terminally differentiated erythroid cells, immature erythroid cells, and megakaryocytes, respectively. Murine bone marrow cells were incubated for 15 minutes on ice with allophycocyanin and phycoerythrin-conjugated antibodies against ter-119 (BD Biosciences Pharmingen) and CD41 (BD Biosciences Pharmingen) to assess erythroid cells and megakaryocytes.
Luciferase assays
Luciferase assays were performed as described previously.14
Quantitative RT-PCR
Quantitative RT-PCR of miR-143, miR-145, snoRNA-142, human Fli-1, and GAPDH was performed using the corresponding TaqMan miRNA expression assays. Syntheses of cDNA and TaqMan were performed as described previously.15,16 All assays were performed in quadruplicate.
Bone marrow transplantation
Viral supernatants for retroviral constructs pMSCV.EGFP, pMSCV.miR-145.EGFP, and pMSCV.Fli-1.EGFP were produced as described previously.17 Similar viral titers were used for each retroviral construct, and transduction efficiencies in primary cells were confirmed for the various constructs by flow cytometric analysis of GFP content. For bone marrow transplantation, 8- to 10-week-old BALB/C or C57BL/6 donor mice were injected with 5-fluorouracil (Sigma-Aldrich) 5 days before bone marrow collection from femurs and tibiae. After an overnight incubation in RPMI 1640 supplemented with 10% FBS, 10 ng/mL mIL-3, 20 ng/mL mIL-6, and 10 ng/mL mSCF, cells were spin-infected with viral supernatants 2 times on days 1 and 2, and 106 cells were injected into the tail vein of lethally irradiated wild-type BALB/C or C57BL/6 recipient mice. Animals were analyzed for BM progenitor cell differentiation 4-6 weeks after transplantation using allophycocyanin-conjugated rat anti–mouse TER119 antibody and PE-conjugated rat anti–mouse CD41 antibody (BD Biosciences PharMingen). Approval for the use of animals in this study was granted by the Children's Hospital Boston Institutional Animal Care and Use Committee.
Colony assays
Mouse megakaryocyte progenitor colony assays were performed according to the MegaCult-C technical manual (StemCell Technologies). Mouse bone marrow cells were added to MegaCult-C medium supplemented with 50 ng/mL murine thrombopoietin, 10 ng/mL human IL-3, 20 ng/mL human IL-6, and 50 ng/mL human IL-11 and plated in double-chamber slides. On day 7, the colonies were fixed and stained according to the manufacturer's instructions. The number of CFU-Mk colonies was assessed on day 8 by a scientist blinded to the experimental conditions. Mouse erythroid and myeloid colony assays were performed according to the MethoCult technical manual (StemCell Technologies). Mouse bone marrow cells were added to Complete MethoCult media and plated in 35-mm dishes. The number of burst-forming unit erythroid, colony-forming unit myeloid, and colony-forming unit granulocyte/erythrocyte/monocyte/megakaryocyte colonies were assessed on day 7 by a scientist blinded to the experimental conditions.
Human colony assays
Human megakaryocyte progenitor colony assays were performed according to the MegaCult-C technical manual (StemCell Technologies). Lentivirally infected human CD34 cells were added to MegaCult-C medium supplements with 50 ng/mL human thrombopoietin, 20 ng/mL human IL-6, and 10 ng/mL human IL-3 and plated in double chamber slides. On day 10, the colonies were fixed and stained according to the manufacturer's instructions. The number of CFU-Mk colonies was assessed on day 11 or 12 by a scientist blinded to the experimental conditions. Human erythroid and myeloid colony assays were performed according to the MethoCult technical manual (StemCell Technologies). Infected human CD34 cells were added to H04434 media and plated in 35-mm dishes. The number of burst-forming unit erythroid, colony-forming unit erythroid, and colony forming unit-granulocyte, macrophage was assessed after 14-16 days by a scientist blinded to the experimental conditions.
MDS sample acquisition and institutional review board approval
MDS patient samples were obtained following informed consent under institutional review board–approved protocols at Massachusetts General Hospital and the University of Massachusetts Medical Center.
