Abstract
An early event in heart valve formation is the epithelial-mesenchymal transformation (EMT) of a subpopulation of endothelial cells in specific regions of the heart tube, the endocardial cushions. The Type III TGFβ receptor (TGFβR3) is required for TGFβ2- or BMP-2-stimulated EMT in atrioventricular endocardial cushion (AVC) explants in vitro but the mediators downstream of TGFβR3 are not well described. Using AVC and ventricular explants as an in vitro assay, we found an absolute requirement for specific TGFβR3 cytoplasmic residues, GAIP-interacting protein, C terminus (GIPC), and specific Activin Receptor-Like Kinases (ALK)s for TGFβR3-mediated EMT when stimulated by TGFβ2 or BMP-2. The introduction of TGFβR3 into nontransforming ventricular endocardial cells, followed by the addition of either TGFβ2 or BMP-2, results in EMT. TGFβR3 lacking the entire cytoplasmic domain, or only the 3 C-terminal amino acids that are required to bind GIPC, fails to support EMT in response to TGFβ2 or BMP-2. Overexpression of GIPC in AVC endocardial cells enhanced EMT while siRNA-mediated silencing of GIPC in ventricular cells overexpressing TGFβR3 significantly inhibited EMT. Targeting of specific ALK’s by siRNA revealed that TGFβR3-mediated EMT requires ALK2 and ALK3, in addition to ALK5, but not ALK4 or ALK6. Taken together, these data identify GIPC, ALK2, ALK3, and ALK5 as signaling components required for TGFβR3-mediated endothelial cell EMT.
1.0 Introduction
Transforming Growth Factor β (TGFβ) controls distinct cellular processes such as cell growth and differentiation, regulating events as diverse as development, wound healing, atherosclerosis, and tumor progression [1]. Although the contributions of the serine-threonine kinase containing Type I (TGFβR1 or ALK5) and Type II (TGFβR2) receptors to TGFβ signaling have been well established [2] there remain significant gaps in our understanding of how the wide array of TGFβ-induced responses are signaled and regulated. The Type III TGFβ receptor (TGFβR3), or betaglycan, contains a short, highly conserved intracellular domain with no enzymatic activity [3–5]. TGFβR3 is required for the high affinity binding of TGFβ2 [3] and can present ligand to TGFβR2, but the cytoplasmic domain is not required for this role [6]. In addition, TGFβR3 can bind and signal in response to BMP-2 [7] and function as an inhibin receptor [8]. We have demonstrated that TGFβR3 is essential for atrioventricular cushion (AVC) transformation in vitro [9], the first step in heart valve formation (reviewed in [10]). Tgfbr3−/− embryos display cardiovascular defects that include double outlet right ventricle, ventricular septal defects, and cushion abnormalities with death due to failed coronary vessel development [11]. These data suggest a unique and nonredundant role for TGFβR3 in mediating the actions of TGFβ during development.
To directly address the role of TGFβR3 in TGFβ signaling we took advantage of the observation that endothelial cell transformation in the AVC requires TGFβR3 and that introduction of TGFβR3 into adjacent ventricular endocardial endothelial cells results in transformation in response to TGFβ [9]. These properties of the endocardium allowed for the development of both loss- and gain-of-function assays to probe the receptor domains and downstream signals required for TGFβR3-mediated endothelial cell transformation (reviewed in [12]). Given that TGFβR3 also binds BMP-2 and induces endothelial cell transformation [13] this approach also allowed for the comparison of TGFβ and BMP signaling through TGFβR3 in mediating cell transformation.
We focused our efforts on determining any requirement for the cytoplasmic domain in signaling and the involvement of potential downstream candidate molecules. We found that the cytoplasmic domain of TGFβR3, and specifically the 3 C-terminal amino acids required to bind GIPC, were required for TGFβ2- and BMP-2-stimulated EMT. Consistent with this finding, GIPC is required for TGFβR3-mediated EMT stimulated by TGFβ2 or BMP-2. Finally, since several ALKs may be activated downstream of TGFβ ligands in addition to the canonical TGFβR1, ALK5 [14–16], we used siRNA to target specific ALKs and revealed that ALK2 and ALK3, in addition to ALK5, are required for TGFβR3-mediated, endocardial cell EMT. These data identify the signaling components required to direct TGFβR3-mediated, endocardial cell EMT.
2.0 Materials and Methods
2.1 Construction of Adenoviral Constructs
Adenoviruses were generated [17] and titered as described [18]. Viral titers ranged from 109 to 1014 pfu/ml. Injections were adjusted to achieve infection of 20–50% of endocardial cells.
2.2 Viral Injections and Collagen Gel Assays
Stage 10 – 12 chick embryos were harvested, injected with adenovirus, incubated, and ventricular or AVC explants excised as described [19]. After 48 h, explants were fixed, and the phenotype of each GFP-expressing cell was scored as described [13, 19, 20].
2.3 Proliferation in AVC Explants
AVC explants were excised from HH Stage 16 chick embryos and incubated on collagen gels for 48 hours, incubated with BrdU (Roche) for 1 hour, and fixed with 4% PFA. Explants were washed 3 times with PBS and permeabilized with 0.5% tritonX-100. Antigen retrieval was accomplished with 2M HCL. Explants were blocked with 5% normal donkey serum and 0.05% PBST for one hour and incubated with monoclonal Alexafluor 594 conjugated BrdU antibody (1:50 dilution, Invitrogen) overnight. Explants were rinsed 3 times with PBS and stained with DAPI (dilution 1:1000) for 5 minutes. Stained explants were imaged with a fluorescent microscope. For each explants approximately a hundred random cells were counted and scored for the presence of BrdU staining in the nucleus. A total of 5 explants were counted for a total of 496 DAPI postive cells and 5 BrdU positive cells. Thus, 1.01% of cells were positive for BrdU staining. AVC explants were excised from HH Stage 16 chick embryos and incubated on collagen gels for 48 hours and then fixed with 4% PFA. Explants were washed 3 times with PBS and permeablized with 0.5% tritonX-100. Explants were blocked with 5% normal donkey serum and 0.05% PBST for one hour and incubated with the primary antibody, monoclonal phospho-histone H3 (Serine 10) antibody (monoclonal, 1:300, Sigma), overnight at 4°C. Explants were then washed 3 times with PBST and incubated with the secondary antibody, Sheep anti-mouse IgG conjugated with Cy3 (1:50, Sigma), overnight at 4°C. Explants were rinsed 3 times with PBS and stained with DAPI (dilution 1:1000) for 5 minutes. Stained explants were imaged with a fluorescent microscope. For each explants approximately a hundred random cells were counted and scored for the presence of phospho-histone H3 staining in the nucleus. A total of 5 explants were counted with 13 phospho-histone H3 positive cells counted out of 535 total cells. Thus, 2.43% of cells were positive for phosphor-histone H3 staining.
2.4 Ligand Addition
Recombinant human (rh) TGFβ2 and BMP-2 (R & D Systems, Minneapolis, MN USA) addition occurred 12 h post placement of explants on collagen pads.
2.5 siRNA Treatment of AVC and Ventricular Explants
AVC and ventricular explants were harvested and siRNA was introduced as described [18]. Target sequences for Smad4, ALK5, and TGFβR3 are published [20]. Target sequences for GIPC, ALK2, ALK3, ALK4, and ALK6 are below. For control siRNA, three scrambled 21 oligonucleotide templates with varying GC content that did not blast to any gene in the chicken genome were designed [20]. The control with the most similar GC content to the target siRNA was used.
| siRNA Construct Sequences | |
|---|---|
| Target: | siRNA Construct Sequences: |
| GIPC-2 (A) | 5′(GCCUAUGAAGUCAUUUGAAtt)3′ |
| GIPC-2 (B) | 5′(GCAGGAAGAGACAAGAAAAtt)3′ |
| GIPC-2 (C) | 5′(GGACAACGAAAAGAAGUGGtt)3′ |
| ALK2 (A) | 5′(GCAGAUUUAUUGG ACCAUUtt)3′ |
| ALK2 (B) | 5′(GGUUAGCAAUGGUAUAGUAtt)3′ |
| ALK3 (A) | 5′(GAUUAACAGUGAACAAUGAtt)3′ |
| ALK3 (B) | 5′(GGAGGAAGCUUGAAGUACAtt)3′ |
| ALK4 (A) | 5′(GGGUUGGAACCAAACGAUAtt)3′ |
| ALK4 (B) | 5′(GAAACAAUCAGAAACCUUUtt)3′ |
| ALK5 (A) | 5′(GCUACGACAUGAAAACAUUtt)3′ |
| ALK5 (B) | 5′(GGAUAUUGCUGCCUUUUAAtt)3′ |
| ALK6 (A) | 5′(GGAUAUACAGUUUUAGUAAtt)3′ |
| ALK6 (B) | 5′(GAAGAUUUCUGAAAAUUGAtt)3′ |
| TGFβR3 (A) | 5′(GGAAGUAAAUCUACAUGAAtt)3′ |
| TGFβR3 (B) | 5′(GACUUUUCCUUUUCACAUUtt)3′ |
2.5 RNA isolation and RT-PCR
Chick Embryonic Fibroblasts were used to confirm knockdown of genes targeted with siRNA as described [18, 20]. Primers used for RT-PCR of Smad4, ALK5, TGFβR3, and GAPDH were as described [20]. Additional primers used for GIPC, ALK2, ALK3, ALK4, and ALK6 are below and used as described [21]. RT-PCR data was analyzed using the 2−ΔΔCT method [22].
| Primers Used to Confirm Gene Knockdown | ||
|---|---|---|
| Gene | Forward Primer | Reverse Primer |
| Smad4 | 5′(GTGCCACAGACAGATGCAACAACA)3′ | 5′(TTTGACGAAGCTCATGCGGAGGAT)3′ |
| GIPC-2 | 5′(AAGGGAGCCTTATGGACCAAACCA)3′ | 5′(AGCTTCAGCTTAAACACCCGACCT)3′ |
| TGFβR3 | 5′(CTTCCCAATAGCACATGCGCAGAA)3′ | 5′(AGAGAGGTGCAAGCTTCATCAGGA)3′, |
| ALK2 | 5′(AGCACCCAGCTGTGGCTAATTACT)3′ | 5′(TCTATGTGCAAATGTGCAAGGCCG)3′ |
| ALK3 | 5′(AGACAACAGGGCTCTCCTCAAACT)3′ | 5′(ACTGCGAGACCCAAGTCAGCAATA)3′ |
| ALK4 | 5′(ACGAGCACGGATCTCTCTTTGACT)3′ | 5′(TCTCTGTGAGCAATCCCAGGCTTT)3′ |
| ALK5 | 5′(ACCAGAGTGGCGTGTTAAGAAGGT)3′ | 5′(TGCACAGAAAGGACCCAAAGCAAC)3′ |
| ALK6 | 5′(AGTACCCAAGGCAAACCGGCTATT)3′ | 5′(AAGCTTTCATCCAGCACCTCAGGA)3′ |
2.6 Co-Immunoprecipitation
COS7 were plated at 1.25 × 105 cells in six-well dishes and allowed to recover overnight. The cells were transfected using Fugene at a 1:2 ratio (Roche Applied Science Indianapolis, IN). Forty eight hours post transfection cells were harvested using CO-IP Lysis Buffer (20 mM Hepes pH 7.4, 0.5% NP-40, 2 mM EDTA, 0.15 M NaCl, 10 mM NaF, 10% Glycerol) and immunoprecipitated using 5μg FLAG antibody + PGS overnight and the entirety loaded onto a sodium dodecyl sulfate-polyacrylamide gel and immunoblotted for the TGFβR3 (Anti-human TGFβR3 [cat#AF-242-PB; R&D Systems (Minneapolis, MN)) at a 1:2000 dilution (0.25 mg/mL). 100μl of lysate was retained for control blots. 36μl of total cell lysate was used for TGFβR3/FLAG (Sigma [cat#F3165] Saint Louis, Missouri) control blot and 6μl was used for β-actin (Sigma [A5441] Saint Louis, Missouri) control blot.
