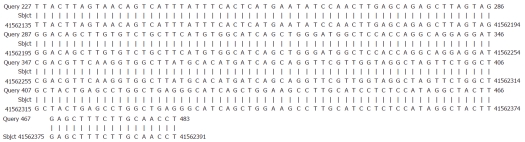Abstract
AIM: To detect human papillomavirus (HPV) DNA in esophageal carcinoma (EC) 109 cells and investigate the relationship between HPV and EC.
METHODS: Genomic DNA and total RNA from EC109 cells were isolated. HPV DNA was detected by polymerase chain reaction (PCR) with the general primer sets of My09/11 and GP5 +/6 + for the HPV L1 gene and type-specific primer sets for HPV18 E6 and HPV18 E6-E7. Reverse transcription (RT) of mRNA isolated from EC109 cells was performed to produce a cDNA. And then a PCR-based protocol for the amplification of papillomavirus oncogene transcripts was used to analyze HPV18 DNA and integrated transcripts of HPV18 in the chromosomes of EC109 cells. The final nested PCR products were cloned into a pMD-18T vector and sequenced to analyze the chromosomal location of HPV integration.
RESULTS: HPV18 DNA was detected in EC109 cells by PCR using the general primer sets of My09/11 and GP5 +/6 + for HPV L1 and the type-specific primer sets for HPV18 E6 and E6-E7 to generate products of 450 bp, 150 bp, 335 bp and 944 bp, respectively. Approximately 600 bp of integrated HPV18-specific transcript was identified. The final nested PCR product of integrated HPV18 DNA was cloned into a pMD-18T vector and sequenced to analyze the chromosomal location of HPV integration. Sequence alignment showed that the HPV18 sequence from EC109 cells was identical to that of the encoded early protein E7-E1 of the standard HPV18 strain X05015, and another partial gene sequence was identical to a partial sequence of human chromosome 8.
CONCLUSION: Integration of the HPV genome into the host cell chromosome suggests that persistent HPV infection is vital for malignant cell transformation and carcinogenesis.
Keywords: Esophageal carcinoma, Human papillomavirus, Integration, Infection, Genome
INTRODUCTION
The esophageal carcinoma (EC) cell line EC109 was established in 1976 by the Cell Biology Research Group at the Chinese Academy of Medical Sciences Institute of Cancer Research. The cell line was derived from esophageal cells that were surgically removed from a patient with a pathological diagnosis of EC[1]. The cells were determined to be positive for human papillomavirus (HPV) type 18[2].
EC is one of the major cancers in China. The etiology of EC has yet to be established despite extensive investigation of the contribution of environmental factors, lifestyle, and low levels of chemical elements. In 1982, Syrjanen formulated a hypothesis on the relationship between HPV infection and the development of EC; the hypothesis was based on the presence of papilloma-like tissues in EC specimens and other molecular evidence[3]. Thereafter, numerous clinical studies have supported this hypothesis[4-13]. However, the link between HPV infection and the etiology of EC remains inconclusive.
A link between HPV infection and squamous cell cancer of the cervix has been identified[14]. Currently, several oncogenic types of HPV are regarded as the etiological agents responsible for the development of cervical squamous cell carcinoma[15,16].
We further studied the association between HPV infection and carcinogenesis using HPV18 E6-E7–transfected stable cell lines derived from fetal esophageal epithelial tissue. Our results strongly supported the conclusion that the expression of HPV18 proteins E6 and E7 induced the transformation of the esophageal cells[17]. HPV18 was also detected in EC109 cells[2]. These results support a link between HPV infection and esophageal carcinogenesis.
Persistent infection with high-risk HPV and the integration of viral genomes into the host genome have been implicated in the etiology of malignant and premalignant disease of the female lower genital tract[18,19]. HPV is divided into low-risk (LR) and high-risk (HR) types according to the presumed degree of risk for the development of cancer. HR HPV types such as HPV16, HPV18, and HPV31 are associated with cancer, while LR HPV types such as HPV6, HPV11, and HPV40 are the causative agents of benign warts[20]. Episomal and integrated HPV can be distinguished using a polymerase chain reaction (PCR)-based protocol for the amplification of papillomavirus oncogene transcripts (APOT), developed by Klaes and his colleagues[21]. The same group hypothesized that HPV transcripts derived from the integrated HPV genome represent suitable molecular markers for a pre-neoplastic lesion at risk for progression to carcinoma. For EC, however, few data are available concerning HPV status and integration patterns. The aim of the present study was to assess high-risk HPV infection and HPV18 DNA integration into the host cell genome in EC.