Statistical analysis
The significance of experimental results was determined by Student t test unless otherwise noted.
Results
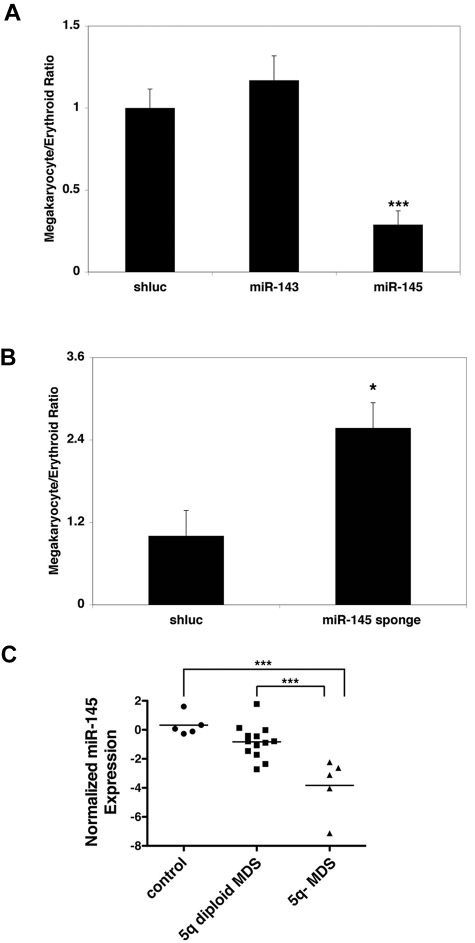
To examine a potential role for miR-143 and miR-145 in the 5q− syndrome and hematopoietic differentiation, we generated lentiviral vectors to overexpress them (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). When introduced into human CD34+ hematopoietic progenitor cells triggered to undergo differentiation in vitro, overexpression of miR-145, but not miR-143, led to a significant decrease in the ratio of megakaryocytic to erythroid cells, even though both miRNAs were overexpressed (Figure 1A; supplemental Figure 1). Notably, these effects appear largely to be the result of defective megakaryocyte differentiation, as miR-145–transduced cells have reduced numbers of megakaryocytic colonies in in vitro progenitor assays (supplemental Figure 1). Conversely, miR-145 expression caused no significant change in erythroid or erythroid blast colony formation, in contrast to knockdown of RPS14, which we have previously described as a regulator of erythroid differentiation (supplemental Figure 1).3 Because the 5q− syndrome manifests in thrombocytosis, a relative decrease in megakaryocyte production in response to miR-145 overexpression is consistent with a role for miR-145 in the pathogenesis of the 5q− syndrome. Therefore, we focused our subsequent analysis on miR-145.
Figure 1.
miR-145 regulates erythroid-megakaryocytic differentiation. (A) Relative to an shRNA-targeting Renilla luciferase (shluc), overexpression of miR-145, but not overexpression of miR-143, promotes erythroid relative to megakaryocytic differentiation in CD34+ progenitors. The ratios of cells from the megakaryocytic and erythroid lineages, indicated on the y-axis, were assessed by flow cytometry with antibodies against CD41 (for the megakaryocytic lineage) and GlyA (for the erythroid lineage), respectively. Similar results were obtained with the early erythroid marker CD71. (B) Relative to an shRNA-targeting Renilla luciferase (shluc), expression of an miRNA sponge targeting miR-145 promotes megakaryocytic relative to erythroid differentiation in CD34+ progenitors as assessed in Figure 1A. (C) Quantitative RT-PCR was performed for miR-145 in CD34+ progenitors from healthy persons (control), MDS CD34+ progenitors wild-type for 5q (5q diploid MDS), and in CD34+ progenitors from patients with the 5q− syndrome (5q− MDS). Expression levels were normalized to snoRNA-142 and then log2 normalized. Values represent the mean of 4 replicates. (A-B) Values are mean ± SEM with propagated error. These experiments were performed in triplicate and replicated twice. *P < .05. ***P < .0005.