2.6 Toxicity to siRNA in AVC Explants
Explants were harvested from stage 16 chick embryos and placed on collagen gels. Explants were incubated with control, Smad4A, and Smad4B siRNA as described in 2.5. BrdU incorporation and analysis was as described in 2.3. After 48 hours explants were incubated with 2 μM lysotracker for 15 minutes, washed for 15 minutes with phosphate buffered saline, incubated with DAPI (1:1000) for 5 minutes, and washed again. Explants were fixed in formaldehyde/gluteraldehye and mounted on a slide. Explants were photographed in brightfield and darkfield to image lysotracker or DAPI and the photomicrographs were overlaid. The total number of lysotracker postitive mesenchymal cells were counted and expressed as a percentage of the total number of mesenchymal cells.
3.0 Results
3.1 Specific Residues in the cytoplasmic domain of TGFβR3 are required for endocardial cell EMT
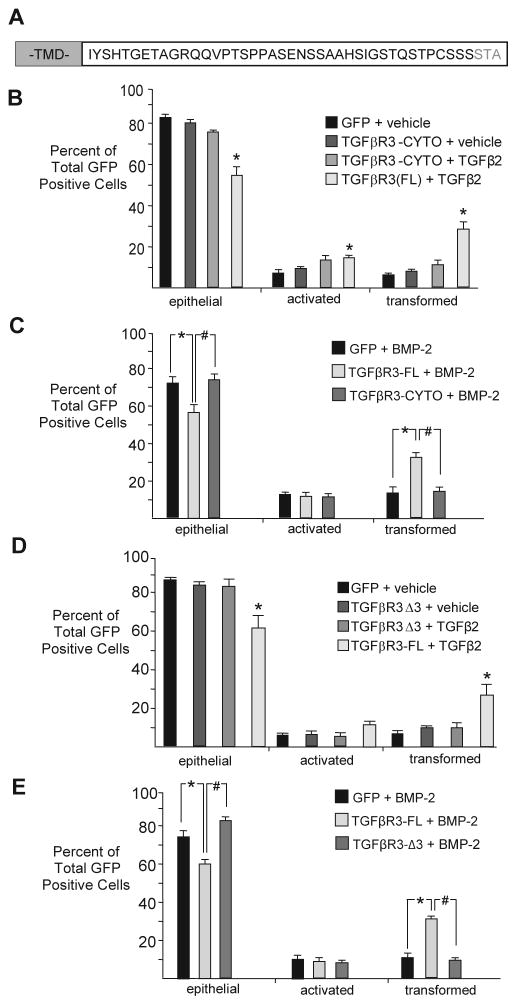
To address the role of the cytoplasmic domain of TGFβR3 (Fig 1A) in mediating endocardial cell EMT we overexpressed GFP alone, GFP and full length TGFβR3 (TGFβR3-FL), or GFP and TGFβR3 lacking the entire cytoplasmic domain (TGFβR3-CYTO) (Fig 1B) in ventricular endocardial cells as described [18, 19]. Infection with adenovirus expressing GFP alone defined the basal distribution of cells as epithelial, activated, or transformed and the addition of vehicle or TGFβ2 do not alter this distribution ([18, 19, 23]). Cells infected with TGFβR3-FL and GFP [23] or TGFβR3-CYTO and GFP had a distribution comparable to cells infected with GFP alone (Fig 1B). Consistent with previous studies, while the addition of 200 pM TGFβ2 to TGFβR3-FL infected ventricular explants resulted in a significant increase in the percent of transformed cells and a concomitant decrease in the percent of cells scored as epithelial (Fig 1B), the addition of 200 pM TGFβ2 to TGFβR3-CYTO infected ventricular explants yielded results similar to infection with GFP adenovirus alone (Fig 1B). These data demonstrate that, while the cytoplasmic domain of TGFβR3 is not required for ligand presentation [6], it is required for TGFβ2-stimulated, TGFβR3-mediated endocardial cell EMT.
Fig 1. Specific Residues in the Cytoplasmic domain of TGFBR3 are required for endocardial cell EMT.
A Amino acid residue depiction of TGFβR3 cytoplasmic domain. (TMD) transmembrane domain. B–G: Average percent of total GFP-expressing cells scored as epithelial, activated or transformed. Means are derived from 3 separate experiments. GFP adenovirus alone served as a negative control to define basal levels of transformed cells. B: In ventricular endothelial cells that overexpress TGFβR3-FL, TGFβ2 (200 pM) induced statistically significant increases in transformed cells with a concomitant decrease in epithelial cells. Overexpression of TGFβR3 lacking the entire cytoplasmic domain (TGFβR3-CYTO) did not support ligand-dependent transformation. GFP, vehicle: epithelial 84±1.4%; (mean±SEM), activated 8±1.1%, transformed 8±0.3%. TGFβR3-CYTO, vehicle: epithelial 80±1.5%, activated 10±1.0%, transformed 10±0.6%. TGFβR3-CYTO, 200 pM TGFβ2: epithelial 75±1.1%, activated 13±2.1%, transformed 13±2.3%. TGFβR3-FL, TGFβ2: epithelial 56±3.8%**, activated 15±1.1%**, transformed 30±3.1%*. Two-tailed Student’s t-tests: *P<0.05, **P<0.01, ***P<0.001 versus control. The number of ventricular explants examined and cells in each category were as follows: GFP control adenovirus, (n=30; total number of cells, 1886; epithelial, 1577; activated, 161; transformed, 148), n=number of explants, TGFβR3-CYTO, vehicle (n=40; total number of cells, 2568; epithelial, 2062; activated, 258; transformed, 248), TGFβR3-CYTO, TGFβ2 (n=38; total number of cells, 2381; epithelial, 1763; activated, 321; transformed, 297), TGFβR3-FL, TGFβ2 (n=36; total number of cells, 1828; epithelial, 1005; activated, 272; transformed, 551).C: Similar results were obtained with the addition of 5 nM BMP2. GFP, 5 nM BMP-2: epithelial 72±3.0%, activated 13±0.8%, transformed 15±2.3%. TGFβR3-FL, BMP-2, vehicle: epithelial 57±3.9%*, activated 11±1.5%, transformed 33±2.4%**. TGFβR3-CYTO, BMP-2: epithelial 74±3.1%, activated 10±1.4%, transformed 15±1.9%. The number of ventricular explants examined and cells in each category were as follows: GFP control adenovirus, (n=43; total number of cells, 3026; epithelial, 2158; activated, 389; transformed, 479), n=number of explants, TGFβR3-FL, BMP-2 (n=33; total number of cells, 1540; epithelial, 854; activated, 170; transformed, 516), TGFβR3-CYTO, BMP-2 (n=32; total number of cells, 1765; epithelial, 1286; activated, 192; transformed, 287).D: Overexpression of TGFβR3 lacking the three C-terminal amino acids (TGFβR3-Δ3) in ventricular endothelial cells did not support TGFβ2-dependent transformation. GFP, vehicle: epithelial 87±0.8%, activated 6±0.6%, transformed 7±1.2%. TGFβR3-Δ3, vehicle: epithelial 84±1.3%, activated 6±1.4%, transformed 10±0.6%. TGFβR3-Δ3, 200 pM TGFβ2: epithelial 83±3.1%, activated 6±1.6%, transformed 10±2.1%. TGFβR3-FL, TGFβ2: epithelial 61±5.5%*, activated 11±2.0%, transformed 28±4.3%*. The number of ventricular explants examined and cells in each category were as follows: GFP control adenovirus, (n=33; total number of cells, 2511; epithelial, 2190; activated, 144; transformed, 177), n=number of explants, TGFβR3-Δ3, vehicle (n=35; total number of cells, 2115; epithelial, 1780; activated, 124; transformed, 211), TGFβR3-Δ3, TGFβ2 (n=37; total number of cells, 2271; epithelial, 1903; activated, 144; transformed, 224), TGFβR3-FL, TGFβ2 (n=36; total number of cells, 1783; epithelial, 1057; activated, 203; transformed, 523). E: Similar results were obtained with the addition of BMP2 (5 nM). GFP, BMP-2: epithelial 76±2.1%, activated 11±1.1%, transformed 12±1.0%. TGFβR3-FL, BMP-2: epithelial 60±2.9%**, activated 9±1.8%, transformed 31±1.3%***. TGFβR3-Δ3, BMP-2: epithelial 83±1.8%, activated 7±1.0%, transformed 10±0.9%. The number of ventricular explants examined and cells in each category were as follows: GFP control adenovirus, (n=42; total number of cells, 1771; epithelial, 1344; activated, 202; transformed, 225), n=number of explants, TGFβR3-FL, BMP-2 (n=36; total number of cells, 681; epithelial, 409; activated, 60; transformed, 212), TGFβR3-Δ3, BMP-2 (n=28; total number of cells, 879; epithelial, 724; activated, 67; transformed, 88).
As TGFβR3 also mediates the effects of BMP-2 on endocardial cell EMT, we assessed the effects of the cytoplasmic domain of TGFβR3 on BMP-2 induced endocardial cell EMT. The addition of 5 nM BMP-2 to GFP expressing ventricular endocardial cells had no effect on the distribution of epithelial, activated or transformed cells when compared to vehicle alone [13, 23]. However, the addition of BMP-2 to ventricular endocardial cells expressing TGFβR3-FL and GFP, but not TGFβR3-CYTO and GFP, increased the percent of transformed cells and a concomitant decrease in the percent of cells scored as epithelial (Fig 1C). These data support a requirement for the cytoplasm domain of TGFβR3 in mediating both TGFβ2 and BMP-2 induced endothelial cell EMT.