MATERIALS AND METHODS
Cell lines
EC109 cells, the human embryonic kidney (HEK) 293 cell line, and the HeLa cell line were maintained by our laboratory. HeLa cells served as the HPV18-positive control, and HEK293 cells served as the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-positive control. The cell lines were cultivated in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and antibiotics (penicillin and streptomycin, the concentration of each antibiotic was 100 U/mL) under standard conditions. Cells were maintained as a subconfluent monolayer at 37 °C in a humidified atmosphere of 5% CO2/95% air. Exponentially proliferating cells were harvested with 0.25% trypsin and 0.02% EDTA, resuspended in fresh medium, and seeded in new flasks. The cells (2 × 106) were collected and washed with phosphate-buffered saline. Then, the cells were separated into two cryotubes, immediately frozen in liquid nitrogen, or stored at -70 °C until further use.
Detection of HPV DNA
Genomic DNA was isolated from each cell line using the Easy-DNA kit (“BioTake”, China) according to the supplier’s instructions. HPV DNA was detected by PCR with the general primer sets of My09/11 (My09: 5′-CGTCCMARRGGAWACTGATC-3′, MY11: 5′-GCMCAGGGWCATAAYAATGG-3′, amplicon size 450 bp) and GP5 +/6 + (GP5 +: 5′-TTTGTTACTGTGGTAGATACTAC-3′, GP6 +: 5′-GAAAAATAAACTGTAAATCATATTC-3′, amplicon size 150 bp) for the HPV L1 gene[22] and type-specific primer sets for HPV18 E6 (forward: 5′-GCGCTTTGAGGATCCAACAC-3′, reverse: 5′-ATTCAACGGTTTCTGGCAC-3′, amplicon size 335 bp) and HPV18 E6-E7 (forward: 5′-AACACACCACAATACTATGGCGCG-3′, reverse: 5′-GCATTTTCGTCCTCGTCATCTG-3′, amplicon size 944 bp). The type-specific primer sets for HPV18 E6 and HPV18 E6-E7 were designed according to the GenBank-provided HPV18 gene sequences of X05015 (http://www.ncbi.nlm.nih.gov/nuccore/X05015).
PCR was conducted in a final volume of 25 μL containing 1× PCR buffer (“BioTake”, China), 0.2 mmol/L dNTPs, 1 μL complexed recombinant Taq DNA polymerase, 0.2 mmol/L of each primer, and 100 ng DNA. An initial 5-min denaturation step at 95 °C was followed by 30 amplification cycles of 30 s at 95 °C, 30 s at 55 °C, and 1 min at 72 °C, with a final extension step of 5 min at 72 °C using a block thermocycler (PeQLab Biotechnologie, Germany). A negative control (HEK293 cell DNA template) was included in each amplification step. DNA from HeLa cells was included as an HPV18-positive control. The PCR products were resolved on a 1.0% agarose gel.
Reverse transcription
Total RNA from the cell lines was isolated using the Micro-to-Midi Total RNA Purification System kit (“BioTake”, China) according to the manufacturer’s instructions. The RNA was quantified by spectrophotometry. Reverse transcription (RT) of mRNA isolated from EC109 cells was performed to produce a double-stranded DNA product that was then amplified by PCR. The final concentrations for the RT reaction were RNase-free H2O (9.2 μL), 0.2 mmol/L dNTP mixture (1 μL), 10 pmol (dT)17-p3 (oligonucleotide primer: GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT; 1 μL), 5 × first-strand buffer (“BioTake”, China; 4 μL), 200 U SuperScript reverse transcriptase (“BioTake”, China; 1 μL), 40 U RNase inhibitor (1 μL), and total RNA (1-2 μg) in a total volume of 20 μL. The RNA in each reaction was reverse transcribed by heating at 42 °C for 50 min and inactivated by heating at 70 °C for 15 min. Samples were stored at 4 °C.