To assess the functional consequences of decreased miR-145 activity in human CD34+ cells, we generated a lentiviral vector encoding an miRNA sponge, which consists of a synthetic concatemer of miR-145 binding sites to sequester the miRNA from its endogenous mRNA targets.18 The sponge functioned as an inhibitor of miR-145 in the context of a synthetic miR-145 reporter (supplemental Figure 1). When transduced into human CD34+ progenitors, inhibition of miR-145 by its sponge caused a significant increase in the ratio of megakaryocyte to erythroid cells, as expected for a factor whose loss contributes to 5q− syndrome (Figure 1B). Taken together, these results suggest that miR-145 regulates the megakaryocyte-erythroid lineage decision in a manner consistent with its role in 5q− syndrome.
To ensure that the functional effects of miR-145 on hematopoiesis were relevant to 5q− MDS, we examined expression of miR-145 in MDS patient samples. Compared with bone marrow from both healthy persons (control) and MDS patient samples, wild-type for 5q (5q diploid MDS), samples from 5q− syndrome patients (5q− MDS) had significantly reduced levels of miR-145 (Figure 1C). This reduction in miR-145 expression showed a modest correlation with platelet counts in 5q− MDS while showing little correlation in 5q diploid MDS patient samples (supplemental Figure 1). We further examined the MIR-145 locus in CD34+ progenitors from 5q− MDS patients to determine whether there are secondary mutations of the miRNA gene. Specifically, we sequenced ∼ 500 bp of flanking sequence containing MIR-145 in 72 MDS patient samples either wild-type or deleted for 5q (53 and 19 samples, respectively). However, we observed no mutations in the remaining allele of MIR-145, in line with previous sequence analysis of RPS14 in 5q− syndrome patients.3 In sum, these findings indicate that miR-145 is down-regulated in 5q− MDS in the absence of secondary loss of function mutations.
To determine the mechanism by which miR-145 regulates hematopoiesis, we examined the predicted targets of miR-145 via the miRNA target prediction program TargetScan.19 Fli-1 was one of the highest scoring targets, with multiple miR-145 bindings sites conserved across mammalian species (supplemental Figure 2). Fli-1, an ETS-family transcription factor originally found as an insertion site of the Friend leukemia virus, has been previously shown to play a role in megakaryocytic and erythroid differentiation. Overexpression of Fli-1 in leukemia cells induces megakaryocyte differentiation while suppressing erythroid differentiation.20,21 Moreover, Fli1−/− embryos exhibit a severe defect in megakaryocyte differentiation, resulting in thrombocytopenia.22 In addition, work has shown that loss of Fli-1 specifically affects both megakaryopoiesis and causes negative regulation of committed hematopoietic progenitor cells.23 Patients harboring terminal deletions of 11q (which contains the FLI1 locus) develop Jacobsen syndrome, which manifests in dysmegakaryopoiesis and thrombocytopenia. Overexpression of Fli-1 in Jacobsen syndrome CD34+ progenitors restores megakaryopoiesis in vitro.24 Finally, a recent report has described Fli-1 as a target of miR-145 in endothelial cells.25 Overall, these data support elevated Fli-1 expression altering megakaryocytic differentiation in the 5q− syndrome.
We therefore performed a series of experiments to determine the interaction of miR-145 and Fli-1. When miR-145 was overexpressed in either HEL cells or CD34+ progenitors, we observed a substantial decrease in endogenous Fli-1 protein, whereas inhibition of miR-145 by its sponge caused a substantial increase in endogenous Fli-1 protein (Figure 2A; supplemental Figure 2). To determine whether miR-145 regulated Fli-1 via its 3′- untranslated region (UTR), we generated a reporter in which the Fli-1 3′-UTR is positioned after Renilla luciferase. As shown in Figure 2B, overexpression of miR-145 was able to repress the Fli-1 reporter, similar to previous reports.25 Conversely, inhibition of miR-145 by an miR-145 sponge caused a significant derepression of the Fli-1 3′-UTR (supplemental Figure 2). Notably, neither overexpression nor inhibition of miR-145 affected expression of a similar reporter targeting the c-Myc 3′-UTR, even though miR-145 has recently been suggested to target c-Myc.26 This may be because of differences in the expression systems and modes of miRNA inhibition, which could potentially affect miRNA activity.