The cytoplasmic domain of TGFβR3 binds the PDZ domain containing protein GIPC via a Type I PDZ domain comprised of the three C-terminal amino acids of the receptor [24]. Deletion of these three C-terminal amino acids (TGFβR3-Δ3) abolishes TGFβR3, GIPC interaction [24]. To assess whether this domain was required for TGFβR3-mediated EMT we used adenoviral constructs that co-express TGFβR3-Δ3 and GFP, TGFβR3-FL and GFP, or GFP alone in ventricle endocardial cells. The addition of 200 pM TGFβ2 (Fig 1D), or 5 nM BMP-2 (Fig 1E), to TGFβR3-FL expressing ventricular endothelial cells resulted in EMT. In contrast, the addition of 200 pM TGFβ2 (Fig 1D), or 5 nM BMP-2 (Fig 1E), to TGFβR3-Δ3 expressing ventricular endocardial cells failed to increase EMT and yielded results similar to infection with GFP adenovirus alone (Fig 1D, E). These data demonstrate a requirement for the three C-terminal amino acids for TGFβR3-stimulated EMT.
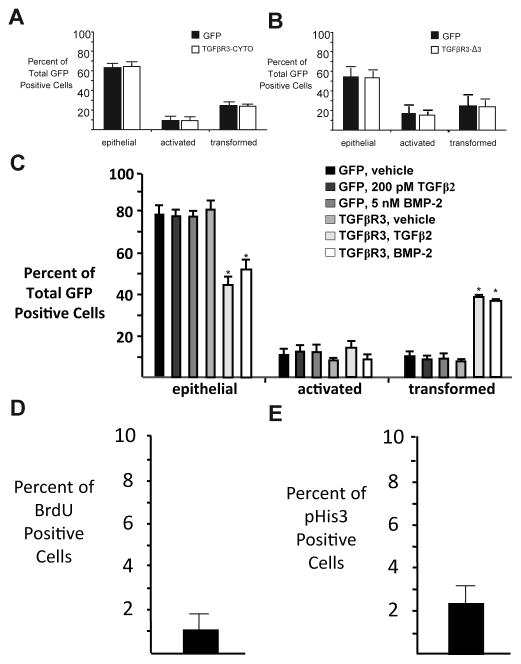
Athough native ventricular endocardial cells do not express TGFβR3 [25], we next addressed the possibility that the TGFβR3-CYTO or TGFβR3-Δ3 form of the receptor could have dominant negative activity. We took advantage of the fact that endocardial cells in AVC explants express TGFβR3 and undergo transformation. The percentage of epithelial, activated, and transformed cells after the overexpression of TGFβR3-CYTO or TGFβR3-Δ3 in AVC endocardial cells was indistinguishable from those seen after infection with adenovirus expressing GFP alone. These data suggest that TGFβR3-CYTO or TGFβR3-Δ3 do not act in a dominant negative manner to regulate EMT (Fig 2A, B). We also confirmed that infection with adenovirus expressing GFP alone defined the basal distribution of cells as epithelial, activated, or transformed and the addition of vehicle or TGFβ2 do not alter this distribution (Fig 2C). Cell proliferation in explants is low and alterations in proliferation cannot explain the decreases in cell invasion noted in these assays ([26], and Fig 2D, E).
Fig 2. Truncated Receptors do not function as dominant negatives.
A–B Adenoviral mediated overexpression of (A) TGFβR3-CYTO or (B) TGFβR3-Δ3 does not alter AVC endothelial cell transformation when compared to GFP alone. GFP control adenovirus (A): epithelial 63±3.6%, activated 10±1.5%, transformed 28±2.1%. TGFβR3-CYTO: epithelial 63±2.0%, activated 10±1.1%, transformed 27±1.0%. GFP control adenovirus (B): epithelial 54±6.6%, activated 19±5.4%, transformed 27±11.0%. TGFβR3-Δ3: epithelial 54±5.6%, activated 19±3.4%, transformed 27±8.2%. The number of ventricular explants examined and cells in each category were as follows: GFP (A) (n=26; total number of cells, 2537; epithelial, 1573; activated, 249; transformed, 715), n=number of explants, TGFβR3-CYTO (n=27; total number of cells, 2578; epithelial, 1638; activated, 245; transformed, 695), GFP (B) (n=38; total number of cells, 1238; epithelial, 665; activated, 234; transformed, 339), n=number of explants, TGFβR3-Δ3 (n=39; total number of cells, 2783; epithelial, 1511; activated, 524; transformed, 748).C: GFP control ventricular explants do not undergo EMT as a response to TGFβ2 or BMP-2. GFP, vehicle: epithelial 77±6.0%, activated 12±2.8%, transformed 11±3.3%. GFP, TGFβ2: epithelial 76±4.5%, activated 14±2.1%, transformed 10±2.3%. GFP, BMP-2: epithelial 76±3.9%, activated 14±3.1%, transformed 10±1.2%. TGFβR3-FL, vehicle: epithelial 81±4.8%, activated 10±1.4%, transformed 9±1.2%. TGFβR3-FL, TGFβ2: epithelial 44±4.8%**, activated 17±3.4%, transformed 39±1.5%***. TGFβR3-FL, BMP-2: epithelial 52±3.6%**, activated 10±2.8%, transformed 38±1.9%***. The number of ventricular explants examined and cells in each category were as follows: GFP control adenovirus, vehicle (n=20; total number of cells, 1251; epithelial, 959; activated, 151; transformed, 141), n=number of explants, GFP control adenovirus, TGFβ2 (n=25; total number of cells, 1435; epithelial, 1093; activated, 202; transformed, 140) GFP control adenovirus, BMP-2, (n=22; total number of cells, 1350; epithelial, 1022; activated, 192; transformed, 136), TGFβR3-FL, vehicle (n=17; total number of cells, 903; epithelial, 714; activated, 98; transformed, 91), TGFβR3-FL, TGFβ2 (n=19; total number of cells, 1189; epithelial, 511; activated, 210; transformed, 468), TGFβR3-FL, BMP-2 (n=18; total number of cells, 1163; epithelial, 612; activated, 114; transformed, 437). D–E: BrdU and phospho-histone H3 staining suggest a low level of proliferation in AVC explants. Immunostaining demonstrated 1.01% of cell that were BrdU positive (D) whereas 2.43% of cells are pHis-H3 (E) positive.
3.2 GIPC is required for TGFβR3 mediated endocardial cell EMT
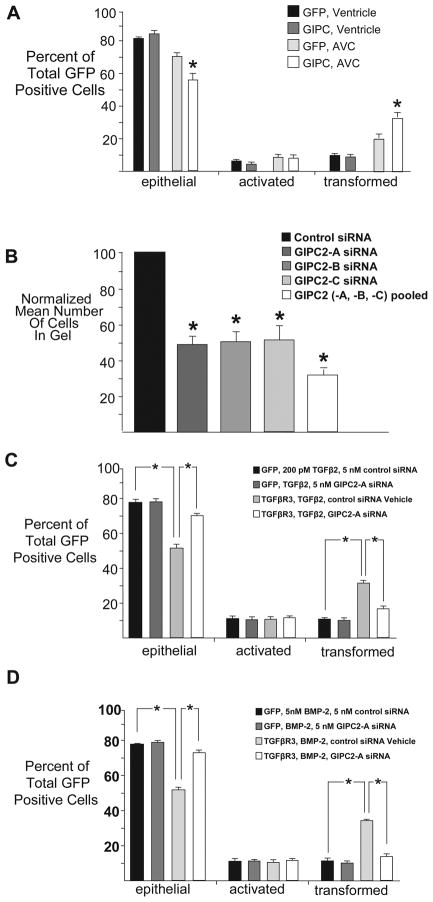
As the three C-terminal amino acids of TGFβR3 mediating binding to GIPC were required for TGFβR3-mediated EMT stimulated by either TGFβ2 or BMP-2, we assessed the role of GIPC. Overexpression of GIPC in AVC endocardial cells, which express endogenous TGFβR3, resulted in a statistically significant enhancement of EMT (Fig 3A). In contrast, overexpression of GIPC had not effect on EMT in ventricular endocardial cells which lack TGFβR3 expression (Fig 3A). These data suggest that GIPC levels regulate EMT in a TGFβR3-dependent manner, and further, that the levels of GIPC may be limiting in AVC endocardial cells. To further assess the role of GIPC in EMT, we assessed the effects of siRNA-mediated silencing of GIPC isoforms exist (three in the human and mouse genomes [27]) to date only GIPC-2 has been identified in the chicken genome and we have demonstrated the presence of this isoform in the endothelium (data not shown). The addition of 5 nM of each of three independent siRNA constructs targeting GIPC-2 in AVC endocardial cells inhibited EMT, with a further reduction noted in the presence of 5 nM of each (15 nM total) siRNA (Fig 3B). These data confirm that GIPC-2 is required for AVC transformation, consistent with the hypothesis that the TGFβR3-GIPC-2 interaction mediates endocardial cell EMT.
Fig 3. GIPC Interaction with TGFβR3 is required for endocardial cell EMT.
