Abstract
Insights into cancer genetics can lead to therapeutic opportunities. By cross-referencing chromosomal changes with an unbiased genetic screen we identify the ephrin receptor A7 (EPHA7) as a tumor suppressor in follicular lymphoma (FL). EPHA7 is a target of 6q deletions and inactivated in 72 % of FLs. Knockdown of EPHA7 drives lymphoma development in a murine FL model. In analogy to its physiological function in brain development, a soluble splice variant of EPHA7 (EPHA7TR) interferes with another Eph-receptor and blocks oncogenic signals in lymphoma cells. Consistent with this drug-like activity, administration of the purified EPHA7TR protein produces anti-tumor effects against xenografted human lymphomas. Further, by fusing EPHA7TR to the anti-CD20 antibody (rituximab) we can directly target this tumor suppressor to lymphomas in vivo. Our study attests to the power of combining descriptive tumor genomics with functional screens and reveals EPHA7TR as tumor suppressor with immediate therapeutic potential.
Introduction
Insights into the molecular pathogenesis of cancer have led to successful therapies. Recent technological advances greatly facilitate the genome-wide detection of genetic and epigenetic changes in cancer cells. For example, sequencing studies have catalogued somatic mutations that occur in several cancers and paired-end sequencing and array-CGH studies provide a genome-level view of complex genomic aberrations that occur in tumorigenesis (Velculescu, 2008). These studies have revealed a diversity of genetic changes that likely account for some of the clinical heterogeneity seen in pathologically similar tumors. Analyses of larger numbers of patient samples have also uncovered common and recurrent changes that may be considered as key drivers of malignant transformation (Chin and Gray, 2008). Notably, tumors often acquire complex genomic aberrations including gains and losses of large sections or even entire chromosomes. Identifying the target gene(s) from such complex genomic changes remains a significant challenge. RNA-interference (RNAi) technology has greatly facilitated loss-of-function studies in mammalian cells and even in animal models (Dickins et al., 2005; McCaffrey et al., 2002). Moreover, the adaptation of RNAi technology to genetic screens enables rapid and unbiased studies of gene inactivation. Such screens offer a powerful tool to identify important cancer genes based on their biological function. Insights into the genetics of diffuse-large B-cell lymphoma (DLBCL) are successful examples of this strategy (Bidere et al., 2009; Lenz et al., 2008; Shaffer et al., 2008). The combination of tumor genomics with functional screens can distinguish “passenger” from “driver” mutations and unmask haplo-insufficient tumor suppressors that may not be obvious from the genomic data alone. Therefore, cross-referencing genomic data with genetic screens can facilitate the discovery of important cancer genes that may be missed based on genomics alone (Ngo et al., 2011; Oricchio et al., 2010; Zender et al., 2008).
Follicular lymphomas (FL) pose a significant clinical problem because they are among the most common Non-Hodgkin’s lymphoma and are considered incurable by standard chemotherapy approaches (Relander et al., 2010). Clinically, FLs are characterized by slow and persistent growth with eventual progression to an aggressive and rapidly spreading disease that resembles DLBCL. The inclusion of the anti-CD20 antibody (rituximab) has improved the outcome of chemotherapy (Maloney, 2003). However, in eligible patients bone marrow transplantation still remains the only curative option (Barr and Lazarus, 2008). Genetically, FLs are characterized by the translocation t(14;18)(q32;q21) that causes increased expression of the anti-apoptotic BCL2 protein. BCL2 expression in germinal center B-lymphocytes is considered the initiating lesion of FL (Bende et al., 2007). Increased BCL2 is not sufficient for tumor development or progression and additional genetic events are required (Bende et al., 2007). Recurrent lesions in FL include amplification of c-MYC, loss of p53, and frequent deletions affecting large segments of chromosome 6q. These changes have also been associated with early transformation into an aggressive disease and shortened patient survival (Gaidano et al., 1992; Nanjangud et al., 2007; Offit et al., 1993). Deletions affecting chromosome 6q also occur in other lymphoid cancers, for example in DLBCL, where some of targets of the 6q deletion have been identified. These include the BLIMP1/PRDM1 gene that controls terminal lymphocyte differentiation and TNFAIP3/A20, a regulator of NFkB signaling (Calado et al., 2010; Compagno et al., 2009; Kato et al., 2009; Mandelbaum et al., 2010; Pasqualucci et al., 2006). The molecular events and genetic targets of 6q deletions in FLs are not understood and potentially insight into the pathogenesis of FL can inform mechanism-based therapies.