Figure 2.
miR-145 suppresses Fli-1. (A) HEL cells were infected with control lentiviruses, lentiviruses overexpressing miR-143 or miR-145, or lentiviruses expressing miRNA sponges targeting miR-143 and miR-145. Western blotting was performed for Fli-1, actin, p38alpha, and GSK3beta. Fold expression was determined by the ratio of Fli-1 expression to p38alpha. These experiments were replicated twice. (B) Luciferase assays were performed in 293T cells cotransfected with control lentiviruses or lentiviruses overexpressing miR-143 or miR-145 and pRL-TK containing binding sites or murine Fli-1 and c-Myc 3′UTRs. pGL3 was used as a transfection control. Expression was normalized to pGL3 levels and subsequently normalized relative to pRL-CXCR4 expression. Values are mean ± SEM (n = 6) with propagated error. These experiments were replicated 3 times. (C) Luciferase assays were performed in 293T cells cotransfected with control lentiviruses (empty FLI1 wt) or lentiviruses overexpressing miR-145 and pRL-TK containing either the wild-type murine Fli-1 3′-UTR (FLI1 wt) or the Fli-1 3′UTR with individual or combined point mutations of miR-145 binding sites (FLI1 mut1-mut4). pGL3 was used as a transfection control. Expression was first normalized to pGL3 levels and subsequently normalized relative to FLI1 wt expression. Values are mean ± SEM (n = 6) with propagated error. These experiments were replicated 3 times. (D) Quantitative RT-PCR was performed for Fli-1 in CD34+ progenitors as described in Figure 1C. Expression levels were normalized to GAPDH. Values are mean (n = 4). These experiments were performed in quadruplicate and replicated twice. (E) Relative to a control cDNA (LacZ), overexpression of Fli-1 promotes megakaryocytic relative to erythroid differentiation in CD34+ progenitors as assessed in Figure 1A. Values are mean ± SEM with propagated error. These experiments were performed in triplicate and replicated 2 times. *P < .05. ***P < .0005.
Finally, we studied the function of predicted miR-145 binding sites in the Fli-1 3′-UTR by generating mutations of person or combined miR-145 binding sites in our Fli-1 3′-UTR reporter. Systematic mutation of the predicted miR-145 binding sites in the Fli-1 3′-UTR led to progressively greater derepression, with mutation of all 4 sites achieving a complete restoration of reporter expression (Figure 2C). To assess the relevance of Fli-1 regulation by miR-145 in MDS, we compared the expression of Fli-1 in bone marrow from healthy persons and in bone marrow from MDS patients with and without 5q deletions. As shown in Figure 2D, there was a significant increase in Fli-1 mRNA in 5q− MDS relative to controls, consistent with previous findings that miRNAs alter both mRNA and protein levels.27–31 Moreover, there is a modest correlation in Fli-1 expression with platelet counts in 5q− MDS patients, with little correlation in 5q diploid MDS samples (supplemental Figure 1). Taken together, these results indicate that miR-145 directly regulates Fli-1 through its 3′-UTR and that deletion of the CDR of 5q in MDS results in decreased miR-145 expression with a consequent increase in Fli-1 levels.
To analyze the functional effects of Fli-1, we expressed its cDNA without its 3′-UTR in primary human hematopoietic progenitor cells and examined the effect on megakaryocytic and erythroid differentiation. Comparable with inhibition of miR-145, overexpression of Fli-1 caused an increase in megakaryocytes relative to erythroid cells in response to differentiating cytokines (Figure 2E). Notably, Fli-1 appears to operate particularly via altered megakaryocytic differentiation, as Fli-1 overexpression significantly increases megakaryocyte colony numbers in in vitro progenitor assays, with notable but not significant effects on erythroid or erythroid blast colony formation (supplemental Figure 1). Moreover, Fli-1 overexpression increased megakaryocytic differentiation in the absence of cytokines when cocultured on stromal cells (supplemental Figure 3). Conversely, decreased expression of Fli-1 with multiple short hairpin RNAs (shRNAs) targeting Fli-1 (supplemental Figures 1 and 3) decreased the number of megarkyocytes relative to erythroid cells (supplemental Figure 3), as seen with overexpression of miR-145. In aggregate, although it is probable that additional predicted miR-145 targets contribute to its effects on megakaryocyte and erythroid differentiation, these results suggest that regulation of altered expression of Fli-1 by miR-145 plays a critical role in megakaryocytic differentiation.
