Abstract
Objective
Corticotropin - releasing hormone receptor 2 (CRHR2) participates in smooth muscle relaxation response and may influence acute airway bronchodilator response to short – acting β2 agonist treatment of asthma. We aim to assess associations between genetic variants of CRHR2 and acute bronchodilator response in asthma.
Methods
We investigated 28 single nucleotide polymorphisms in CRHR2 for associations with acute bronchodilator response to albuterol in 607 Caucasian asthmatic subjects recruited as part of the Childhood Asthma Management Program (CAMP). Replication was conducted in two Caucasian adult asthma cohorts – a cohort of 427 subjects enrolled in a completed clinical trial conducted by Sepracor Inc. (MA, USA) and a cohort of 152 subjects enrolled in the Clinical Trial of Low-Dose Theopylline and Montelukast (LODO) conducted by the American Lung Association Asthma Clinical Research Centers.
Results
Five variants were significantly associated with acute bronchodilator response in at least one cohort (p-value ≤ 0.05). Variant rs7793837 was associated in CAMP and LODO (p-value = 0.05 and 0.03, respectively) and haplotype blocks residing at the 5’ end of CRHR2 were associated with response in all three cohorts.
Conclusion
We report for the first time, at the gene level, replicated associations between CRHR2 and acute bronchodilator response. While no single variant was significantly associated in all three cohorts, the findings that variants at the 5’ end of CRHR2 are associated in each of three cohorts strongly suggest that the causative variants reside in this region and its genetic effect, although present, is likely to be weak.
Keywords: Asthma, genetics, corticotrophin releasing hormone receptor 2, CRHR2, bronchodilator response, polymorphism, β2 adrenergic receptor agonist
INTRODUCTION
β2 – adrenergic receptor (β2 AR) agonists are the most widely used class of medication in the treatment of asthma worldwide [1], [2]. Variability of the acute bronchodilator response in asthmatic individuals exists [3]. Findings from genetic studies of candidate genes (e.g. beta-2 adrenergic receptor (ADRB2) [4], [5], [6], [7], [8], [9], [10] and adenylate cyclase 9 (ADCY9) [11]) and bronchodilator response have provided evidence of a genetic component in inter-individual variability.
In this study, we hypothesize that genetic variants of CRHR2 are associated with acute airway bronchodilator response upon β2 AR agonists administration in asthmatic individuals. The logic in looking at the CRHR2 gene as an indicator of bronchodilator response to beta agonists comes from 2 lines of evidence: (1) Similar smooth muscle relaxation mechanism between the beta-2 agonist and CRHR2 pathways. Agonists produce bronchodilation in asthmatics by binding to and directly simulating the β2 AR on airway smooth muscle cells. The interaction of agonist and receptor leads to multiple downstream signals, one of which is the activation of adenylyl cyclase via stimulatory G proteins, which in turn increases cyclic adenosine monophosphate (cAMP) production and activates protein kinase A (PKA). PKA phosphorylates several target proteins, decreases intracellular calcium cations, and leads to smooth muscle relaxation [2]. Similar to β2 AR, CRHR2 belongs to the family of G protein coupled – receptors (GPCRs). CRHR2 and its homologue, corticotropin releasing hormone receptor 1 (CRHR1), signal mainly through G proteins to increase adenylyl cyclase activity and cAMP levels. In mice, CRHR1 and CRHR2 have been found to be expressed in airway epithelium and smooth muscle relaxation response was induced by urocrotin (UCN) III – CRHR2 mediated cAMP elevation [12]. In rats, UCN mRNA was expressed in lung tissues in non-allergically sensitized animals and was expressed more pronouncedly in their sensitized counterparts [13]. By interacting with its ligands (UCN-I, II, III and CRH), CRHR2 may initiate various downstream signaling pathways (e.g. cAMP/PKA and MLC20 phosphorylation) to elicit contractile/relaxation responses in airway smooth muscles. (2) Interactions between beta-2 agonist and corticosteroid pathways. In vitro study has shown that β2AR mRNA level is elevated in the presence glucocorticoid [11]. CRHR2 signaling pathways regulate the hypothalamic-pituitary-adrenocortical axis [14]. Animal models have suggested an important role of the corticotropin-releasing hormone (CRH) system in airways [12], [13]. Since CRHR2 modulates the release of corticosterone [14], [15], asthmatic individuals with different genetic variants of CRHR2 may have different endogenous levels of corticosterone, leading to different degrees of interaction with beta-2 agonist in the airways.
The CRHR2 gene is located on genomic region 7p21-p15 [16] and spans roughly 50 thousand base-pairs (Kbp) [17]. The gene consists of 12 exons; alternate splicing of exon 1 gives rise to 3 isoforms (CRHR2α, β and γ)[18]. Animal and human expression studies suggested that CRHR2α and CRHR2β are expressed in both the brain and the periphery such as smooth muscle and CRHR2γ predominantly expressed in the brain (reviewed in [19]). Our aim is to identify genetic variants of CRHR2 contributing to variability in acute bronchodilator response in asthmatic individuals.