Our study produces actionable insight into the genetics of FL. By cross-referencing tumor genomic data with an unbiased loss-of-function screen we identify EPHA7 as a tumor suppressor that is shed from germinal center B-cells and lost in FL. We confirm its tumor suppressive function in a genetically and pathologically accurate mosaic model of FL. Moreover, we define the molecular mechanism of EPHA7 action and develop an antibody-targeted delivery system for the restitution of EPHA7 in a xenograft system. Hence, our study highlights the power of functional genomics in cancer gene discovery, and we demonstrate how genetic insights can be translated into therapies.
Results
A functional genomics study of follicular lymphoma
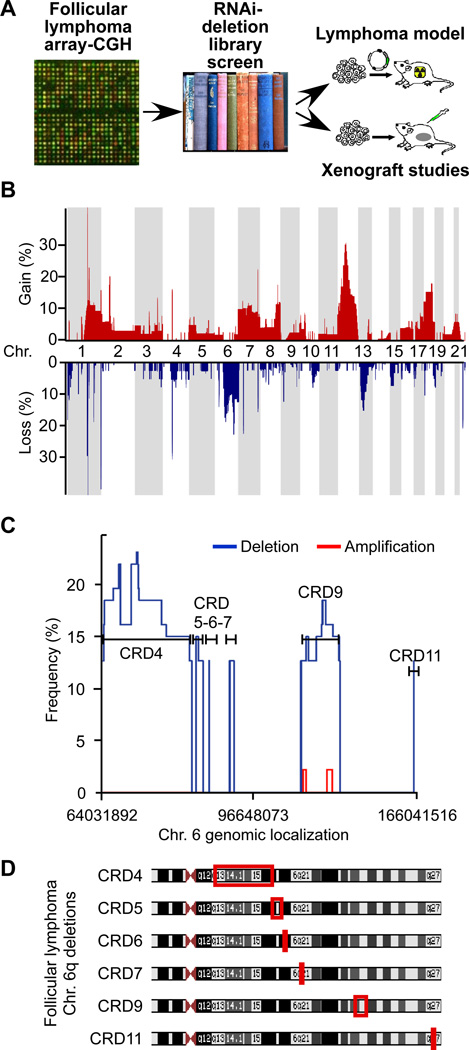
We conducted a systematic functional genomics study into the molecular pathogenesis of FL (Figure 1A). First, we analyzed 64 FLs representing all pathological grades I–III (Grade I: n=21; II: n=20, IIIa: n=16, IIIb: n=7) by array-CGH. Overall, we observed 92 common regions of change. We defined a common change as gains or losses occurring in > 10% of cases. In detail, 38 common changes were tumor specific and not found in the reference DNA, four represented physiological changes (e.g. B-cell receptor rearrangements), and 50 were previously identified common copy number variations (CNVs) (Figure 1B). Deletions affecting chromosome 6q11-27 were the most common losses and occurred in 23% of FLs (15/64 cases). This is consistent with previous cytogenetic studies (Gaidano et al., 1992; Hauptschein et al., 1998; Offit et al., 1993). Moreover, 6q deletions have been linked to patient survival in FL indicating the presence of fundamentally important tumor suppressor(s) in this region (Gaidano et al., 1992; Nanjangud et al., 2007; Offit et al., 1993). Individual cases showed a heterogeneous pattern and typically had large, hemizygous losses of 6q affecting many genes (Suppl. Figure 1). Cumulative analyses of all cases revealed common regions of deletion (CRDs) that ranged from 5kb (CRD11) to 27Mb (CRD4) and harbored between 1 and 78 genes (Figure 1C and 1D). Almost all deletions were hemizygous and two small regions of apparent homozygous loss within the CRD4 and CRD11 regions did not affect any genes directly (Suppl. Table 1–3). Thus, we find a complex pattern of large and hemizygous deletions affecting 6q in FL, which suggests the presence of multiple tumor suppressor genes, however an analysis of the genomic data does not directly pinpoint any specific target gene.
Figure 1. Oncogenomic study to identify tumor suppressor genes in follicular lymphoma (FL).
A, The study design combines genomic tumor analyses with an RNAi screen and validation in murine models and in xenografts; B, Array-CGH analysis of 64 follicular lymphomas showing frequencies of genomic gain (red) and loss (blue) across the genome; C, High resolution depiction of recurrent gains (red) and losses (blue) affecting chromosome 6q, indicated are common regions of deletion (CRDs found in > 10%); D, Mapping of CRDs. The observed 6q deletions are typically large and hemizygous and do not readily identify a target gene. See also Figure S1.