A, C, D Average percent of total GFP-expressing cells scored as epithelial, activated or transformed. Means are derived from 3 separate experiments. GFP adenovirus alone served as a negative control to define basal levels of transformed cells. A: Overexpression of GIPC caused a statistically significant increase in transformation in AVC endothelial cells with no alteration in transformation in ventricular endothelial cells. GFP, ventricle: epithelial 82±0.3%; (mean±SEM), activated 7±0.5%, transformed 11±0.4%. GIPC, ventricle: epithelial 85±1.6%, activated 5±0.9%, transformed 9±0.7%. GFP, AVC: epithelial 71±2.9%, activated 9±1.3%, transformed 20±2.1%. GIPC, AVC: epithelial 57±3.9%*, activated 8±1.6%, transformed 35±2.3%**. Two-tailed Student’s t-tests: *P<0.05, **P<0.01, ***P<0.001 versus control. The number of ventricular explants examined and cells in each category were as follows: GFP control adenovirus, ventricle (n=43; total number of cells, 2335; epithelial, 1918; activated, 160; transformed, 348), n=number of explants, GIPC, ventricle (n=44; total number of cells, 1452; epithelial, 1245; activated, 75; transformed, 132), GFP, AVC (n=43; total number of cells, 3170; epithelial, 2251; activated, 283; transformed, 636), GIPC, AVC (n=42; total number of cells, 1319; epithelial, 753; activated, 106; transformed, 160).B: Targeting of GIPC by siRNA in AVC endothelial cells. Quantification of cells migrated into collagen gel. Means are derived from 3 separate experiments and are normalized to control siRNA. GIPC-A (5 nM), GIPC-B (5 nM), GIPC-C (5 nM), or pooled (15 nM) siRNAs significantly decreased the number of cells in the gel when compared to control siRNA. Control: Normalized to 100%. GIPC-A siRNA: 49±4.7% (mean±SEM), GIPC-B siRNA: 51±6.1%), GIPC-C siRNA: 52±9.6% Pooled siRNA: 34%. Two-tailed Student’s t-test: GIPC-A vs. negative control P=0.005 (* P<0.05), GIPC-B vs. negative control P=0.010 (* P<0.05), GIPC-C vs. negative control P=0.026 (* P<0.05). The number of AVC explants examined and cells in each category were as follows: Control (n=42; total number of cells in gel, 6432), n=number of explants. GIPC-A siRNA (n=40; total number of cells in gel, 3006). GIPC-B siRNA (n=42; total number of cells in gel, 3339). GIPC-C siRNA (n=44; total number of cells in gel, 3437). Pooled siRNA (n=14; total number of cells in gel, 963). C–D: GIPC is required for TGFβR3-mediated EMT. C: Ventricular endocardial cells were infected with an adenovirus expressing GFP alone or TGFβR3-FL and GFP, and control or targeted siRNA against GIPC was delivered. GIPC2-A siRNA (5nM) abolished the ability of TGFβR3-FL to cause TGFβ2 (200 pM)- mediated gain-of-function. GFP, TGFβ2, control siRNA: epithelial 77±1.7%; (mean±SEM), activated 11±1.3%, transformed 11±0.9%. GFP, TGFβ2, GIPC-A siRNA: epithelial 78±1.8%, activated 11±1.2%, transformed 11±1.0%. TGFβR3-FL, TGFβ2, control siRNA: epithelial 54±2.4%***, activated 11±1.3%, transformed 35±1.2%***. TGFβR3-FL, TGFβ2, GIPC-A siRNA: epithelial 69±1.1%, activated 12±1.1%, transformed 18±1.0%. Two-tailed Student’s t-tests: *P<0.05, **P<0.01, ***P<0.001 versus control. The number of ventricular explants examined and cells in each category were as follows: GFP control adenovirus, TGFβ2, control siRNA (n=28; total number of cells, 1006; epithelial, 779; activated, 114; transformed, 113), n=number of explants, GFP control adenovirus, TGFβ2, GIPC-A siRNA (n=27; total number of cells, 912; epithelial, 714; activated, 99; transformed, 99), TGFβR3, TGFβ2, control siRNA (n=26; total number of cells, 972; epithelial, 528; activated, 110; transformed, 334), TGFβR3, TGFβ2, GIPC-A siRNA (n=27; total number of cells, 974; epithelial, 674; activated, 119; transformed, 181).D: GIPC2-A siRNA (5nM) abolished BMP-2 (5 nM)- stimulated, TGFβR3-mediated ventricular endothelial cell transformation. GFP, BMP-2, control siRNA: epithelial 77±0.1%, activated 11±0.4%, transformed 12±0.5%. GFP, BMP-2, GIPC-A siRNA: epithelial 79±0.7%, activated 11±1.0%, transformed 10±0.5%. TGFβR3-FL, BMP-2, control siRNA: epithelial 52±0.5%***, activated 10±0.4%, transformed 38±0.2%***. TGFβR3-FL, BMP-2, GIPC-A siRNA: epithelial 83±1.8%, activated 7±1.0%, transformed 10±0.9%. The number of ventricular explants examined and cells in each category were as follows: GFP control adenovirus, BMP-2, control siRNA (n=24; total number of cells, 891; epithelial, 689; activated, 99; transformed, 103), n=number of explants, GFP control adenovirus, BMP-2, GIPC-A siRNA (n=24; total number of cells, 861; epithelial, 677; activated, 96; transformed, 88), TGFβR3, BMP-2, control siRNA (n=24; total number of cells, 850; epithelial, 443; activated, 86; transformed, 321), TGFβR3, BMP-2, GIPC-A siRNA (n=24; total number of cells, 644; epithelial, 473; activated, 75; transformed, 96).
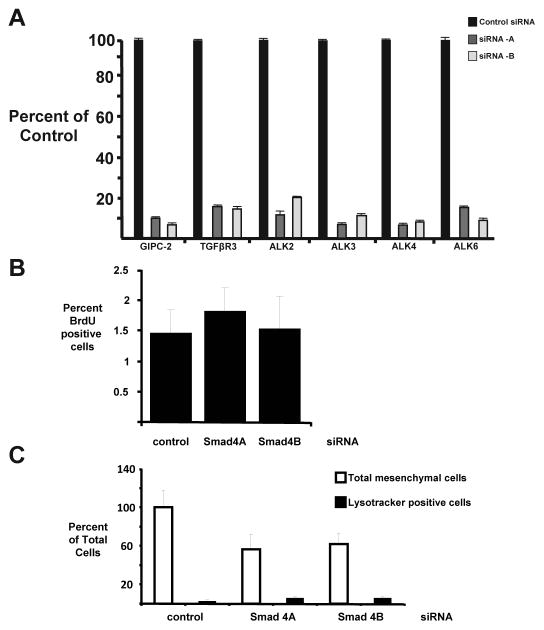
To directly test whether GIPC functions downstream of TGFβR3-mediated, TGFβ2- or BMP-2-stimulated EMT we used adenovirus to overexpress either GFP alone or TGFβR3-FL and GFP in ventricular endothelial cells, cultured ventricular explants in the presence of siRNA, and delivered ligand. The percentage of GFP positive transformed cells after infection with adenovirus expressing GFP alone defined basal levels. As expected, the overexpression of TGFβR3-FL and GFP led to a significant increase in the percentage of GFP positive transformed cells in response to either TGFβ2 (Fig 3C) or BMP-2 (Fig 3D) that was unaffected by control siRNA. However, the addition of 5 nM siRNA targeted against GIPC-2 abolished TGFβR3-mediated EMT in response to either TGFβ2 (Fig 3C) or BMP-2 (Fig 3D). Knockdown of GIPC-2, and all genes targeted by siRNA, was confirmed via RT-PCR (Fig 4A). Incubation of explants with siRNA did not alter cell proliferation rate as demonstrated by a representative experiment targeting Smad4 (Fig 4B) and no toxicity was seen with siRNA addition (Fig 4C). Taken together, these data demonstrate that GIPC is required for TGFβR3-mediated, ligand-dependent EMT.
Fig 4. Incubation of explants with siRNA is effective and nontoxic.
A Quantification of siRNA Knockdown. The levels of expression of GIPC, ALK2, ALK3, ALK4, and ALK6, with TGFβR3 as a positive control, were assayed by quantitative real time (RT)-PCR after inhibition of mRNA by siRNA treatment of CEFs. All specific siRNAs decreased mRNA levels of the target by >30%. The levels of expression of GIPC-2, TGFβR3, ALK2, ALK3, ALK4, and ALK6 were assayed by quantitative real time (RT)-PCR after inhibition of mRNA by siRNA treatment of CEFs. GIPC-2 mRNA expression level was significantly reduced by 84% and 93% after GIPC-2A and GIPC-2B and GIPC-2C siRNA treatment versus control siRNA treatment respectively. TGFβR3 mRNA expression level was significantly reduced by 84% and 93% after TGFβR32-A and TGFβR3-B siRNA treatment versus control siRNA treatment respectively. ALK2 mRNA expression level was significantly reduced by 84% and 93% after ALK2-A and ALK2-B siRNA treatment versus control siRNA treatment respectively. ALK3 mRNA expression level was significantly reduced by 84% and 93% after ALK3-A and ALK3-B siRNA treatment versus control siRNA treatment respectively. ALK4 mRNA expression level was significantly reduced by 84% and 93% after ALK4-A and ALK4-B siRNA treatment versus control siRNA treatment respectively. ALK6 mRNA expression level was significantly reduced by 84% and 93% after ALK6-A and ALK6-B siRNA treatment versus control siRNA treatment respectively. Methods were as described previously where ALK5 and Smad4 mRNA expression levels were previously shown to be significantly reduced by 95%, 86%, 95% and 86% by siRNA treatment with ALK5-A, ALK5-B, Smad4-A and Smad4-B siRNA treatment respectively. B–C: Lack of toxicity to siRNA in AVC explants. B: BrdU incorporation is unchanged between control (scrambled siRNA, n=12), Smad4A (n=11), and Smad4B siRNA (n=13). C: There are no significant differences in the percentage of lysotracker positive cells between control (scrambled siRNA, n=21, 2.8±1.4% lysotracker positive), Smad4A (n=22), and Smad4B siRNA (n=28).
3.3 TGFβR3-mediated endocardial cell EMT requires ALK2 and ALK3
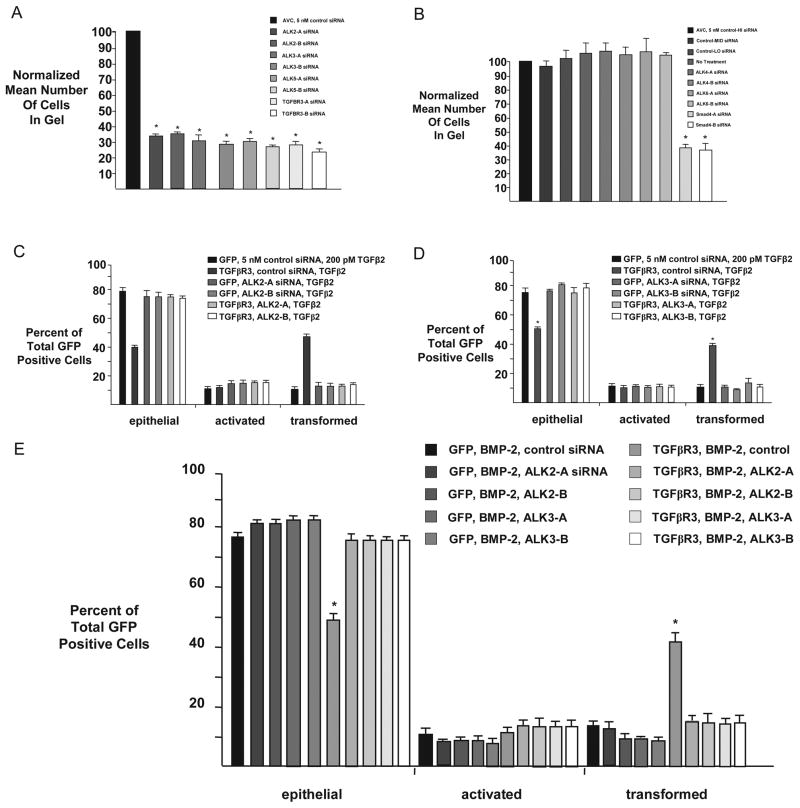
We had previously defined a role for ALK5 in TGFβR3-mediated endocardial cell EMT [20]. However, TGFβR3 also interacts with other ALKs, including ALK3 and ALK6[28]. Given the potential for several ALK’s to interact with TGFβR3, we assessed whether ALK2, ALK3, ALK 4, or ALK6 were required for endocardial cell EMT in vitro and if any of these ALKs act downstream of TGFβR3. Initially, we delivered siRNA constructs against ALK2, ALK3, ALK5 and TGFβR3 to AVC explants. As compared to control siRNA, two independent siRNA constructs to each target resulted in a 60–70% decrease in the number of transformed cells (Fig 5A). TGFβR3 and ALK5 knockdown served as a positive control for these experiments [18, 29]. In contrast siRNA constructs targeting either ALK4 or ALK6 did not alter EMT (Fig 5B) where Smad4 knockdown served as a positive control [29]. These data demonstrate that endocardial cell EMT requires ALK2 and ALK3, as well as ALK5, while ALK4 and ALK6 are dispensable for endocardial cell EMT.