METHODS
Primary study population
The study population consisted of asthma patients enrolled in the Childhood Asthma Management Program (CAMP), a multicenter, randomized, double-blinded clinical trial testing the safety and efficacy of inhaled steroid versus non-steroid anti-inflammatory agent versus placebo over a mean of 4.3 years ([20], [21]). Briefly, children ages 5 to 12 years who suffered from mild to moderate asthma were recruited based on criteria including (1) asthma symptoms and/or (2) medication use for ≥ 6 months in the previous year and (3) at least a 20% reduction in FEV1 after the administration of methacholine at a concentration less than or equal to 12.5 mg/ml (PC20 ≤ 12.5mg/ml). Pulmonary function including bronchodilator responses and airway responsiveness was measured before randomization. For this study, acute bronchodilator response at baseline (i.e. before randomization) was the phenotype of interest, measured as the percent change in FEV1 after the administration of 2 puffs of albuterol, compared with FEV1 measured immediately prior to albuterol administration. Subjects were asked to cease all bronchodilator medications for at least 12 hours before the measurement of responsiveness. Data on acute bronchodilator response prior to randomization to different treatment arms from 607 Caucasian subjects were analyzed for genetic association. Approval by the Institutional Review Board was obtained and informed consents from all subjects were received.
Replication study populations
The first replication cohort consisted of asthma patients enrolled in an adult study of β2 – agonist conducted by Sepracor Inc (Marlborough, MA), hereafter named Sepracor. A total of 427 subjects, who were over the age of 18, Caucasians, and had the full range of asthma severity were assessed for associations [22], including spirometry and bronchodilator responsiveness. The second replication cohort consisted of individuals enrolled in a completed trial conducted by the American Lung Association Asthma Clinical Research Centers (ALA-ACRC) to evaluate the effectiveness of low dose Theophylline as add-on treatment in asthma (the LODO trial), as described previously [23]. Briefly, the LODO trial was a multi-center, randomized, double-masked and placebo-controlled trial. Inclusion criteria include 15 years of age of older, a history of physician-diagnosed asthma, daily asthma medications use for 1 year or more, an FEV1 of 50% or more of predicted value, and poor asthma control as defined by a score of 1.5 of greater on the Asthma Control Questionnaire [24], [25]. Exclusion criteria include use of oral corticosteroid, leukotriene antagonists, or theophylline within 4 weeks before enrollment, and current or former smokers with 20 packyears or more smoking history. For the purpose of replication, data from 152 Caucasian subjects were analyzed for associations. Similar to the dosage used to measure acute bronchodilator response in CAMP, acute bronchodilator response at baseline prior to randomization were obtained by measuring the change in FEV1 after administration of 2 inhalations (90μg) of albuterol by metered dose inhaler. Approval by the Institutional Review Board was obtained and informed consents from all subjects in the two replication cohorts were received.
SNPs genotyping and statistical analyses
We investigated 39kb of genomic DNA harboring CRHR2, spanning from chromosome 7 position 30697689 to 30658511 on build 36.1 genome assembly released by the National Center for Biotechnology Information. A total of 28 SNPs were selected from public databases, prior to the availability of the HapMap database, for investigation in CAMP in order to cover the genomic region of CRHR2 based on physical location and compatibility with genotyping methods. Subsequent investigation of the HapMap Release 22 data showed that there are 7 SNPs which tag (tagging SNPs) the region with r2≥0.8 in the CEU population, and these tagging SNPs were in the original 28 SNPs panel. SNP genotyping was performed using standard protocol for the iplex assay on Sequenom MassARRAY MALDI-TOF mass spectrometer (26) (Sequenom, CA, USA) and TaqMan assays (27) (Applied Biosystems, CA, USA).
Statistical analyses were performed using SAS statistical software (SAS Institute Inc., Cary, NC). Multivariate associations between individual SNP and acute bronchodilator response in the presence of potential confounders (height, age, gender, and baseline FEV1 pre-bronchodilator administration) were tested using general linear model. Genotypes were coded for association testing under an additive model. SNP characteristics (e.g. allele frequency, Hardy – Weinberg equilibrium) for each SNP were assessed using the software Haploview [28].
Haplotype analyses were conducted using the software haplo.stats [29]. To narrow the genomic region (e.g. lengths of haplotype blocks) within the 32kb region that was associated with the phenotype, the sliding window option was used to test for associations between haplotypes of length 2 to 10 SNPs and acute bronchodilator response, adjusted for potential confounders. All p-values were obtained from simulation with default values of 1000 replicates. Simulated p-value is the number of times the simulated score statistic, calculated from a permutated-reordering of the trait and covariates and the original ordering of the genotyping matrix, exceeds the observed, divided by the total number of simulations.
RESULTS
Populations Baseline characteristics
A total of 607 Caucasian CAMP subjects with the mean age of 8.86 years (standard deviation (sd) = 2.13 years) were analyzed in this study (Table 1). The mean age of onset was 3.06 years (sd = 2.44 years). The male to female ratio was 1.5:1.0. The mean bronchodilator response was 10.75% (sd = 10.13%). In Sepracor, a total of 427 Caucasian asthmatic subjects with the mean age of 32.55 years (sd = 13.72 years) were analyzed. The male to female ratio was 1:1 and the mean bronchodilator response was 40.36% (sd = 20.98%). In LODO, a total of 152 Caucasian asthmatic subjects with the mean age of 42.84 years (sd = 14.77 years) were analyzed. The male to female ratio was 0.33:1 and the mean bronchodilator response was 9.68% (sd = 11.13%). We have confined our analyses to Caucasians to avoid potential confounding due to population substructures. In addition, power to detect associations in other racial groups is low due to small sample sizes.
Table 1.