To confirm that EC109 cells with detectable HPV18 E7 mRNA were indeed harboring HPV, mRNA for a human housekeeping gene was amplified by RT-PCR to ensure that mRNA isolated from EC109 cells was of sufficient integrity to be amplified by PCR. mRNA encoding human GAPDH was used as a target for the RT-PCR. PCR was carried out as described previously[19], using 10 pmol/L of each GAPDH primer (forward 5’-CATCACCATCTTCCAGGA-3’; reverse 5’-GTCTACCACCCTATTGCA-3’) and 2 μL cDNA at a 52 °C annealing temperature for 30 s to generate a GAPDH product of 500 bp.
Detection of viral-cell fusion transcripts by nested PCR
HPV18 PCR reactions were prepared as described by Klaes et al[21] using forward primer p1-18 specific for HPV18 E7 (5’-TAGAAAGCTCAGCAGACGACC-3’) and p3 (5’-GACTCGAGTCGACATCG-3’) as reverse primer, 1 × buffer, 2.5 mmol/L MgCl2, 0.2 mmol/L dNTPs, 10 pmol/L primers, 1 U Ex Taq DNA polymerase, 3 μL cDNA product in a total volume of 25 μL. PCR was conducted as follows: 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 56 °C for 1 min, and elongation at 72 °C for 3 min. The last cycle was followed by a final extension step at 72 °C for 5 min.
The amplification product (5 μL) was used for nested PCR under identical conditions using forward primers p2-18 specific for HPV18 E7 (5’-ACGACCTTCGAGCATTCCAGCAG-3’) and (dT)17-p3 as reverse primer, except that the annealing temperature was 67 °C. The positions of the two primers were 814-835 for p1-18 and 830-853 for p2-18. To control for false-positives, a negative control (HEK293 cell DNA template) was included in each amplification. Electrophoresis was performed using a 1.2% agarose gel.
Cloning and sequence analysis
To confirm specific HPV18 oncogene transcription in EC109 cells, the final nested PCR products were cloned into a pMD-18T vector and sequenced to analyze the chromosomal location of HPV integration.
RESULTS
Detection of HPV DNA in EC109 cells
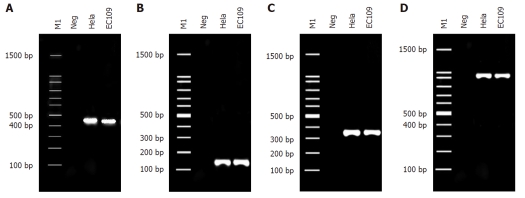
HPV18 DNA was detected in EC109 cells by PCR using the general primer sets of My09/11 and GP5 +/6 + for HPV L1 and the type-specific primer sets for HPV18 E6 and E6-E7 to generate products of 450 bp, 150 bp, 335 bp and 944 bp respectively (Figure 1).
Figure 1.
Detection of HPV18 DNA in EC109 cells. A-D: HPV18 DNA detection in EC109 cells using the primer pairs Y09/11, GP5 +/6 +, HPV18E6, and HPV18E6-E7, respectively. M1: 100 bp DNA ladder; Neg: Negative control (template of HEK 293 cell DNA); HeLa: Positive control (template of HPV18-infected cervical cancer cells; EC109: DNA from EC109 cells. HPV: Human papillomavirus; EC: Esophageal carcinoma.
HPV18 integration–derived transcript in EC109 cells
EC109 cells were used subsequently for HPV18 E7–specific nested PCR followed by gel electrophoresis. Approximately 600 bp of integrated HPV18-specific transcript was identified. The final nested PCR product of integrated HPV18 DNA was cloned into a pMD-18T vector and sequenced to analyze the chromosomal location of HPV integration. Sequence alignment showed that the HPV18 sequence from EC109 cells was identical to that of the encoded early protein E7-E1 of the standard HPV18 strain X05015 (Figure 2). Sequence alignment also showed that another partial gene sequence was identical to a partial sequence of human chromosome 8 (Figure 3).
Figure 2.