Fig 5. ALK2/ALK3 activation is required for TGFBR3-mediated endocardial cell EMT.
A–B Quantification of cells from AVC explants in the collagen gel. Data are derived from three independent experiments normalized to control siRNA. A: siRNA to ALK2 and ALK3 inhibits transformation. AVC endocardial cells incubated with control siRNA transform on collagen gels, whereas siRNA targeted to ALK2, ALK3, ALK5 or TGFβR3 inhibits transformation. Control siRNA: Normalized to 100%. ALK2-A siRNA: 34±1.7%; (mean±SEM), ALK2-B siRNA: 36±1.0%, ALK3-A siRNA: 31±3.9%, ALK3-B siRNA: 28±0.9%, ALK5-A siRNA: 31±1.2%, ALK5-B siRNA: 27±0.5%, TGFβR3-A siRNA: 28±1.1%, TGFβR3-B siRNA: 24±1.0%. Two-tailed Student’s t-test (Control vs. Treatment) ALK2-A: P=0.0007 * (P<0.05), ALK2-B: P=0.0002 * (P<0.05), ALK3-A: P=0.003 * (P<0.05), ALK3-B: P=0.0002 * (P<0.05), ALK5-A: P=0.0003 * (P<0.05), ALK5-B: P=4.2e-05 * (P<0.05), TGFβR3-A: P=0.0002 * (P<0.05), TGFβR3-B: P=0.0002 * (P<0.05). The number of AVC explants examined and cells in each category were as follows: Control (n=30; total number of cells in gel, 4694), n=number of explants. ALK2-A (n=30; total number of cells in gel, 1604). ALK2-B (n=30; total number of cells in gel, 1710). ALK3-A (n=30; total number of cells in gel, 1474). ALK3-B (n=29; total number of cells in gel, 1292). ALK5-A (n=30; total number of cells in gel, 1463). ALK5-B (n=30; total number of cells in gel, 1284). TGFβR3-A (n=30; total number of cells in gel, 1323). TGFβR3-B (n=30; total number of cells in gel, 1117). B: ALK4 and ALK6 are dispensable for endocardial cell EMT. Quantification of cells in the collagen gel. Data are derived from three independent experiments normalized to control (HI) siRNA. Endocardial cells from AVC explants given either no treatment or any of 3 independent scrambled control siRNAs with varying GC content (high (HI), medium (MID), low (LO)) transform on collagen gels, as well as explants given siRNA targeted against ALK4 or ALK6. siRNA targeted against Smad4, serving as a positive control, inhibits AVC endocardial cell transformation. Control-HI siRNA: Normalized to 100%. Control-MID siRNA: 97±4.3%; (mean±SEM), Control-LO siRNA: 103±6.0%, No Treatment: 106±7.7%, ALK4-A siRNA: 107±6.5%, ALK4-B siRNA: 103±4.8%, ALK6-A siRNA: 107±10.5%, ALK6-B siRNA: 104±1.7%, Smad4-A siRNA: 39±2.8%, Smad4-B siRNA: 36±3.7%. Two-tailed Student’s t-test (Control-HI vs. Treatment) Control-MED: P=0.549, Control-LO: P=0.706, No Treatment: P=0.528, ALK4-A: P=0.408, ALK4-B: P=0.563, ALK6-A: P=0.553, ALK6-B: P=0.123, Smad4-A: P=0.002 * (P<0.05), Smad4-B: P=0.003 * (P<0.05). The number of AVC explants examined and cells in each category were as follows: Control-HI (n=24; total number of cells in gel, 3676), n=number of explants. Control-MED (n=24; total number of cells in gel, 3557). Control-LO (n=24; total number of cells in gel, 3763). No Treatment (n=24; total number of cells in gel, 3880). ALK4-A (n=24; total number of cells in gel, 3915). ALK4-B (n=24; total number of cells in gel, 3790). ALK6-A (n=24; total number of cells in gel, 3936). ALK6-B (n=24; total number of cells in gel, 3834). Smad4-A (n=24; total number of cells in gel, 1415). Smad4-B (n=25; total number of cells in gel, 1391). C, D: Average percent of total GFP-expressing cells scored as epithelial, activated or transformed. Means are derived from 3 separate experiments. GFP adenovirus alone served as a negative control to define basal levels of transformed cells. C: ALK2 is required for TGFβ2 stimulated, TGFβR3-mediated ventricular endocardial cell EMT. All explants incubated with TGFβ2 (200 pM). GFP adenovirus was used to define basal levels of transformation. TGFβR3-FL plus control siRNA significantly increased transformation. This effect is abolished by either of two independent siRNA constructs targeted to ALK2. GFP, TGFβ2, control siRNA: epithelial 79±2.6%; (mean±SEM), activated 11±1.2%, transformed 10±1.5%. TGFβR3-FL, TGFβ2, control siRNA: epithelial 40±0.7%***, activated 12±1.6%, transformed 47±1.1%***. GFP, TGFβ2, ALK2-A siRNA: epithelial 74±1.4%, activated 14±1.0%, transformed 12±0.5%. GFP, TGFβ2, ALK2-B siRNA: epithelial 73±1.3%, activated 14±0.6%, transformed 13±1.1%. TGFβR3-FL, TGFβ2, ALK2-A siRNA: epithelial 75±4.7%, activated 13±2.3%, transformed 12±2.4%. TGFβR3-FL, TGFβ2, ALK2-B siRNA: epithelial 74±4.1%, activated 14±2.1%, transformed 12±2.0%. Two-tailed Student’s t-tests: *P<0.05, **P<0.01, ***P<0.001 versus control. The number of ventricular explants examined and cells in each category were as follows: GFP, TGFβ2, control siRNA (n=24; total number of cells, 1504; epithelial, 1184; activated, 167; transformed, 153), n=number of explants, TGFβR3-FL, TGFβ2, control siRNA (n=23; total number of cells, 711; epithelial, 287; activated, 87; transformed, 337), GFP, TGFβ2, ALK2-A siRNA (n=24; total number of cells, 1285; epithelial, 952; activated, 181; transformed, 152), GFP, TGFβ2, ALK2-B siRNA (n=25; total number of cells, 1085; epithelial, 790; activated, 154; transformed, 141), TGFβR3-FL, TGFβ2, ALK2-A siRNA (n=23; total number of cells, 657; epithelial, 506; activated, 80; transformed, 71), TGFβR3-FL, TGFβ2, ALK2-B siRNA (n=23; total number of cells, 569; epithelial, 421; activated, 79; transformed, 69).D: ALK3 is required for TGFβ2 stimulated, TGFβR3-mediated ventricular endocardial cell EMT. In experiments similar to C, the addition of two independent siRNA constructs targeted against ALK3 blocked transformation. GFP, TGFβ2, control siRNA: epithelial 75±3.8%, activated 13±2.1%, transformed 12±2.1%. TGFβR3-FL, TGFβ2, control siRNA: epithelial 51±0.7%***, activated 10±1.7%, transformed 40±1.6%***. GFP, TGFβ2, ALK3-A siRNA: epithelial 74±4.9%, activated 12±1.8%, transformed 14±3.1%. GFP, TGFβ2, ALK3-B siRNA: epithelial 78±3.2%, activated 11±1.3%, transformed 11±2.0%. TGFβR3-FL, TGFβ2, ALK3-A siRNA: epithelial 77±0.6%, activated 12±0.8%, transformed 11±1.1%. TGFβR3-FL, TGFβ2, ALK3-B siRNA: epithelial 81±1.2%, activated 10±1.3%, transformed 9±0.2%. The number of ventricular explants examined and cells in each category were as follows: GFP, TGFβ2, control siRNA (n=23; total number of cells, 780; epithelial, 585; activated, 98; transformed, 97), n=number of explants, TGFβR3-FL, TGFβ2, control siRNA (n=22; total number of cells, 786; epithelial, 397; activated, 75; transformed, 314), GFP, TGFβ2, ALK3-A siRNA (n=24; total number of cells, 942; epithelial, 701; activated, 113; transformed, 128), GFP, TGFβ2, ALK3-B siRNA (n=24; total number of cells, 1062; epithelial, 831; activated, 120; transformed, 111), TGFβR3-FL, TGFβ2, ALK3-A siRNA (n=24; total number of cells, 693; epithelial, 531; activated, 84; transformed, 78), TGFβR3-FL, TGFβ2, ALK2-B siRNA (n=24; total number of cells, 540; epithelial, 439; activated, 53; transformed, 48).E: ALK2 and ALK3 are required for BMP-2 stimulated, TGFβR3-mediated ventricular endocardial cell EMT. All explants were given BMP-2 (5 nM). The addition of two independent siRNA constructs targeted against ALK2 or ALK3 blocked transformation. GFP, BMP-2, control siRNA: epithelial 77±1.2%, activated 11±1.3%, transformed 12±0.4%. GFP, BMP-2, ALK2-A siRNA: epithelial 81±0.9%, activated 9±0.1%, transformed 11±1.0%. GFP, BMP-2, ALK2-B siRNA: epithelial 81±0.7%, activated 10±0.2%, transformed 9±0.9%. GFP, BMP-2, ALK3-A siRNA: epithelial 82±0.4%, activated 9±0.4%, transformed 9±0.1%. GFP, BMP-2, ALK3-B siRNA: epithelial 82±0.2%, activated 8±0.4%, transformed 9±0.2%. TGFβR3-FL, BMP-2, control siRNA: epithelial 49±1.9%***, activated 10±0.4%, transformed 42±2.2%***. TGFβR3-FL, BMP-2, ALK2-A siRNA: epithelial 75±1.6%, activated 12±0.6%, transformed 13±1.0%. TGFβR3-FL, BMP-2, ALK2-B siRNA: epithelial 75±0.6%, activated 12±1.4%, transformed 13±1.6%. TGFβR3-FL, BMP-2, ALK3-A siRNA: epithelial 75±0.4%, activated 12±0.8%, transformed 13±0.4%. TGFβR3-FL, BMP-2, ALK3-B siRNA: epithelial 75±0.8%, activated 12±1.1%, transformed 13±1.3%. The number of ventricular explants examined and cells in each category were as follows: GFP, BMP-2, control siRNA (n=24; total number of cells, 778; epithelial, 603; activated, 83; transformed, 92), n=number of explants, GFP, BMP-2, ALK2-A siRNA (n=24; total number of cells, 862; epithelial, 694; activated, 76; transformed, 92), GFP, BMP-2, ALK2-B siRNA (n=24; total number of cells, 911; epithelial, 741; activated, 88; transformed, 82), GFP, BMP-2, ALK3-A siRNA (n=24; total number of cells, 895; epithelial, 729; activated, 82; transformed, 84), GFP, BMP-2, ALK3-B siRNA (n=24; total number of cells, 908; epithelial, 747; activated, 77; transformed, 84), TGFβR3-FL, BMP-2, control siRNA (n=24; total number of cells, 843; epithelial, 410; activated, 80; transformed, 353, TGFβR3-FL, BMP-2, ALK2-A siRNA (n=24; total number of cells, 495; epithelial, 372; activated, 59; transformed, 64), TGFβR3-FL, BMP-2, ALK2-B siRNA (n=24; total number of cells, 529; epithelial, 399; activated, 62; transformed, 68), TGFβR3-FL, BMP-2, ALK3-A siRNA (n=24; total number of cells, 503; epithelial, 378; activated, 61; transformed, 64), TGFβR3-FL, BMP-2, ALK3-B siRNA (n=24; total number of cells, 508; epithelial, 382; activated, 60; transformed, 66).