Population characteristics
| CAMP | SEPRACOR | LODO | |
|---|---|---|---|
| Number of subjects | 607 | 427 | 152 |
| Age (yrs), mean (sd) | 8.86 (2.13) | 32.55 (13.72) | 42.88 (14.74) |
| Age (yrs), range | 5.17-13.24 | 12-80 | 15-76 |
| Age at first asthma symptoms (yrs), mean (sd) | 3.06 (2.43) | not available | not available |
| Male gender count (frequency) | 364 (0.60) | 214 (0.50) | 38 (0.25) |
| Female gender count (frequency) | 243 (0.40) | 213 (0.50) | 114 (0.75) |
| Baseline pre - bronchodilator FEV1 percent predicted (sd) | 94.52 (14.06) | 54.95 (7.76) | 68.07 (14.31) |
| Baseline post - bronchodilator FEV1 percent predicted (sd) | 103.55 (12.51) | 76.58 (12.65) | 73.91 (13.78) |
| Baseline brochodilator response % (sd) | 10.75 (10.13) | 40.36 (20.98) | 9.67 (11.07) |
CAMP – Childhood Asthma Management Program, LODO – Low Dose Theophylline and Montelukast trial, FEV1 – forced expiratory volume in 1 second, sd – standard deviation
SNP Characteristics
A total of 28 SNPs were investigated in CAMP (Figure 1, Table 2). Minor allele frequencies range from 1.9% to 39.0%. The average frequency of missing genotypes for the 28 SNPs was 4.0% (ranges from 1.15 - 11.37% (rs733453)). In Sepracor, 26 of the 28 SNPs tested in CAMP were analyzed. Minor allele frequencies range from 1.9% to 40.4% (Table 2). The average missing genotype rate was 3.1% (0 – 11.2% (rs2014663)). In LODO, 23 of the 28 SNPs were analyzed. Minor allele frequencies range from 0.70% to 48.4% (Table 2). The average missing genotype rate was 6.1% (0 - 13.1% (rs2284217)). Across all 3 cohorts, only 4 variants (rs733453, rs917195, rs929377 and rs1076291) were marginally out of Hardy-Weinberg equilibrium (0.03 ≤ p-value ≤ 0.05), as expected by chance given the number of SNPs tested.
Figure 1. Genomic organization of CRHR2.
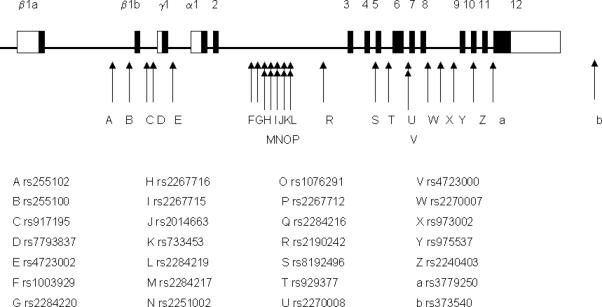
Exons are represented by black boxes connected by straight line representing introns. White boxes representing untranslated regions of exons. Location of exons are described elsewhere [17]. Positions and names of the 28 SNPs analyzed are represented by arrows and letters below the gene structure.
Table 2.
SNP characteristics
| SNP rs# | chromosome position | MA | MAFCAMP | MAFSEPRACOR | MAFLODO | genomic location, downstream of exon |
|---|---|---|---|---|---|---|
| rs255102 | 30697689 | T | 0.33 | 0.30 | 0.38 | intronic, exon β1a |
| rs255100 | 30695433 | A | 0.39 | 0.40 | 0.39 | intronic, exon β1a |
| rs917195 | 30694977 | T | 0.22 | 0.24 | 0.23 | intronic, exon β1b |
| rs7793837 | 30693302 | T | 0.24 | 0.26 | 0.26 | intronic, exon β1b |
| rs4723002 | 30692226 | G | 0.09 | 0.10 | 0.08 | intronic, exon γ1 |
| rs1003929 | 30685574 | T | 0.16 | 0.13 | 0.15 | intronic, exon 2 |
| rs2284220 | 30684628 | G | 0.15 | 0.14 | 0.13 | intronic, exon 2 |
| rs2267716 | 30683168 | C | 0.23 | 0.24 | 0.23 | intronic, exon 2 |
| rs2267715 | 30682612 | G | 0.37 | 0.37 | 0.36 | intronic, exon 2 |
| rs2014663 | 30682098 | C | 0.15 | 0.12 | NT | intronic, exon 2 |
| rs733453 | 30681298 | G | 0.36 | 0.38 | NT | intronic, exon 2 |
| rs2284219 | 30680961 | A | 0.37 | 0.38 | 0.37 | intronic, exon 2 |
| rs2284217 | 30680133 | A | 0.22 | 0.21 | 0.19 | intronic, exon 2 |
| rs2251002 | 30679369 | G | 0.35 | NT | NT | intronic, exon 2 |
| rs1076291 | 30679120 | A | 0.35 | 0.35 | 0.30 | intronic, exon 2 |
| rs2267712 | 30678759 | A | 0.15 | 0.14 | 0.13 | intronic, exon 2 |
| rs2284216 | 30678486 | T | 0.09 | 0.10 | 0.09 | intronic, exon 2 |
| rs2190242 | 30676000 | C | 0.23 | 0.20 | 0.20 | intronic, exon 2 |
| rs8192496 | 30671630 | G | 0.29 | 0.29 | 0.27 | intronic, exon 4 |
| rs929377 | 30670684 | A | 0.31 | 0.31 | 0.29 | intronic, exon 5 |
| rs2270008 | 30668769 | T | 0.12 | 0.13 | 0.14 | intronic, exon 6 |
| rs4723000 | 30668439 | A | 0.12 | 0.12 | 0.13 | intronic, exon 6 |
| rs2270007 | 30666497 | G | 0.17 | 0.16 | 0.15 | intronic, exon 8 |