Alignment sequencing results for esophageal carcinoma109 cells with human papillomavirus18. Query 288-388, sequencing results for esophageal carcinoma109 cells after polymerase chain reaction amplification with P2-18E7-specific primers; Sbjct 844-944, partial sequence of human papillomavirus18E7-E1 from X05015 in GenBank.
Figure 3.
Alignment sequencing results for esophageal carcinoma109 cells with human chromosome. Query 227-483, sequencing results for esophageal carcinoma109 cells after polymerase chain reaction amplification with P2-18E7-specific primers; Sbjct 41562135-41562391, partial sequence of human chromosome 8.
DISCUSSION
The APOT assay is based on the structures of the 3’-ends of oncogenic HPV transcripts[21]. Integration of HPV genomes in carcinoma cells usually result in disruption of both the E1 and E2 open reading frames (ORFs), and transcripts derived from the integrated E6 and E7 oncogenes usually contain viral sequences at their 5’ ends and flanking host cell–derived sequences at their 3’ ends[21,23,24]. Klaes et al[21] applied the APOT assay to clinical samples of cervical dysplasia infected with HR-HPV types 16 and 18 and found a strong correlation between the detection of integration-derived transcripts and the stage of progression of the cervical dysplasia.
In the current study, HPV18 DNA and integration-derived transcripts were detected in EC109 cells, and HPV18 was found to be integrated into chromosome 8. Our results indicate that an episomal viral genome was broken for integration which usually leads to the disruption of the E1 and E2 ORFs. During the viral life cycle, E2-derived proteins act as important regulators of E6 and E7 ORF expression[24]. In most infected epithelia, E2 appears to inhibit transcription from E6 and E7 ORFs which helps to maintain the regulation of cellular proliferation[25,26]. Disruption of the E2 ORF with retention of the E6 and E7 ORFs could result in the unregulated expression of E6 and E7, which would lead to abrogation of cell-cycle activity and uncontrolled cell proliferation[27]. The high-risk HPV E6 and E7 gene are commonly integrated into the genome of cells in malignant tumors[28], when this occurs, longer incubation periods may be required for viral DNA integration into the appropriate host cell genomic location. When HPV DNA is integrated into the host nuclear genome, expression of E6 and E7 is elevated, and this leads to the occurrence of cancer[29]. After HPV infection of the esophageal epithelium, HPV DNA can be randomly integrated into human chromosomal DNA to produce a variety of genetic changes, leading to chromosomal instability and eventually malignant transformation.
COMMENTS
Background
Human papilloma virus (HPV) in patients with esophageal carcinoma (EC) has been studied previously, but the association of HPV with EC has not been firmly established. The authors hypothesized that integration of HPV DNA into host chromosomes is a critical step in the carcinogenesis of EC as a result of altered expression of two viral transforming genes, E6 and E7. The aim of this work was to study the relationship between HPV and esophageal tumors and to determine the chromosomal integration sites of HPV DNA in EC cells.
Research frontiers
EC is one of the major cancers in China, where the incidence and mortality rank first in the world. In 1982, Syrjanen hypothesized a relationship between HPV infection and the development of EC. However, the role of HPV in the carcinogenesis of EC remains unclear. In this study, the authors demonstrate the integration of HPV DNA into host chromosomes, which could be a potential mechanism where by HPV infection leads to the development of EC.
Innovations and breakthroughs
Recent reports have highlighted the importance of HPV infection in EC. This paper is the first study to report that HPV18 integrated into one part of chromosome 8 in a cell line, EC109, derived from human EC cells. This study further suggests that HPV infection may be the cause of EC.
Applications
By understanding how EC is induced after HPV infection, this study may provide a future strategy for the diagnosis and prevention of EC.
Peer review
The authors examined HPV18 integration into one part of chromosome 8 in EC109 cells. Integration of the HPV genome into the host cell chromosome suggests that persistent HPV infection is a key factor in malignant cell transformation and carcinogenesis. The results may represent a molecular mechanism for the development of EC.