To explore whether our analyses of miR-145 and Fli-1 activity in cell culture assays were relevant in vivo, we generated retroviral vectors for expression of miR-145, Fli-1, or a sponge targeting miR-145 and transplanted virally transduced bone marrow into lethally irradiated murine hosts. Consistent with the results of our cell culture studies, we found that expression of the miR-145 sponge led to a significant increase in platelet count in the peripheral blood whereas miR-145 overexpression caused a converse decrease in platelet count (Figure 3A). In contrast, miR-145 overexpression and the miR-145 sponge did not cause changes in the hematocrit or WBC levels in peripheral blood (supplemental Figure 4; and data not shown). Moreover, overexpression of Fli-1 led to an increase in the levels of megakaryocytes, whereas overexpression of miR-145 caused a substantial increase in the levels of total erythroid cells in the bone marrow (Figure 3B; supplemental Figure 4). Histologically, overexpression of Fli-1 led to an elevated number of hypolobated micromegakaryocytes, a key pathologic finding in the 5q− syndrome, relative to controls (Figure 3C-D; supplemental Figure 5).
Figure 3.
miR-145 and Fli-1 regulate megakaryocyte and erythroid differentiation in vivo. (A) Relative to an empty vector, miR-145 overexpression reduces peripheral blood platelet counts, whereas an miR-145 sponge increases platelet counts 5 to 6 weeks after bone marrow transplantation into BALB/C recipient mice. Platelet counts were assessed from peripheral blood of mice transplanted with hematopoietic progenitors infected with an empty vector (empty), a vector overexpressing miR-145 (miR-145), or a vector expressing a sponge targeting miR-145 (miR-145 sponge). (B) Relative to an empty vector (empty), overexpression of Fli-1 promotes total megakaryocyte differentiation 5 to 6 weeks after bone marrow transplantation into C57BL/6 animals. The percentage of GFP-positive mononucleated cells from the megakaryocyte lineage was assessed by flow cytometry with antibodies against CD41 and CD61. Values are mean ± SEM (n = 3). (C) H&E and von Willebrand factor staining of bone marrow from the sternum of C57BL/6 mice transplanted with HSCs infected with control virus (empty), virus overexpressing miR-145 (miR-145), and virus overexpressing Fli-1 (Fli-1). Arrowheads indicate normal megakaryocytes; and arrows, smaller megakaryocytes with hypolobated nuclei. Note the increased numbers of megakaryocytes in Fli-1-overexpressing HSCs seen both by hematoxylin and eosin and von Willebrand factor staining. Original magnifications: H&E ×1000; and von Willebrand factor ×200. The samples were analyzed using an Olympus BX41 microscrope with the objective lens of 100×/0.75 Olympus UPlanFL or 20×/0.75 Olympus UPlanFL (Olympus). The pictures were taken using Olympus QColor5 and analyzed with acquisition software QCapture Pro Version 6.0 (QImaging) and Adobe Photoshop CS3 (Adobe). (D) Quantitation of hypolobated micromegakarycoytes was performed by pathologic analysis of an independent set of transplantations into BALB/C animals. (E) Bone marrow cells from empty, miR-145, or Fli-1–overexpressing C57BL/6 recipient animals were plated in megakaryocyte differentiation media. Megakaryocyte colonies were quantitated from 1.1 × 106 sorted cells. These experiments were performed in triplicate and replicated twice. (F) Bone marrow cells from empty, miR-145, or Fli-1–overexpressing C57BL/6 recipient animals were plated in methylcellulose, and 1 × 105 transduced bone marrow cells were assayed for erythroid blast colonies. Values are mean ± SEM (n = 3) with propagated error. These experiments were performed in triplicate and replicated twice. *P < .05.