| rs973002 | 30665429 | G | 0.17 | NT | NT | intronic, exon 8 |
| rs975537 | 30663882 | T | 0.22 | 0.22 | 0.19 | intronic, exon 8 |
| rs2240403 | 30661727 | T | 0.09 | 0.09 | 0.09 | exonic, exon 10 |
| rs3779250 | 30660785 | C | 0.35 | 0.35 | NT | intronic, exon 11 |
| rs3735430 | 30658511 | T | 0.02 | 0.02 | 0.01 | intronic, exon 12 |
rs – refSNP number, MA – minor allele, MAF – minor allele frequency, CAMP – Childhood Asthma Management Program, LODO – Low Dose Theophylline and Montelukast trial, NT – not tested
Associations between SNPs and acute bronchodilator response
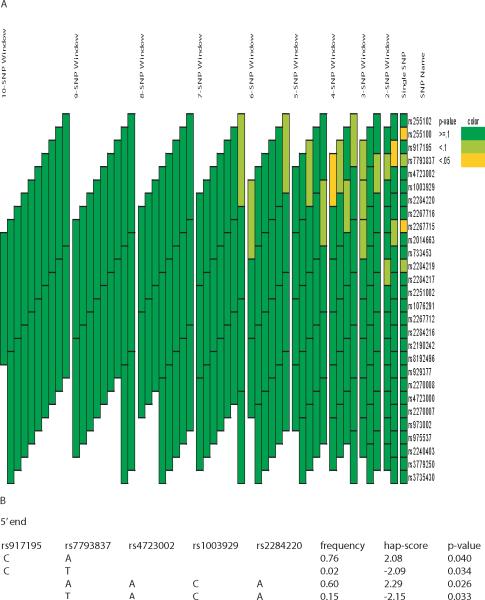
In CAMP, variants rs255100, rs7793837 and rs2267715 were statistically associated with acute bronchodilator response under an additive model (p-value ≤ 0.05) (Table 3). Minor alleles rs255100A, rs7793837T and rs2267715G were associated with reduced bronchodilator response. Haplotypes of length ranging from 2 to 10 SNPs were analyzed for associations and a total of 2 haplotype blocks of lengths 2 and 4 SNPs demonstrated significant associations under an additive model (simulated global p-value < 0.05) (Figure 2A). Of the 3 associated SNPs, rs255100 and rs2267715 are in strong linkage disequilibrium (r2 = 0.88), rs255100 and rs7793837, and rs7793837 and rs2267715 are in moderate linkage disequilibrium (r2 = 0.48 and 0.40, respectively) (data not shown). Haplotypes of each associated blocks were further assessed for associations and a common haplotype (rs7793837A-rs4723002A-rs1003929C-rs2284220A (frequency = 0.60)) demonstrated the most significant association (simulated p-value = 0.026) (Figure 2B).
Table 3.
Significant associations of CRHR2 genotypes and acute bronchodilator response in CAMP and replication cohorts
| CAMP | SEPRACOR | LODO | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variants | Count (freq) | mean (se) | p-value | Count (freq) | mean (se) | p-value | Count (freq) | mean (se) | p-value |
| rs255100 | |||||||||
| TT | 221 (0.36) | 11.70 (0.54) | 0.02 | 151 (0.35) | 40.49 (2.33) | 0.78 | 61 (0.40) | 11.23 (1.35) | 0.38 |
| TA | 287 (0.47) | 10.82 (0.48) | 209 (0.49) | 39.91 (1.32) | 71 (0.47) | 8.43 (1.15) | |||
| AA | 88 (0.15) | 9.22 (0.87) | 67 (0.16) | 40.93 (1.55) | 20 (0.13) | 10.09 (2.13) | |||
| missing | 11 (0.02) | 0 (0.00) | 0 (0.00) | ||||||
| rs7793837 | |||||||||
| AA | 349 (0.57) | 11.33 (0.44) | 0.05 | 235 (0.55) | 39.04 (1.23) | 0.24 | 75 (0.49) | 11.46 (1.08) | 0.03 |
| AT | 207 (0.34) | 10.07 (0.57) | 155 (0.36) | 41.98 (1.52) | 55 (0.36) | 7.34 (1.26) | |||
| TT | 40 (0.07) | 9.62 (1.29) | 31 (0.07) | 40.41 (3.40) | 8 (0.05) | 8.34 (3.35) | |||
| missing | 13 (0.02) | 6 (0.01) | *14 (0.09) | ||||||
| rs2284220 | |||||||||
| AA | 402 (0.66) | 10.82 (0.40) | 0.9 | 311 (0.73) | 41.08 (1.06) | 0.03 | 111 (0.73) | 9.34 (0.92) | 0.52 |
| AG | 141 (0.23) | 11.57 (0.68) | 103 (0.24) | 37.97 (1.84) | 27 (0.18) | 11.88 (1.91) | |||
| GG | 10 (0.02) | 6.67 (2.69) | 5 (0.01) | 20.86 (8.40) | 5 (0.03) | 8.07 (4.37) | |||
| missing | 54 (0.09) | 8 (0.02) | 9 (0.06) | ||||||
| rs2267716 | |||||||||
| TT | 361 (0.60) | 11.23 (0.43) | 0.1 | 246 (0.58) | 40.04 (1.22) | 0.84 | 82 (0.54) | 11.34 (1.11) | 0.05 |
| TC | 203 (0.33) | 10.09 (0.57) | 151 (0.35) | 41.25 (1.56) | 53 (0.35) | 7.11 (1.38) | |||
| CC | 33 (0.05) | 9.91 (1.41) | 26 (0.06) | 38.83 (3.78) | 5 (0.03) | 10.43 (4.71) | |||
| missing | 10 (0.02) | 4 (0.01) | *12 (0.08) | ||||||
rs # - refSNP number, count – genotype count, freq – frequency, means (se) –means bronchodilator response estimate obtained from linear regression (standard error), p – p-value obtained from linear regression under an additive model, CAMP – Childhood Asthma Management Program, LODO – Low Dose of Theophylline and Montelukast trial. Only variants with significant association (p-value ≤ 0.05) in at least 1 cohort are shown.