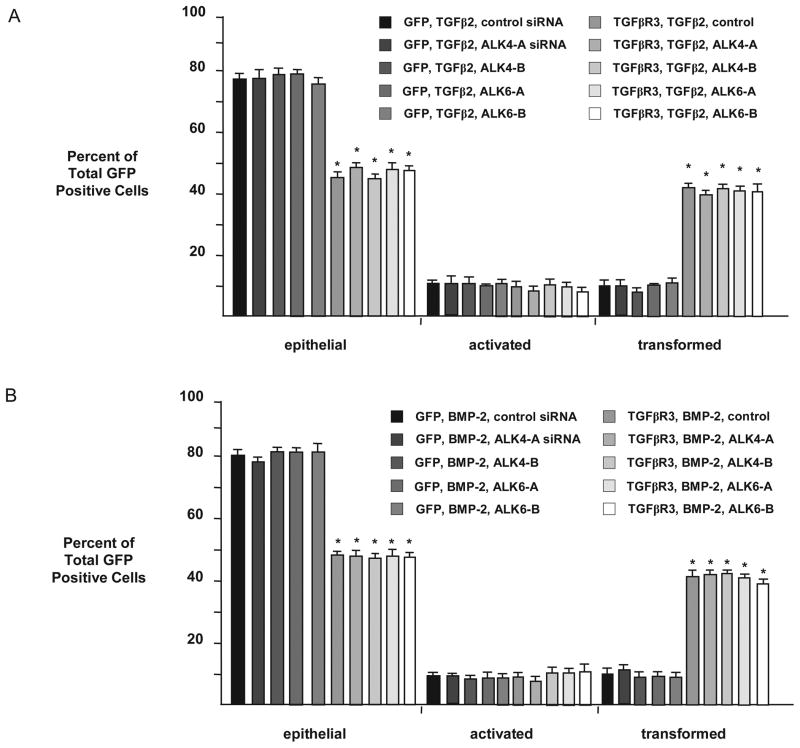
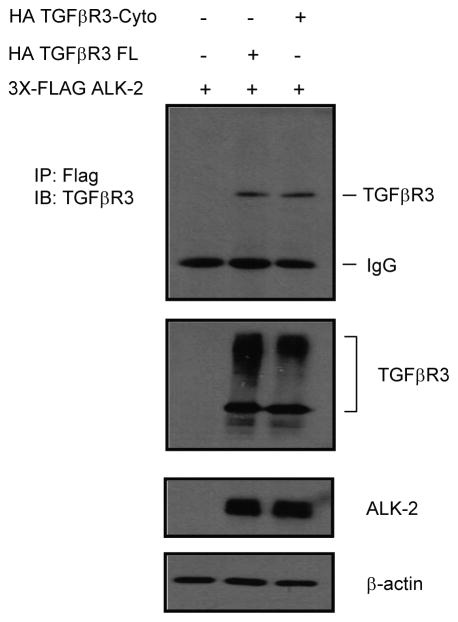
We next explored directly the requirement of these molecules for TGFβR3-mediated EMT. The overexpression of TGFβR3-FL and GFP in ventricular endocardial cells led to a significant increase in the percentage of transformed cells in response to either TGFβ2 (Fig 5C–D) or BMP-2 (Fig 5E). The presence of control siRNA did not alter these percentages, however the addition of 5 nM siRNA targeted against either ALK2 or ALK3 abolished TGFβR3-mediated EMT in response to TGFβ2 (Fig 5C, D) or BMP-2 (Fig 5E). The addition of siRNA targeted against either ALK4 or ALK6 had no effect on TGFβR3-mediated ventricular endocardial cell EMT when stimulated by TGFβ2 (Fig 6A) or BMP-2 (Fig 6B). Both ALK3 and ALK6 have been demonstrated to interact with TGFβR3. However similar data for ALK2 is not available. Therefore we directly tested for the ability of ALK2 to interact with TGFβR3 [30]. When co-expressed in COS cells, ALK2 co-immunoprecipitated with flagged tagged TGFβR3-FL demonstrating that ALK2 can interact with TGFβR3 (Fig 7). These data collectively suggest that ALK2 and ALK3, in addition to ALK5 ([18, 23]), are required for TGFβR3-mediated EMT.
Fig 6. ALK4 and ALK6 are dispensable for TGFβR3-mediated ventricular endocardial cell EMT.
Average percent of total GFP-expressing cells scored as epithelial, activated or transformed. Means are derived from 3 separate experiments. A: All explants were given TGFβ2 (200 pM). GFP served as a negative control to determine the basal level of transformation. TGFβR3 induced statistically significant increases in transformed cells with a concomitant decrease in epithelial cells. The addition of two independent siRNA constructs targeted against either ALK4 or ALK6 had no effect on EMT versus control siRNA. GFP, TGFβ2, control siRNA: epithelial 77±1.4%, activated 12±0.4%, transformed 11±0.9%. GFP, TGFβ2, ALK4-A siRNA: epithelial 77±3.3%, activated 12±2.1%, transformed 11±1.3%. GFP, TGFβ2, ALK4-B siRNA: epithelial 78±1.4%, activated 12±0.7%, transformed 10±1.3%. GFP, TGFβ2, ALK6-A siRNA: epithelial 78±0.4%, activated 11±0.5%, transformed 11±0.1%. GFP, TGFβ2, ALK6-B siRNA: epithelial 75±1.5%, activated 12±1.0%, transformed 12±0.5%. TGFβR3-FL, TGFβ2, control siRNA: epithelial 45±2.0%**, activated 11±1.5%, transformed 44±0.6%***. TGFβR3-FL, TGFβ2, ALK4-A siRNA: epithelial 50±0.5%***, activated 12±1.1%, transformed 41±0.6%***. TGFβR3-FL, TGFβ2, ALK4-B siRNA: epithelial 45±0.9%***, activated 12±1.3%, transformed 43±0.4%***. TGFβR3-FL, TGFβ2, ALK6-A siRNA: epithelial 48±1.5%***, activated 11±0.6%, transformed 42±1.3%***. TGFβR3-FL, TGFβ2, ALK6-B siRNA: epithelial 47±0.9%***, activated 10±1.0%, transformed 42±1.9%***. The number of ventricular explants examined and cells in each category were as follows: GFP, TGFβ2, control siRNA (n=24; total number of cells, 956; epithelial, 737; activated, 111; transformed, 108), n=number of explants, GFP, TGFβ2, ALK4-A siRNA (n=23; total number of cells, 866; epithelial, 664; activated, 103; transformed, 99), GFP, TGFβ2, ALK4-B siRNA (n=24; total number of cells, 912; epithelial, 713; activated, 111; transformed, 88), GFP, TGFβ2, ALK6-A siRNA (n=24; total number of cells, 897; epithelial, 702; activated, 97; transformed, 98), GFP, TGFβ2, ALK6-B siRNA (n=24; total number of cells, 987; epithelial, 743; activated, 124; transformed, 120), TGFβR3-FL, TGFβ2, control siRNA (n=23; total number of cells, 957; epithelial, 431; activated, 107; transformed, 419, TGFβR3-FL, TGFβ2, ALK4-A siRNA (n=24; total number of cells, 966; epithelial, 476; activated, 93; transformed, 397), TGFβR3-FL, TGFβ2, ALK4-B siRNA (n=24; total number of cells, 943; epithelial, 426; activated, 108; transformed, 409), TGFβR3-FL, TGFβ2, ALK6-A siRNA (n=24; total number of cells, 1009; epithelial, 481; activated, 105; transformed, 423), TGFβR3-FL, TGFβ2, ALK6-B siRNA (n=24; total number of cells, 1008; epithelial, 478; activated, 104; transformed, 426). B: All explants were given BMP-2 (5 nM). GFP served as a negative control to determine the basal level of transformation. TGFβR3 induced statistically significant increases in transformed cells with a concomitant decrease in epithelial cells. The addition of two independent siRNA constructs targeted against either ALK4 or ALK6 had no effect on EMT versus control siRNA. GFP, BMP-2, control siRNA: epithelial 80±1.2%, activated 9±0.2%, transformed 11±1.0%. GFP, BMP-2, ALK4-A siRNA: epithelial 78±0.7%, activated 10±0.1%, transformed 12±0.8%. GFP, BMP-2, ALK4-B siRNA: epithelial 81±0.8%, activated 9±0.8%, transformed 10±0.4%. GFP, BMP-2, ALK6-A siRNA: epithelial 81±0.6%, activated 9±0.6%, transformed 10±0.1%. GFP, BMP-2, ALK6-B siRNA: epithelial 81±1.6%, activated 9±0.6%, transformed 10±1.1%. TGFβR3-FL, BMP-2, control siRNA: epithelial 48±0.5%***, activated 10±0.3%, transformed 42±0.1%***. TGFβR3-FL, BMP-2, ALK4-A siRNA: epithelial 48±0.7%***, activated 9±1.1%, transformed 43±1.7%***. TGFβR3-FL, BMP-2, ALK4-B siRNA: epithelial 47±0.8%***, activated 11±1.0%, transformed 43±0.3%***. TGFβR3-FL, BMP-2, ALK6-A siRNA: epithelial 48±1.2%**, activated 11±0.2%, transformed 41±1.0%**. TGFβR3-FL, BMP-2, ALK6-B siRNA: epithelial 48±0.9%***, activated 12±1.2%, transformed 40±2.0%**. The number of ventricular explants examined and cells in each category were as follows: GFP, BMP-2, control siRNA (n=23; total number of cells, 763; epithelial, 609; activated, 71; transformed, 83), n=number of explants, GFP, BMP-2, ALK4-A siRNA (n=24; total number of cells, 873; epithelial, 684; activated, 88; transformed, 101), GFP, BMP-2, ALK4-B siRNA (n=24; total number of cells, 928; epithelial, 752; activated, 87; transformed, 89), GFP, BMP-2, ALK6-A siRNA (n=24; total number of cells, 864; epithelial, 696; activated, 81; transformed, 87), GFP, BMP-2, ALK6-B siRNA (n=24; total number of cells, 873; epithelial, 706; activated, 78; transformed, 89), TGFβR3-FL, BMP-2, control siRNA (n=23; total number of cells, 914; epithelial, 437; activated, 89; transformed, 388, TGFβR3-FL, BMP-2, ALK4-A siRNA (n=24; total number of cells, 882; epithelial, 423; activated, 83; transformed, 376), TGFβR3-FL, BMP-2, ALK4-B siRNA (n=24; total number of cells, 872; epithelial, 409; activated, 92; transformed, 371), TGFβR3-FL, BMP-2, ALK6-A siRNA (n=24; total number of cells, 895; epithelial, 426; activated, 99; transformed, 370), TGFβR3-FL, BMP-2, ALK6-B siRNA (n=24; total number of cells, 871; epithelial, 418; activated, 106; transformed, 347).