Footnotes
Supported by An independent research fund from the National Institute for Viral Disease Control and Prevention, the Chinese Center for Disease Control and Prevention; the State Key Laboratory for Infectious Disease Prevention and Control (Grant No. 2011SKLID103)
Peer reviewers: Piero Marco Fisichella, MD, Assistant Professor of Surgery, Medical Director, Swallowing Center, Loyola University Medical Center, Department of Surgery, Stritch School of Medicine, 2160 South First Avenue, Room 3226, Maywood, IL 60153, United States; Kenichi Goda, MD, PhD, Department of Endoscopy, The Jikei University School of Medicine, 3-25-8 Nishi-shimbashi, Minato-ku, Tokyo 105-8461, Japan
S- Editor Sun H L- Editor Cant MR E- Editor Zhang DN
References
- 1.Establishment of a cell line from human esophageal carcinoma. Chin Med J (Engl) 1976;2:357–364. [PubMed] [Google Scholar]
- 2.Qi ZL, Xu XJ, Zhang B, Shen ZY, Huo X. Esophageal carcinoma 109 cell line is found positive in HPV type 18. Dis Esophagus. 2007;20:362–363. doi: 10.1111/j.1442-2050.2007.00753.x. [DOI] [PubMed] [Google Scholar]
- 3.Syrjänen KJ. Histological changes identical to those of condylomatous lesions found in esophageal squamous cell carcinomas. Arch Geschwulstforsch. 1982;52:283–292. [PubMed] [Google Scholar]
- 4.Benamouzig R, Pigot F, Quiroga G, Validire P, Chaussade S, Catalan F, Couturier D. Human papillomavirus infection in esophageal squamous-cell carcinoma in western countries. Int J Cancer. 1992;50:549–552. doi: 10.1002/ijc.2910500409. [DOI] [PubMed] [Google Scholar]
- 5.Chang F, Syrjänen S, Shen Q, Ji HX, Syrjänen K. Human papillomavirus (HPV) DNA in esophageal precancer lesions and squamous cell carcinomas from China. Int J Cancer. 1990;45:21–25. doi: 10.1002/ijc.2910450106. [DOI] [PubMed] [Google Scholar]
- 6.Lavergne D, de Villiers EM. Papillomavirus in esophageal papillomas and carcinomas. Int J Cancer. 1999;80:681–684. doi: 10.1002/(sici)1097-0215(19990301)80:5<681::aid-ijc8>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 7.Li T, Lu ZM, Chen KN, Guo M, Xing HP, Mei Q, Yang HH, Lechner JF, Ke Y. Human papillomavirus type 16 is an important infectious factor in the high incidence of esophageal cancer in Anyang area of China. Carcinogenesis. 2001;22:929–934. doi: 10.1093/carcin/22.6.929. [DOI] [PubMed] [Google Scholar]
- 8.Suzuk L, Noffsinger AE, Hui YZ, Fenoglio-Preiser CM. Detection of human papillomavirus in esophageal squamous cell carcinoma. Cancer. 1996;78:704–710. doi: 10.1002/(SICI)1097-0142(19960815)78:4<704::AID-CNCR2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 9.Togawa K, Jaskiewicz K, Takahashi H, Meltzer SJ, Rustgi AK. Human papillomavirus DNA sequences in esophagus squamous cell carcinoma. Gastroenterology. 1994;107:128–136. doi: 10.1016/0016-5085(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 10.Toh Y, Kuwano H, Tanaka S, Baba K, Matsuda H, Sugimachi K, Mori R. Detection of human papillomavirus DNA in esophageal carcinoma in Japan by polymerase chain reaction. Cancer. 1992;70:2234–2238. doi: 10.1002/1097-0142(19921101)70:9<2234::aid-cncr2820700903>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Chen SH, Liu ZH, Zhang WD, Li LZ, Cen S, Tan LZ, Shen ZY, Zeng Y. The relationship between human papillomavirus and esophageal and cardia carcinoma in the Jieyang area. Chin J Exp Clin Virol. 1998;12:382–383. [Google Scholar]
- 12.Chen HB, Chen L, Zhang JK, Shen ZY, Su ZJ, Cheng SB, Chew EC. Human papillomavirus 16 E6 is associated with the nuclear matrix of esophageal carcinoma cells. World J Gastroenterol. 2001;7:788–791. doi: 10.3748/wjg.v7.i6.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen ZY, Hu SP, Lu LC, Tang CZ, Kuang ZS, Zhong SP, Zeng Y. Detection of human papillomavirus in esophageal carcinoma. J Med Virol. 2002;68:412–416. doi: 10.1002/jmv.10219. [DOI] [PubMed] [Google Scholar]