The mean bronchodilator response of subjects with missing genotypes is similar to the mean of those with genotypes (10.10% for rs7793837 and 9.46% for rs2267716 versus 9.68%); hence data can be considered missing at random.
Figure 2. Association analyses of CRHR2 variants and acute bronchodilator response in CAMP.
(A) Significance of association, as indicated by simulated global p-values, between genetic variants of CRHR2 (individual SNPs and haplotypes) and phenotype is represented by color and constructed using the graphical tool GRASP[33]. The SNPs making up each haplotype block can be found by matching the upper and lower boundaries of each block to the SNP name on the ‘SNP Name’ column. (B) Haplotypes within the associated blocks were further assessed for association. Hap-scores are the score statistics calculated from Haplo.stats. Only haplotypes with significant (simulated) p-values were listed.
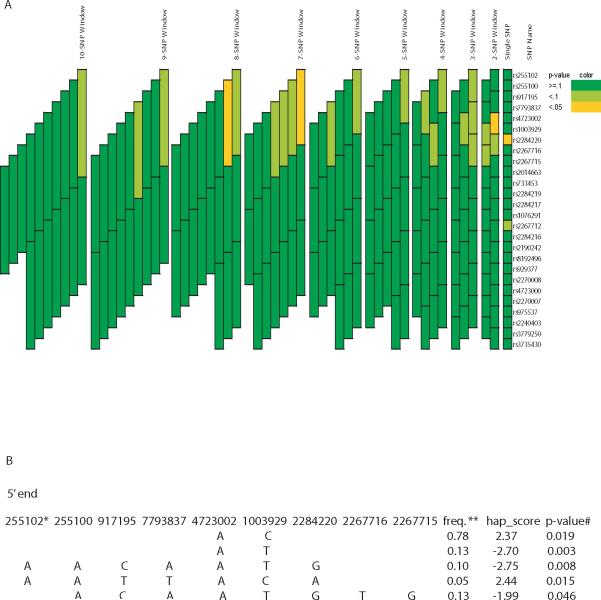
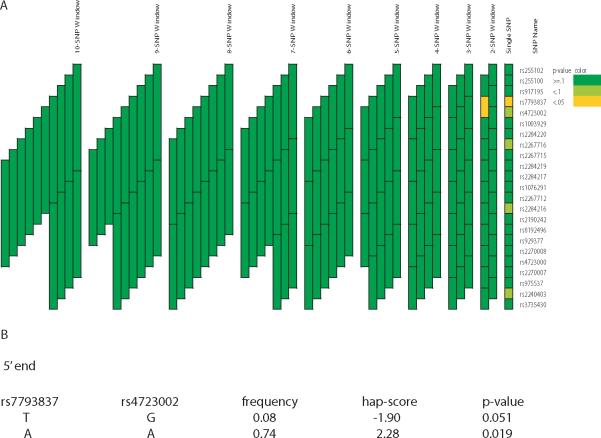
In Sepracor, none of the associated variants in CAMP was associated with acute bronchodilator response. A different variant, rs2284220, was associated with the phenotype (p-value = 0.03) (Figure 3A). The minor allele (rs2284220G) was associated with reduced bronchodilator response (Table 3). A total of 3 haplotype blocks of length 2, 7 and 8 SNPs demonstrated significant associations (simulated global p-value < 0.05) (Figure 3A). Further assessment for associations within each block showed that haplotype rs4723002A-rs1003929T (frequency = 0.13) was associated with the phenotype with the most statistical significance (simulated p-value = 0.003), followed by haplotype rs255102A-rs255100A-rs917195C-rs7793837A-rs4723002A-rs1003929T-rs2284220G (frequency = 0.10, simulated p-value = 0.008) (Figure 3B). In LODO, variants rs7793837 and rs2267716 were associated with acute bronchodilator response (p-value ≤ 0.05) (Figure 4A). The minor alleles (rs7793837T and rs2267716C) were associated with reduced bronchodilator response (Table 3). The two SNPs are in linkage disequilibrium (r2 = 0.87). A 2-SNP haplotype block was associated with the phenotype (simulated global p-value = 0.05) and further assessment of individual haplotypes within this block showed that haplotype rs4723002A-rs7793837A (frequency = 0.75) was the most significantly associated (simulated p-value = 0.02, respectively) with acute bronchodilator response (Figure 4B).
Figure 3. Association analyses of CRHR2 variants and acute bronchodilator response in SEPRACOR.
See legend of figure 2. *SNP rs number **Frequency
Figure 4. Association analyses of CRHR2 variants and acute bronchodilator response in LODO.
See legend of figure 2.