Bone marrow progenitor cells overexpressing miR-145 produced significantly fewer megakaryocyte colonies than control cells, whereas Fli-1 overexpression increased megakaryocyte colony formation (Figure 3E). Conversely, bone marrow progenitors overexpressing miR-145 had increased erythroid colony formation, whereas overexpression of Fli-1 suppressed erythroid colony formation (Figure 3F; supplemental Figure 4). We observed no significant change in hematopoietic stem cell (HSC) numbers in the bone marrow (supplemental Figure 4). We also noted no significant changes in total colony number, granulocyte-macrophage, or multipotent progenitor colony formation (supplemental Figure 4). There was a modest increase in spleen weight in Fli-1-overexpressing animals (supplemental Figure 4). Overall, these findings demonstrate that altered expression of both miR-145 and Fli-1 affects megakaryocyte production in vivo.
We have previously demonstrated that hemizygous deletion of a protein-coding gene, RPS14, in the 5q− syndrome potently suppresses erythroid differentiation.3 We next sought to determine whether combined depletion of RPS14 and inhibition of miR-145 cooperate in the development of 5q− MDS. In contrast to knockdown of any other protein-coding gene in the 5q− CDR, the miR-145 sponge caused a significant increase in the ratio of megakaryocyte to erythroid cells when combined with shRNAs targeting RPS14 (Figure 4; supplemental Figure 6).3 Furthermore, overexpression of Fli-1 in tandem with shRNAs targeting RPS14 promoted megakaryocyte versus erythroid differentiation, suggesting that Fli-1 up-regulation by miR-145 specifically cooperates with loss of RPS14 (supplemental Figure 6). Overall, these findings suggest that miR-145 loss and subsequent overexpression of Fli-1 can cooperate with loss of RPS14 in hematopoietic progenitors.
Figure 4.
miR-145–mediated suppression of Fli-1 cooperates with RPS14 to suppress 5q− syndrome. Relative both to a lentivirus for an shRNA-targeting Renilla luciferase (shluc) and to a control lentivirus (LacZ) coinfected with an shRNA against RPS14 (shRPS14 #1), expression of a miRNA sponge targeting miR-145 promotes megakaryocytic relative to erythroid differentiation in CD34+ progenitors as assessed in Figure 1A. These experiments were performed in triplicate and replicated 3 times. *P < .05. ***P < .0005.
Based on this ability of miR-145 inhibition to cooperate with RPS14 loss and the antiproliferative effects of RPS14 knockdown, we examined the consequences of altered miR-145 activity on the general proliferation and survival of hematopoietic progenitor cells. When miR-143 or miR-145 were overexpressed in CD34+ progenitor cells, we observed a reduction in cellular number, and RPS14 shRNAs caused an even greater degree of inhibition (Figure 5A). In contrast, both the miR-145 sponge and overexpression of Fli-1 increased the number of CD34+ progenitor cells (Figure 5B). Notably, the miR-145 sponge also increased the number of CD34+ progenitor cells that simultaneously expressed shRNAs against RPS14 (Figure 5C). Notably, these effects on progenitor number tracked closely with effects on apoptosis, with miR-143 and miR-145 overexpression and knockdown of RPS14 causing an increase in CD34+ cell death and expression of the miR-145 sponge conversely reducing cell death (supplemental Figure 6).
Figure 5.
miR-145 loss provides a clonal advantage to hematopoietic progenitors. (A) CD34+ progenitors were infected with lentiviruses expressing a control shRNA (shluc), miR-143, miR-145, or 3 independent shRNAs targeting RPS14. Total cell number of CD34+ progenitors was assessed. Values are mean ± SEM (n = 3) with propagated error. P values are calculated via comparison with the shluc value for a given day. (B) CD34+ progenitors were infected with lentiviruses expressing a control cDNA (LacZ), Fli-1, or a sponge targeting miR-145, and total cell number was assessed as in panel A. P values are calculated via comparison with the LacZ value for a given day. (C) CD34+ progenitors were coinfected with (1) a control shRNA (shluc) or 3 independent shRNAs targeting RPS14 and (2) a control cDNA (LacZ) or the miR-145 sponge. Total cell number was assessed as in panel A. P values are calculated via comparison with the shRNA/LacZ value for a given day. **P < .005. ***P < .0005.