- 14.zur Hausen H. Condylomata acuminata and human genital cancer. Cancer Res. 1976;36:794. [PubMed] [Google Scholar]
- 15.zur Hausen H. Immortalization of human cells and their malignant conversion by high risk human papillomavirus genotypes. Semin Cancer Biol. 1999;9:405–411. doi: 10.1006/scbi.1999.0144. [DOI] [PubMed] [Google Scholar]
- 16.Bosch FX, Lorincz A, Muñoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen ZY, Cen S, Xu LY, Cai WJ, Chen MH, Shen J, Zeng Y. E6/E7 genes of human papilloma virus type 18 induced immortalization of human fetal esophageal epithelium. Oncol Rep. 2003;10:1431–1436. [PubMed] [Google Scholar]
- 18.van Beurden M, ten Kate FJ, Smits HL, Berkhout RJ, de Craen AJ, van der Vange N, Lammes FB, ter Schegget J. Multifocal vulvar intraepithelial neoplasia grade III and multicentric lower genital tract neoplasia is associated with transcriptionally active human papillomavirus. Cancer. 1995;75:2879–2884. doi: 10.1002/1097-0142(19950615)75:12<2879::aid-cncr2820751214>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 19.Remmink AJ, Walboomers JM, Helmerhorst TJ, Voorhorst FJ, Rozendaal L, Risse EK, Meijer CJ, Kenemans P. The presence of persistent high-risk HPV genotypes in dysplastic cervical lesions is associated with progressive disease: natural history up to 36 months. Int J Cancer. 1995;61:306–311. doi: 10.1002/ijc.2910610305. [DOI] [PubMed] [Google Scholar]
- 20.Dell G, Gaston K. Human papillomaviruses and their role in cervical cancer. Cell Mol Life Sci. 2001;58:1923–1942. doi: 10.1007/PL00000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, Schneider A, Lotz B, Melsheimer P, von Knebel Doeberitz M. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 1999;59:6132–6136. [PubMed] [Google Scholar]
- 22.Karlsen F, Kalantari M, Jenkins A, Pettersen E, Kristensen G, Holm R, Johansson B, Hagmar B. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J Clin Microbiol. 1996;34:2095–2100. doi: 10.1128/jcm.34.9.2095-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadaja M, Sumerina A, Verst T, Ojarand M, Ustav E, Ustav M. Genomic instability of the host cell induced by the human papillomavirus replication machinery. EMBO J. 2007;26:2180–2191. doi: 10.1038/sj.emboj.7601665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker CC, Phelps WC, Lindgren V, Braun MJ, Gonda MA, Howley PM. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells SI, Aronow BJ, Wise TM, Williams SS, Couget JA, Howley PM. Transcriptome signature of irreversible senescence in human papillomavirus-positive cervical cancer cells. Proc Natl Acad Sci USA. 2003;100:7093–7098. doi: 10.1073/pnas.1232309100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee D, Kim HZ, Jeong KW, Shim YS, Horikawa I, Barrett JC, Choe J. Human papillomavirus E2 down-regulates the human telomerase reverse transcriptase promoter. J Biol Chem. 2002;277:27748–27756. doi: 10.1074/jbc.M203706200. [DOI] [PubMed] [Google Scholar]
- 27.Scheffner M, Romanczuk H, Münger K, Huibregtse JM, Mietz JA, Howley PM. Functions of human papillomavirus proteins. Curr Top Microbiol Immunol. 1994;186:83–99. doi: 10.1007/978-3-642-78487-3_5. [DOI] [PubMed] [Google Scholar]
- 28.Baker CC, Phelps WC, Lindgren V, Braun MJ, Gonda MA, Howley PM. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillemanns P, Wang X. Integration of HPV-16 and HPV-18 DNA in vulvar intraepithelial neoplasia. Gynecol Oncol. 2006;100:276–282. doi: 10.1016/j.ygyno.2005.10.003. [DOI] [PubMed] [Google Scholar]