Discussions
This is the first study to report gene-based replicated associations between the CRHR2 gene and acute bronchodilator response upon short-acting β2 agonist administration in asthmatic subjects. In our primary and two replication cohorts, different SNPs of CRHR2 were found to be associated with acute bronchodilator response to albuterol, however, the physical proximity of these SNPs with each other (between exon β1a and exon 3) suggests that the causative variant(s) may reside towards the 5’ end of the gene. By comparing the haplotypes showing the most significant associations in each cohort, the associated region spans approximately 8.7kb between introns β1b and 2 (Table 4). The haplotype analyses detected the common haplotypes (frequency between 60% - 78%) to be associated with acute bronchodilator response in the three cohorts. The haplotypes are of different length, yet in CAMP and LODO, allele rs7793837A resides in almost all the associated common haplotypes. In Sepracor, the associated haplotype with a frequency of 78% consisted of 2 neighboring SNPs (rs4723002 and rs1003929) 1 and 7 kb downstream of variant rs7793837. We speculated that variant rs7793837 was associated in Sepracor because (1) it is not the causative variant but in LD with the causative variant, and (2) Sepracor is phenotypically different from CAMP and LODO. This cohort has a higher mean bronchodilator response due to the recruitment requirement of subjects with >15% bronchodilator response (Sepracor: -40.98% (standard deviation of 20.98), CAMP:10.75 (sd 10.13) and LODO: 9.67 (sd 11.07)). A different causative variant, also residing at 5’ end of CRHR2 may be responsible for the association observed in Sepracor. This 5’ region encodes the N-terminal extracellular domain of CRHR2, where interactions with ligands occur (reviewed in [19]). In addition to encoding the N-terminal, this region houses multiple exons 1 (β1b, γ1 and α1), where various isoforms of CRHR2 are translated depending on the splicing transcripts being translated [19]. Additional sequencing of the 5’ end of CRHR2 was carried out to identify the causative variant(s). Approximately 32 kb of genomic regions (exons β1a, β1b, γ1, α1, 2 and flanking intronic regions) were sequenced in 48 CAMP subjects, revealing 3 novel intronic variants located 199bp upstream and 26 and 74bp downstream of exon β1a, respectively. Two of the 3 novel variants were chosen for their potential functions and were genotyped in a subset of CAMP subjects, but no association was observed with acute bronchodilator response (data not shown). Although the original 28 SNPs panel did not cover exon β1a, subsequent sequencing and genotyping efforts ensured exon β1a and surrounding intronic regions were investigated.
Table 4.
Haplotypes demonstrating the most significant association with acute bronchodilator response in CAMP, SEPRACOR and LODO
| cohort | rs7793837 | rs4723002 | rs1003929 | rs2284220 | frequency | hap-score | simulated p-value |
|---|---|---|---|---|---|---|---|
| CAMP | A | A | C | A | 0.6 | 2.29 | 0.026 |
| SEPRACOR | A | T | 0.13 | -2.7 | 0.003 | ||
| LODO | A | A | 0.74 | 2.28 | 0.019 |
CAMP – Childhood Asthma Management Program, LODO – Low Dose of Theophylline and montelukast trial, Hap-score – the score statistics calculated from Haplo.stats.
The rationale for testing for associations in both pediatric and childhood asthma cohorts is to establish generalization of the associations between CRHR2 genetic variants and acute bronchodilator response across different age groups; to determine whether the association was a developmental, an aging, or both a developmental and aging phenomenon in the lungs. Our findings that different SNPs were associated with acute bronchodilator response in the three cohorts and that these SNPs reside at the 5’ end of CRHR2 suggest that the associations observed are likely due to linkage disequilibrium between associated SNPs and the true phenotype causing variant(s), in both childhood and adult asthma.
The 5’ end of CRHR2 is of great biological interest. The CRHR2 protein has 3 isoforms (CRHR2α, CRHR2β, and CRHR2γ) which differ at the N termini, and are encoded by the 5’ end exons of CRHR2. Translations of exon β1a and exon β1b encode the N terminus of the CRHR2β isoform, and exon 1α and 1γ encode the N termini of CRHR2α and CRHR2γ, respectively [18]. In vitro study has demonstrated that the three isoforms have different downstream signaling capacities [18]. We speculate that the causative variant(s) may influence acute bronchodilator response through regulating translations of CRHR2 into various isoforms.
How genetic variations in CRHR2 function contribute to differential responses to β2-agonist remains to be investigated. Literature has presented complex interactions between β2 AR and other signaling pathways, based on which we propose three molecular mechanisms that could potentially explain the association of CRHR2 variants with bronchodilator response. First, CRHR2 signaling may desensitize β2 AR function in asthmatics. β2 AR signaling can be desensitized not only by its own activation but also by signaling through other G protein-coupled receptors. This cross-talk, which is referred to as heterologous desensitization (versus homologous desensitization by its own signaling), has been observed between β2 AR and PGE2 receptors [30]. This mechanism predicts that individuals with higher CRHR2 signaling capacity may have reduced bronchodilator response due to stronger cross-desensitization. A second mechanism takes into consideration the documented anti-inflammatory role of CRHR2 [12]. In this case, reduced CRHR2 function may lead to increased production of pro-inflammatory cytokines, many of which will down-regulate the signaling through β2 ARs [31]. The last mechanism concerns the possible synergistic interaction between CRHR2 and β2 AR since both of them relax smooth muscles by stimulating cAMP production. If synergistic contribution from CRHR2 is pivotal for β2 agonists to achieve effective bronchoprotection in asthmatics, patients carrying CRHR2 alleles that have lower biological activities will have dampened therapeutic response to β2 agonists. Synergy between G-protein coupled receptors (GPCRs) has been demonstrated in many biological systems where activation of one GPCR can amplify the signaling events in a parallel but separate pathway [32], [33]. To determine which one of these three proposed paradigms is valid, we need to perform physiological studies on mice that were engineered to be CRHR2-deficient as well as biochemical analysis of signaling interaction between CRHR2 and β2 AR in primary airway smooth muscle cells.