To validate the functional relevance of miR-145 in the 5q− syndrome, we sought to determine whether overexpression of miR-145 could rescue the megakaryocytic differentiation defect in bone marrow cells from patients with MDS and 5q deletions. CD34+ cells from viably frozen bone marrow mononuclear cells obtained from MDS patients with and without 5q deletions were induced to undergo differentiation in vitro. FACS analysis showed that, relative to a control shRNA, lentiviral expression of miR-145 promoted erythroid differentiation specifically in samples from patients with 5q− MDS and not in patients with the 5q diploid form of the disease (Figure 6). These results indicate that loss of miR-145 cooperates with loss of RPS14 in the megakaryocyte-erythroid differentiation effects seen in the 5q− syndrome.
Figure 6.
miR-145 promotes differentiation specifically in 5q− MDS hematopoietic progenitors. CD34+ cells from bone marrow aspirates of patients with 5q− syndrome and MDS patients without 5q deletions were infected with a lentivirus expressing miR-145 or an shRNA-targeting Renilla luciferase (shluc). In patients with 5q deletions (shown in red), miR-145 overexpression increased erythroid relative to megakaryocytic differentiation. No consistent changes in erythroid relative to megakaryocytic differentiation were observed with miR-145 overexpression in patients lacking 5q deletions (shown in blue). The difference between the groups was statistically significant (P = .0005 by 2-way ANOVA), and the pairwise differences in erythroid relative to megakaryocytic differentiation in the 5q− MDS samples were statistically significant by the Student t test. Values are mean ± SEM (n = 3) with propagated error. *P < .05.
Discussion
The results presented here support a model in which the hemizygous deletion of a microRNA (MIR-145) and a protein-coding gene (RPS14) act coordinately to alter hematopoiesis in the 5q− syndrome. Decreased expression of miR-145 causes a relative increase in the production of megakaryocytes, whereas increased expression of miR-145 decreases the relative production of megakaryocytes. We found that a critical target of miR-145 is the FLI1 gene, encoding a transcription factor that is known to play a central role in megakaryopoiesis. In addition, Fli-1 overexpression can lead to myeloid malignancies.32 As expected, in MDS patients with del(5q), the expression of miR-145 is decreased and the expression of Fli-1 is increased. The effects of decreased expression of miR-145 and RPS14 are additive, indicating that the 5q− syndrome is a coordinate gene deletion syndrome, the first such syndrome to be described that involves an miRNA and a protein-coding gene.
A recent study reported that decreased expression of miR-145 (in combination with miR-146a) in murine hematopoietic progenitors, when transplanted into irradiated hosts, caused thrombocytosis and hypolobated micromegakaryocytes.13 Whereas miR-145 lies within the reported 5q− syndrome CDR, and miR-146a does not, both miRNAs are deleted in most patients with del(5q). A mechanism was identified in which the thrombocytosis is caused by paracrine signaling via up-regulation of TIRAP and TRAF6 (which are regulated by miR-145 and miR-146a, respectively), causing secretion of IL-6. In combination with our findings, this study indicates that deletion miR-145 and miR-146a alters megakaryopoiesis via complementary cell autonomous and non–cell-autonomous mechanisms.
We found that MDS patients with del(5q) have a loss of miR-145 expression that is greater than the 50% decrease that might be expected because of heterozygous deletion. A similar expression pattern for miR-145 was reported by Starczynowski et al.13 We did not identify any mutations in MIR-145 in 5q− MDS patient samples. Recent work in a mesenchymal stem cell model of Ewing sarcoma, which is driven by an activating translocation of FLI1, has shown that Fli-1 can directly bind and repress transcription of the primary transcript for miR-145.33 Thus, it is possible that hemizygous loss of miR-145 generates a positive feedback loop via Fli-1-mediated repression of miR-145 transcription, allowing for enhanced overexpression of Fli-1 in the 5q− syndrome.
Large, recurrent heterozygous deletions are common in many tumor types, and it is probable that reduced expression of more than one gene has phenotypic consequences within many of these deletions.34 Future genomic analyses of cytogenetic deletions should examine the integrated effects of copy number changes of both coding and noncoding genes.