Fig. 7. ALK2 associates with TGFβR3.
Immunoprecipitations performed in COS7 cells transfected with empty vector (pcDNA 3.1) + 3X-Flag ALK2, TGFβR3-Fl + 3X-Flag ALK2 or TGFβR3-Δ3 + 3x-Flag ALK2 using anti-FLAG antibody, and analyzed by immunoblotting using anti-TGFβR3. Total cell lysates were immunoblotted with either anti-TGF-βR3, anti-Flag, or β-actin antibodies to serve as loading controls.
4.0 Discussion
Here we demonstrate that although the cytoplasmic domain of TGFβR3 is not required for ligand presentation to TGFβR2 and augmentation of TGFβ signaling via TGFβR2/TGFβR1 [6], this domain is required for endocardial cell EMT. Further analysis revealed that loss of the 3 C-terminal amino acids of the cytoplasmic domain phenocopy loss of the entire cytoplasmic domain. Since GIPC is known to bind to the 3 C-terminal amino acids of TGFβR3 and stabilizes it on the plasma membrane [24], this identified GIPC as a putative downstream mediator of TGFβR3 function. We demonstrate that GIPC is required for AVC endothelial cell transformation and specifically TGFβR3-mediated endothelial cell EMT in response to either TGFβ2 or BMP-2. As BMP-2 or TGFβ2 stimulated, TGFβR3-dependent EMT requires both the C-terminal amino acids of TGFβR3 and GIPC, these data support a model in which TGFβ2 and BMP-2 binding to TGFβR3 activates a common downstream pathway that results in endothelial cell EMT.
Although we demonstrate a requirement for GIPC in TGFβR3-mediated endothelial cell EMT, the role of TGFβR3/GIPC interaction in regulating cell behavior is not fully understood. TGFβR3 signaling has been implicated in the regulation of proliferation, migration and adhesion of several cancer cell lines (reviewed in [31]). However, unlike endocardial cell EMT, data in cancer cell lines suggest that TGFβR3 inhibits cell migration [32] as the loss of expression of TGFβR3 is common in human breast, ovarian, pancreatic, prostate and non-small cell lung cancers [33–37]. Further, it has been shown that TGFβR3 suppressed breast cancer progression through GIPC-mediated inhibition of TGFβ signaling [38]. It should be noted that unlike data in cancer cell lines [39], there is no apparent role for Cdc42 in mediating migration or invasion in endocardial cells [29], suggesting that these developmental and pathophysiologic pathways downstream of TGFβR3 may be divergent. These data collectively suggest that interaction of GIPC and TGFβR3 has important developmental and pathophysiologic implications and that the balance of GIPC/TGFβR3, as well as the physiologic context at which TGFβR3 signaling occurs, may regulate distinct aspects of TGFβR3 signaling. In addition to GIPC, ALK5 is required for TGFβR3-mediated EMT stimulated by either TGFβ2 or BMP-2, although ALK5 is not sufficient for endocardial cell EMT [19]. ALK5 is known to play a role in the activation of the Par6/Smurf1/RhoA pathway where Par6 functions downstream of TGFβ to recruit Smurf1, an E3 ubiquitin ligase, to target RhoA for degradation to control apical-basal polarity and tight junction dissolution [40]. We have recently shown that this pathway is operative in the regulation of endocardial cell EMT [18, 23] and our current studies suggest that both TGFβ2 and BMP-2 may access this pathway to cause the disassembly of tight junctions via ALK5 after binding TGFβR3.
Several ALKs, in addition to ALK5, have been shown to mediate endocardial cell EMT. We have previously shown that ALK2 is required and sufficient for endocardial cell EMT in vitro [19] [19, 41] while others have demonstrated that both ALK2 [42] and ALK3 [43, 44] are required for endocardial cell EMT in vivo. In humans, a dominant negative form of ALK2 has been associated in a patient with AVC defects [45] and a second mutant form has been described in a Down’s Syndrome patient with congenital heart defects [46] highlighting the importance of ALK2 signaling in human cardiac cushion morphogenesis. We therefore used a siRNA knockdown approach to test for the requirement of several ALKs downstream of TGFβ2 or BMP-2 stimulated, TGFβR3-mediated endocardial cell EMT. Using this approach we demonstrated a requirement for ALK2 and ALK3 downstream of TGFβR3, while targeting of ALK4 and ALK6 had no effect on TGFβR3-mediated EMT. Previous data has shown that ALK3 and ALK6 can interact with TGFβR3 [28]. Here we show directly that ALK2 can interact with TGFβR3 although the C-terminal amino acids of TGFβR3 are not required for this interaction. Our data suggesting that ALK6 is not required for AVC endocardial cell EMT is in contrast to reports demonstrating that dominant negative ALK6 inhibits endocardial cell EMT and constitutively active ALK6 stimulates endocardial cell EMT [47, 48]. We cannot demonstrate the presence of ALK6 in AVC endocardial cells in the developing heart either by in situ hybridization or RT-PCR of RNA isolated from AVC explants and we suggest that the results obtained by the previous study might be the consequence of off target effects of dominant negative and constitutively active ALK6 when introduced into endothelial cells. Taken together, our experiments support a model whereby TGFβR3, through interactions between the cytoplasmic domain of the receptor and GIPC, activates several ALKs that support endothelial cell EMT.
The observation that ALK’s associated with both TGFβ and BMP signaling that may activate either Smads 2, 3 or Smads 1, 5, 8 are required for endocardial cell EMT is consistent with prior studies of Smad signaling. A requirement for Smad signaling in endocardial cell EMT was demonstrated by targeting Smad4, the common mediator Smad, in AVC explant assays[20], a result confirmed in mice[49]. Experiments targeting the receptor-regulated Smads (1, 2, 3, & 5) demonstrated that all are required for EMT [20]. However, overexpression of Smad1 or Smad3 does not induce EMT in ventricular endocardial cells suggesting that Smad signaling, although required for endocardial cell EMT, is not sufficient for EMT. The role of the inhibitory Smad, Smad6, has also been examined in endocardial cell EMT and valve formation. Smad6 null mice have valvular hyperplasia that is consistent with either enhanced EMT or mesenchymal cell proliferation in the cushions [50]. Experiments in the chick revealed that overexpression of Smad6 in the AVC decreased EMT [19]. Since ALK2 activates Smad1 and Smad6 blocks Smad1 signaling [51], these data are consistent with the known role of ALK218, 40 and Smad1[20] in endocardial cell EMT [19, 42]. Overall, these data indicate that the coordinated activation of several ALK’s and their respective downstream Smads are necessary for endocardial cell EMT.
The finding that BMP-2 signals endocardial cell EMT via TGFβR3 suggests a re-evaluation of the actions of BMP-2 with respect to which receptors are signaling BMP-2-mediated responses is required. Targeting of BMP-2 in the mouse embryo abrogates EMT in vivo [52]. BMP-2 induces endocardial cell EMT in vitro [53], accompanied by the expression of twist and Id1, as well as the expression of the marker of mesenchymal cell maturation in the valves, periostin [47]. BMP-2 has also been reported to induce the expression of TGFβ2 [47] demonstrating a possible sequential mode of action of these growth factors where TGFβ2 may regulate later stages of EMT and mesenchymal cell maturation. This role for TGFβ2 is consistent with the cushion phenotype seen in Tgfb2 null mice which is characterized by altered EMT and mesenchymal cell maturation [54–56]. Although, the relative contribution of TGFβR3 or the canonical BMP and TGFβ receptors to these BMP-2 and TGFβ2 dependent events is currently not known, our data suggests a role for TGFβR3 downstream of BMP-2 and TGFβ2 in mediating both early and late events in valve development.
Highlights.
TGFβ2 and BMP-2 signals endothelial transformation via TGFβ2R3
TGFβ2R3 requires GIPC to signal in response to each ligand
The Type I Receptors, ALK2, ALK3, and ALK5 are also required for TGFβ2R3 signaling
Acknowledgments
The authors thank Dr. Andries Ziljstra and Tyson Foods, Inc. for chicken eggs and Dr. Christopher B. Brown for critical review of the manuscript. Overall project support via Systems based Consortium for Organ Design and Engineering, HL09551. T.A.T. was supported via GM007628 & HL09551, G.C.B. via CA135006 and CA136786, JYR via GM062459 & HL09551, and J.V.B. via HL09551.
Abbreviations
- ALK
Activin Receptor-Like Kinases
- AVC
Atrioventricular Endocardial Cushion
- BMP
Bone Morphogenetic Protein
- EMT
Epithelial-Mesenchymal Transformation
- GFP
Green Fluorescent Protein
- GIPC
GAIP-interacting protein, C terminus
- TGFβ
Transforming Growth Factor Beta
- TGFβR1
Type I TGFβ receptor
- TGFβR2
Type II TGFβ receptor
- TGFβR3
Type III TGFβ receptor
- TGFβR3-FL
Type III TGFβ receptor-full length
- TGFβR3-CYTO
Type III TGFβ receptor-lacking the entire cytoplasmic domain
- TGFβR3
Type III TGFβ receptor-lacking the 3 C-terminal amino acids
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Todd A. Townsend, Email: Todd.Townsend@Vanderbilt.edu.