This study has limitations. First, we did not detect a single SNP that was associated across the 3 cohorts. Second, the significant associations detected would not reach significance level when corrected for multiple testing. However, in spite of these limitations, we felt that by detecting associations with SNPs covering a region within the gene across 3 populations, the findings that CRHR2, as a gene, is associated with acute bronchodilator response is likely to be valid. The lack of any single SNP being associated in all three cohorts suggests that the genetic effect of CRHR2, although present, is likely to be weak. Being a complex trait, the number of genetic variants explaining inter-individual variations in bronchodilator response is likely to be high, with each variant exerts low to moderate effect. Our findings suggests that genetic variants of CRHR2, in addition to the widely studied β2AR and many yet to be characterized genetic variants, influence acute bronchodilator response to short acting β2A. Despite our subsequent sequencing and genotyping effort of exons at the 5’ end of CRHR2, we did not detect any causative variants, hence, the causative variants are likely to be in untranslated regions. Additional exploration of the region will be needed to identify the causative variants affecting acute bronchodilator response so that subsequent screening tests can be developed to predict in advance the efficacy of administering β2 agonists as an asthma treatment in asthmatic individuals.
Acknowledgements
We thank all families for their enthusiastic participation in the CAMP Genetics Ancillary Study, supported by the National Heart, Lung, and Blood Institute (NHLBI), NO1-HR-16049. We also acknowledge the CAMP investigators and research team, supported by NHLBI, for collection of CAMP Genetic Ancillary Study data. All work on data was conducted at the Channing Laboratory, Brigham and Women's Hospital under appropriate CAMP policies and human subjects protections. We acknowledge the American Lung Association (ALA) and the ALA's Asthma Clinical Research Centers investigators and research teams for use of the LODO data.
Funding sources: UO1 HL65899: Pharmacogenetics of Asthma Therapy (PI:Weiss), PO1 HL67664: the CAMP Genetics Ancillary Study from the National Heart, Lung, and Blood Institute, the American Lung Association Asthma Clinical Research Centers, The American Lung Association, HL-071394 (JJL); Nemours Children's Clinic, the Croucher Foundation.
References
- 1.Raby BA, Weiss ST. Beta2-adrenergic receptor genetics. Curr Opin Mol Ther. 2001;3(6):554–66. [PubMed] [Google Scholar]
- 2.Johnson M. Molecular mechanisms of beta(2)-adrenergic receptor function, response, and regulation. J Allergy Clin Immunol. 2006;117(1):18–24. doi: 10.1016/j.jaci.2005.11.012. quiz 25. [DOI] [PubMed] [Google Scholar]
- 3.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56(4):1054–70. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 4.Silverman EK, Kwiatkowski DJ, Sylvia JS, Lazarus R, Drazen JM, Lange C, et al. Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J Allergy Clin Immunol. 2003;112(5):870–6. doi: 10.1016/s0091-6749(03)02023-2. [DOI] [PubMed] [Google Scholar]
- 5.Drysdale CM, McGraw DW, Stack CB, Stephens JC, Judson RS, Nandabalan K, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci U S A. 2000;97(19):10483–8. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lima JJ, Thomason DB, Mohamed MH, Eberle LV, Self TH, Johnson JA. Impact of genetic polymorphisms of the beta2-adrenergic receptor on albuterol bronchodilator pharmacodynamics. Clin Pharmacol Ther. 1999;65(5):519–25. doi: 10.1016/S0009-9236(99)70071-8. [DOI] [PubMed] [Google Scholar]
- 7.Choudhry S, Ung N, Avila PC, Ziv E, Nazario S, Casal J, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am J Respir Crit Care Med. 2005;171(6):563–70. doi: 10.1164/rccm.200409-1286OC. [DOI] [PubMed] [Google Scholar]
- 8.Tsai HJ, Shaikh N, Kho JY, Battle N, Naqvi M, Navarro D, et al. Beta 2-adrenergic receptor polymorphisms: pharmacogenetic response to bronchodilator among African American asthmatics. Hum Genet. 2006;119(5):547–57. doi: 10.1007/s00439-006-0169-2. [DOI] [PubMed] [Google Scholar]
- 9.Woszczek G, Borowiec M, Ptasinska A, Kosinski S, Pawliczak R, Kowalski ML. Beta2-ADR haplotypes/polymorphisms associate with bronchodilator response and total IgE in grass allergy. Allergy. 2005;60(11):1412–7. doi: 10.1111/j.1398-9995.2005.00869.x. [DOI] [PubMed] [Google Scholar]
- 10.Martinez FD, Graves PE, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J Clin Invest. 1997;100(12):3184–8. doi: 10.1172/JCI119874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tantisira KG, Small KM, Litonjua AA, Weiss ST, Liggett SB. Molecular properties and pharmacogenetics of a polymorphism of adenylyl cyclase type 9 in asthma: interaction between beta-agonist and corticosteroid pathways. Hum Mol Genet. 2005;14(12):1671–7. doi: 10.1093/hmg/ddi175. [DOI] [PubMed] [Google Scholar]