Supplementary Material
Acknowledgments
The authors thank D. Wilpitz for experimental assistance.
This work was supported by the National Cancer Institute (grant 2-PO1-CA42063), in part by the National Cancer Institute (Cancer Center Support grant P30-CA14051), and in part by the National Heart, Lung, and Blood Institute (grant R01-HL082945). B.L.E. was supported by a Burroughs Wellcome Fund Career Award in the Medical Sciences. Animal research was approved by Animal Welfare Assurance (A33033-01). M.S.K. was an NSF Graduate Research Fellow. D.G.G. and T.J. are investigators of the Howard Hughes Medical Institute. T.J. is a Ludwig Scholar.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.S.K. and B.L.E. conceived the project; M.S.K., A. Narla, A. Nonami, A.M., N.D., B.B., L.P., J.R.M., and J.L.K. carried out all the experiments; N.G., A.R., and E.A. provided experimental materials; D.G.G., T.J., and B.L.E. supervised the experimental work and interpretation of data; and M.S.K., A. Narla, T.J., and B.L.E. prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Benjamin L. Ebert, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Karp Research Bldg, 1 Blackfan Circle, 5th Fl, CHRB 05.211, Boston, MA 02115; e-mail: benjamin_ebert@dfci.harvard.edu.
References
- 1.Ebert BL. Deletion 5q in myelodysplastic syndrome: a paradigm for the study of hemizygous deletions in cancer. Leukemia. 2009;23(7):1252–1256. doi: 10.1038/leu.2009.53. [DOI] [PubMed] [Google Scholar]
- 2.Boultwood J, Fidler C, Strickson AJ, et al. Narrowing and genomic annotation of the commonly deleted region of the 5q− syndrome. Blood. 2002;99(12):4638–4641. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 3.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q− syndrome gene by RNA interference screen. Nature. 2008;451(7176):335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 6.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98(12):1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 9.Wang X, Tang S, Le SY, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3(7):e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xin M, Small EM, Sutherland LB, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23(18):2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 13.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q− syndrome phenotype. Nat Med. 2010;16(1):49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 14.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17(4):438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercher T, Wernig G, Moore SA, et al. JAK2T875N is a novel activating mutation that results in myeloproliferative disease with features of megakaryoblastic leukemia in a murine bone marrow transplantation model. Blood. 2006;108(8):2770–2779. doi: 10.1182/blood-2006-04-014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Athanasiou M, Mavrothalassitis G, Sun-Hoffman L, Blair DG. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia. 2000;14(3):439–445. doi: 10.1038/sj.leu.2401689. [DOI] [PubMed] [Google Scholar]
- 21.Athanasiou M, Clausen PA, Mavrothalassitis GJ, Zhang XK, Watson DK, Blair DG. Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell Growth Differ. 1996;7(11):1525–1534. [PubMed] [Google Scholar]
- 22.Hart A, Melet F, Grossfeld P, et al. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13(2):167–177. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- 23.Kruse EA, Loughran SJ, Baldwin TM, et al. Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc Natl Acad Sci U S A. 2009;106(33):13814–13819. doi: 10.1073/pnas.0906556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raslova H, Komura E, Le Couedic JP, et al. FLI1 monoallelic expression combined with its hemizygous loss underlies Paris-Trousseau/Jacobsen thrombopenia. J Clin Invest. 2004;114(1):77–84. doi: 10.1172/JCI21197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsson E, Fredlund Fuchs P, Heldin J, et al. Discovery of microvascular miRNAs using public gene expression data: miR-145 is expressed in pericytes and is a regulator of Fli1. Genome Med. 2009;1(11):108. doi: 10.1186/gm108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sachdeva M, Zhu S, Wu F, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106(9):3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 29.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 30.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 31.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torchia EC, Boyd K, Rehg JE, Qu C, Baker SJ. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol Cell Biol. 2007;27(22):7918–7934. doi: 10.1128/MCB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riggi N, Suva ML, De Vito C, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24(9):916–932. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.