Jamille Y. Robinson, Email: Jamille.Robinson@Vanderbilt.edu.
Tam How, Email: How00001@mc.duke.edu.
Daniel M. DeLaughter, Email: Daniel.M.Delaughter@Vanderbilt.edu.
Gerard C. Blobe, Email: Blobe001@mc.duke.edu.
References
- 1.Jessell TM, Melton DA. Cell. 1992;68:257–270. doi: 10.1016/0092-8674(92)90469-s. [DOI] [PubMed] [Google Scholar]
- 2.Kretzschmar M, Massague J. Curr Opin Genet Dev. 1998;8:103–111. doi: 10.1016/s0959-437x(98)80069-5. [DOI] [PubMed] [Google Scholar]
- 3.Lopez-Casillas F, Cheifetz S, Doody J, Andres JL, Lane WS, Massague J. Cell. 1991;67:785–795. doi: 10.1016/0092-8674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- 4.Wang XF, Lin HY, Ng-Eaton E, Downward J, Lodish HF, Weinberg RA. Cell. 1991;67:797–805. doi: 10.1016/0092-8674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- 5.Cheifetz S, Bellon T, Cales C, Vera S, Bernabeu C, Massague J, Letarte M. J Biol Chem. 1992;267:19027–19030. [PubMed] [Google Scholar]
- 6.Blobe GC, Schiemann WP, Pepin MC, Beauchemin M, Moustakas A, Lodish HF, O’Connor-McCourt MD. J Biol Chem. 2001;276:24627–24637. doi: 10.1074/jbc.M100188200. [DOI] [PubMed] [Google Scholar]
- 7.Kirkbride KC, Townsend TA, Bruinsma MW, Barnett JV, Blobe GC. J Biol Chem. 2008:M704883200. doi: 10.1074/jbc.M704883200. [DOI] [PubMed] [Google Scholar]
- 8.Wiater E, Harrison CA, Lewis KA, Gray PC, Vale WW. J Biol Chem. 2006;281:17011–17022. doi: 10.1074/jbc.M601459200. [DOI] [PubMed] [Google Scholar]
- 9.Brown CB, Boyer AS, Runyan RB, Barnett JV. Science. 1999;283:2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 10.Barnett JV, Desgrosellier JS. Birth Defects Res Part C Embryo Today. 2003;69:58–72. doi: 10.1002/bdrc.10006. [DOI] [PubMed] [Google Scholar]
- 11.Compton LA, Potash DA, Brown CB, Barnett JV. Circ Res. 2007;101:784–791. doi: 10.1161/CIRCRESAHA.107.152082. [DOI] [PubMed] [Google Scholar]
- 12.Delaughter DM, Saint-Jean L, Baldwin HS, Barnett JV. Birth Defects Res A Clin Mol Teratol. 2011;91:511–525. doi: 10.1002/bdra.20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirkbride KC, Townsend TA, Bruinsma MW, Barnett JV, Blobe GC. J Biol Chem. 2008;283:7628–7637. doi: 10.1074/jbc.M704883200. [DOI] [PubMed] [Google Scholar]
- 14.Goumans MJ, Valdimarsdottir G, Itoh S, Lebrin F, Larsson J, Mummery C, Karlsson S, ten Dijke P. Mol Cell. 2003;12:817–828. doi: 10.1016/s1097-2765(03)00386-1. [DOI] [PubMed] [Google Scholar]
- 15.Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Embo J. 2002;21:1743–1753. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward SM, Desgrosellier JS, Zhuang X, Barnett JV, Galper JB. J Biol Chem. 2002;277:50183–50189. doi: 10.1074/jbc.M209668200. [DOI] [PubMed] [Google Scholar]
- 17.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Townsend TA, Wrana JL, Davis GE, Barnett JV. J Biol Chem. 2008;283:13834–13841. doi: 10.1074/jbc.M710607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desgrosellier JS, Mundell NA, McDonnell MA, Moses HL, Barnett JV. Dev Biol. 2005;280:201–210. doi: 10.1016/j.ydbio.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 20.Townsend TA, Robinson JY, Deig CR, Hill CR, Misfeldt A, Blobe GC, Barnett JV. Cells Tissues Organs. 2011;194:1–12. doi: 10.1159/000322035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bushdid PB, Chen CL, Brantley DM, Yull F, Raghow R, Kerr LD, Barnett JV. Dev Biol. 2001;237:107–115. doi: 10.1006/dbio.2001.0356. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Townsend TA, Robinson JY, Deig CR, Hill CR, Misfeldt A, Blobe GC, Barnett JV. Cells Tissues Organs. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blobe GC, Liu X, Fang SJ, How T, Lodish HF. J Biol Chem. 2001;276:39608–39617. doi: 10.1074/jbc.M106831200. [DOI] [PubMed] [Google Scholar]
- 25.Brown CB, Drake CJ, Barnett JV. Dev Dyn. 1999;215:79–85. doi: 10.1002/(SICI)1097-0177(199905)215:1<79::AID-DVDY9>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 26.Lencinas A, Broka DM, Konieczka JH, Klewer SE, Antin PB, Camenisch TD, Runyan RB. Toxicol Sci. 2010;116:273–285. doi: 10.1093/toxsci/kfq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh M. Int J Mol Med. 2002;9:585–589. [PubMed] [Google Scholar]
- 28.Lee NY, Kirkbride KC, Sheu RD, Blobe GC. Mol Biol Cell. 2009;20:4362–4370. doi: 10.1091/mbc.E09-07-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Townsend TA, Robinson JY, Deig CR, Hill CR, Misfeldt A, Blobe GC, Barnett JV. Cells Tissues Organs. doi: 10.1159/000322035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee NY, Haney JC, Sogani J, Blobe GC. Faseb J. 2009;23:3712–3721. doi: 10.1096/fj.09-131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatza CE, Oh SY, Blobe GC. Cell Signal. 22:1163–1174. doi: 10.1016/j.cellsig.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert KE, Huang H, Mythreye K, Blobe GC. Mol Biol Cell. 22:1463–1472. doi: 10.1091/mbc.E10-11-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong M, How T, Kirkbride KC, Gordon KJ, Lee JD, Hempel N, Kelly P, Moeller BJ, Marks JR, Blobe GC. J Clin Invest. 2007;117:206–217. doi: 10.1172/JCI29293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hempel N, How T, Dong M, Murphy SK, Fields TA, Blobe GC. Cancer Res. 2007;67:5231–5238. doi: 10.1158/0008-5472.CAN-07-0035. [DOI] [PubMed] [Google Scholar]
- 35.Gordon KJ, Blobe GC. Biochim Biophys Acta. 2008;1782:197–228. doi: 10.1016/j.bbadis.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Turley RS, Finger EC, Hempel N, How T, Fields TA, Blobe GC. Cancer Res. 2007;67:1090–1098. doi: 10.1158/0008-5472.CAN-06-3117. [DOI] [PubMed] [Google Scholar]
- 37.Finger EC, Lee NY, You HJ, Blobe GC. J Biol Chem. 2008;283:34808–34818. doi: 10.1074/jbc.M804741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JD, Hempel N, Lee NY, Blobe GC. Carcinogenesis. 31:175–183. doi: 10.1093/carcin/bgp271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mythreye K, Blobe GC. Cell Cycle. 2009;8:3069–3070. doi: 10.4161/cc.8.19.9419. [DOI] [PubMed] [Google Scholar]
- 40.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 41.Lai YT, Beason KB, Brames GP, Desgrosellier JS, Cleggett MC, Shaw MV, Brown CB, Barnett JV. Dev Biol. 2000;222:1–11. doi: 10.1006/dbio.2000.9698. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Sridurongrit S, Dudas M, Thomas P, Nagy A, Schneider MD, Epstein JA, Kaartinen V. Dev Biol. 2005;286:299–310. doi: 10.1016/j.ydbio.2005.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song L, Fassler R, Mishina Y, Jiao K, Baldwin HS. Dev Biol. 2007;301:276–286. doi: 10.1016/j.ydbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Park C, Lavine K, Mishina Y, Deng CX, Ornitz DM, Choi K. Development. 2006;133:3473–3484. doi: 10.1242/dev.02499. [DOI] [PubMed] [Google Scholar]
- 45.Smith KA, Joziasse IC, Chocron S, van Dinther M, Guryev V, Verhoeven MC, Rehmann H, van der Smagt JJ, Doevendans PA, Cuppen E, Mulder BJ, Ten Dijke P, Bakkers J. Circulation. 2009;119:3062–3069. doi: 10.1161/CIRCULATIONAHA.108.843714. [DOI] [PubMed] [Google Scholar]
- 46.Joziasse IC, Smith KA, Chocron S, van Dinther M, Guryev V, van de Smagt JJ, Cuppen E, Ten Dijke P, Mulder BJ, Maslen CL, Reshey B, Doevendans PA, Bakkers J. Eur J Hum Genet. 19:389–393. doi: 10.1038/ejhg.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inai K, Norris RA, Hoffman S, Markwald RR, Sugi Y. Dev Biol. 2008;315:383–396. doi: 10.1016/j.ydbio.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okagawa H, Markwald RR, Sugi Y. Dev Biol. 2007;306:179–192. doi: 10.1016/j.ydbio.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moskowitz IP, Wang J, Peterson MA, Pu WT, Mackinnon AC, Oxburgh L, Chu GC, Sarkar M, Berul C, Smoot L, Robertson EJ, Schwartz R, Seidman JG, Seidman CE. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4006–4011. doi: 10.1073/pnas.1019025108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galvin KM, Donovan MJ, Lynch CA, Meyer RI, Paul RJ, Lorenz JN, Fairchild-Huntress V, Dixon KL, Dunmore JH, Gimbrone MA, Jr, Falb D, Huszar D. Nat Genet. 2000;24:171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- 51.Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma L, Lu MF, Schwartz RJ, Martin JF. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- 53.Sugi Y, Yamamura H, Okagawa H, Markwald RR. Dev Biol. 2004;269:505–518. doi: 10.1016/j.ydbio.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 54.Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartram U, Molin DG, Wisse LJ, Mohamad A, Sanford LP, Doetschman T, Speer CP, Poelmann RE, Gittenberger-de Groot AC. Circulation. 2001;103:2745–2752. doi: 10.1161/01.cir.103.22.2745. [DOI] [PubMed] [Google Scholar]
- 56.Azhar M, Runyan RB, Gard C, Sanford LP, Miller ML, Andringa A, Pawlowski S, Rajan S, Doetschman T. Dev Dyn. 2009;238:431–442. doi: 10.1002/dvdy.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]