- 12.Moffatt JD, Lever R, Page CP. Activation of corticotropin-releasing factor receptor-2 causes bronchorelaxation and inhibits pulmonary inflammation in mice. Faseb J. 2006;20(11):1877–9. doi: 10.1096/fj.05-5315fje. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y, Zhou H, Xu Y, Li S. Enhanced expression of urocortin in lung tissues of rats with allergic asthma. Biochem Biophys Res Commun. 2006;341(2):532–40. doi: 10.1016/j.bbrc.2005.12.214. [DOI] [PubMed] [Google Scholar]
- 14.Coste SC, Kesterson RA, Heldwein KA, Stevens SL, Heard AD, Hollis JH, et al. Abnormal adaptations to stress and impaired cardiovascular function in mice lacking corticotropin-releasing hormone receptor-2. Nat Genet. 2000;24(4):403–9. doi: 10.1038/74255. [DOI] [PubMed] [Google Scholar]
- 15.Bale TL, Contarino A, Smith GW, Chan R, Gold LH, Sawchenko PE, et al. Mice deficient for corticotropin-releasing hormone receptor-2 display anxiety-like behaviour and are hypersensitive to stress. Nat Genet. 2000;24(4):410–4. doi: 10.1038/74263. [DOI] [PubMed] [Google Scholar]
- 16.Meyer AH, Ullmer C, Schmuck K, Morel C, Wishart W, Lubbert H, et al. Localization of the human CRF2 receptor to 7p21-p15 by radiation hybrid mapping and FISH analysis. Genomics. 1997;40(1):189–90. doi: 10.1006/geno.1996.4521. [DOI] [PubMed] [Google Scholar]
- 17.Liaw CW, Lovenberg TW, Barry G, Oltersdorf T, Grigoriadis DE, de Souza EB. Cloning and characterization of the human corticotropin-releasing factor-2 receptor complementary deoxyribonucleic acid. Endocrinology. 1996;137(1):72–7. doi: 10.1210/endo.137.1.8536644. [DOI] [PubMed] [Google Scholar]
- 18.Catalano RD, Kyriakou T, Chen J, Easton A, Hillhouse EW. Regulation of corticotropin-releasing hormone type 2 receptors by multiple promoters and alternative splicing: identification of multiple splice variants. Mol Endocrinol. 2003;17(3):395–410. doi: 10.1210/me.2002-0302. [DOI] [PubMed] [Google Scholar]
- 19.Hillhouse EW, Grammatopoulos DK. The molecular mechanisms underlying the regulation of the biological activity of corticotropin-releasing hormone receptors: implications for physiology and pathophysiology. Endocr Rev. 2006;27(3):260–86. doi: 10.1210/er.2005-0034. [DOI] [PubMed] [Google Scholar]
- 20.Childhood Asthma Management Program Research Group The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials. 1999;20(1):91–120. [PubMed] [Google Scholar]
- 21.The Childhood Asthma Management Program Research Group Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med. 2000;343(15):1054–63. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 22.Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13(13):1353–9. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 23.Clinical trial of low-dose theophylline and montelukast in patients with poorly controlled asthma. Am J Respir Crit Care Med. 2007;175(3):235–42. doi: 10.1164/rccm.200603-416OC. [DOI] [PubMed] [Google Scholar]
- 24.Juniper EF, O'Byrne PM, Guyatt GH, Ferrie PJ, King DR. Development and validation of a questionnaire to measure asthma control. Eur Respir J. 1999;14(4):902–7. doi: 10.1034/j.1399-3003.1999.14d29.x. [DOI] [PubMed] [Google Scholar]
- 25.Juniper EF, Bousquet J, Abetz L, Bateman ED. Identifying ‘well-controlled’ and ‘not well-controlled’ asthma using the Asthma Control Questionnaire. Respir Med. 2006;100(4):616–21. doi: 10.1016/j.rmed.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Sun X, Ding H, Hung K, Guo B. A new MALDI-TOF based mini-sequencing assay for genotyping of SNPS. Nucleic Acids Res. 2000;28(12):E68. doi: 10.1093/nar/28.12.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee LG, Connell CR, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21(16):3761–6. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finney PA, Belvisi MG, Donnelly LE, Chuang TT, Mak JC, Scorer C, et al. Albuterol-induced downregulation of Gsalpha accounts for pulmonary beta(2)-adrenoceptor desensitization in vivo. J Clin Invest. 2000;106(1):125–35. doi: 10.1172/JCI8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shore SA. Cytokine regulation of beta-adrenergic responses in airway smooth muscle. J Allergy Clin Immunol. 2002;110(6 Suppl):S255–60. doi: 10.1067/mai.2002.129947. [DOI] [PubMed] [Google Scholar]
- 32.Selbie LA, Hill SJ. G protein-coupled-receptor cross-talk: the fine-tuning of multiple receptor-signalling pathways. Trends Pharmacol Sci. 1998;19(3):87–93. doi: 10.1016/s0165-6147(97)01166-8. [DOI] [PubMed] [Google Scholar]
- 33.Werry TD, Wilkinson GF, Willars GB. Mechanisms of cross-talk between G-protein-coupled receptors resulting in enhanced release of intracellular Ca2+. Biochem J. 2003;374(Pt 2):281–96. doi: 10.1042/BJ20030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathias RA, Gao P, Goldstein JL, Wilson AF, Pugh EW, Furbert-Harris P, et al. A graphical assessment of p-values from sliding window haplotype tests of association to identify asthma susceptibility loci on chromosome 11q. BMC Genet. 2006;7:38. doi: 10.1186/1471-2156-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]