A deletion-specific RNAi screen to identify candidate tumor suppressors at chromosome 6q
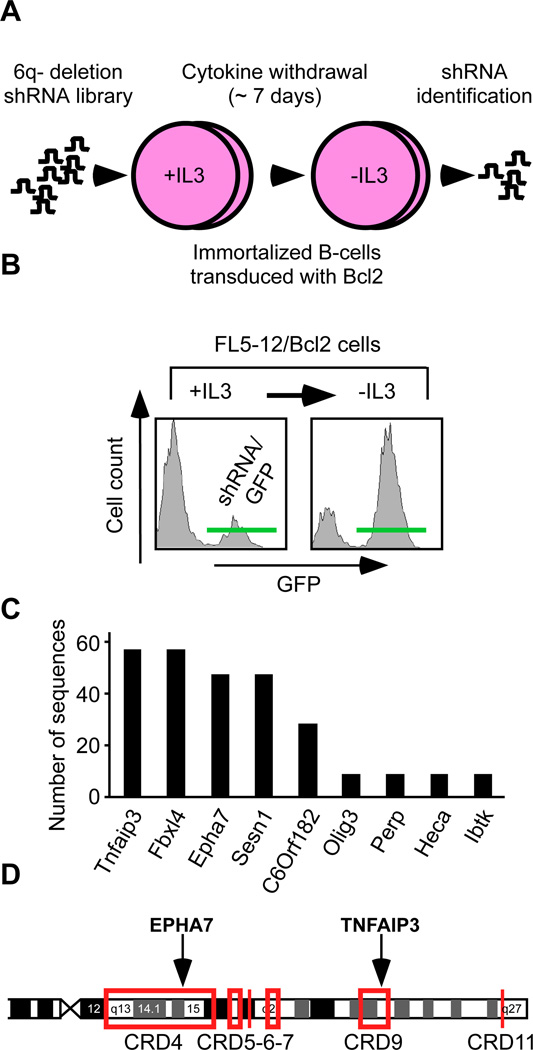
Given the complexity of 6q deletions in FL, we wondered whether an unbiased deletion-specific loss-of-function screen could point us to potential tumor suppressor genes. We constructed a library of 260 shRNAs against 84 genes (1–7 shRNAs per gene) in an MSCV-based, GFP-expressing vector (Suppl. Table 4). For our screen we used non-transformed murine pro-B lymphocytes engineered to express increased levels of Bcl2 as a tractable surrogate in vitro system. We screened the 6q deletion library for shRNAs that could protect the lymphocytes from cytokine (IL-3) depletion (Mavrakis et al., 2010) (Figure 2A). Briefly, we partially transduced the FL5-12/Bcl2 cells with the pooled 6q-deletion library or empty vector and monitored for enrichment of GFP/shRNA expressing cells (Figure 2B). We identified the shRNAs in the enriched population by subcloning and sequencing. A count of clones representing each shRNA revealed a distribution consistent with an enrichment of specific shRNAs (Figure 2C). We then individually re-tested each shRNA that was identified in the enriched cell population using the same assay and confirmed a protective effect for shRNAs targeting Tnfaip3/A20 and EphA7 (Suppl. Figures 2A). Three additional EphA7 shRNAs reduced EPHA7 protein levels and re-produced this protective effect, which was reversible by expressing the human EPHA7 cDNA that is not recognized by the murine shRNA (Suppl. Figures 2B–D). EphA7 knockdown also facilitated faster recovery of cell cycle progression following depletion and re-addition of IL-3 in vitro (Suppl. Fig. 2E). Tnfaip3/A20 has been implicated as a tumor suppressor in the activated B-cell (ABC) type of DLBCL (Compagno et al., 2009; Kato et al., 2009; Novak et al., 2009; Schmitz et al., 2009). The Eph-receptor A7 (EphA7) is a surprising and new candidate gene. Looking back at the genomic data, we find that EPHA7 and TNFAIP3 are affected by common deletions in FL, and fall into the CRD4 and CRD9 region, respectively (Figure 2D, Suppl. Fig. 1B–C). BLIMP1/PRDM1 has been implicated in DLBCL (Calado et al., 2010; Mandelbaum et al., 2010; Pasqualucci et al., 2006), but fell outside the common region of deletion and did not emerge from our screen (Figure 2D, Suppl. Figure 2A). Hence, our RNAi screen identifies a known tumor suppressor TNFAIP3/A20 (Compagno et al., 2009), and points to the ephrin receptor A7 (EPHA7) as a candidate tumor suppressor gene.
Figure 2. A 6q-deletion specific RNAi screen functionally identifies EPHA7 as a candidate tumor suppressor gene.
A, Design of a pooled, deletion-specific shRNA library screen in a surrogate model (immortalized FL5-12/Bcl2 cells); B, FACS profiles for GFP showing enrichment of cells expressing the shRNA library (and the GFP reporter) following IL-3 depletion. See also Figure S2; C, Absolute number and identity of shRNA sequences retrieved from the enriched population; D, EPHA7 and TNFAip3 map to the CRD4 and CRD9 in FL. See also Figure S1.
EphA7 acts as a tumor suppressor in a murine model of follicular lymphoma
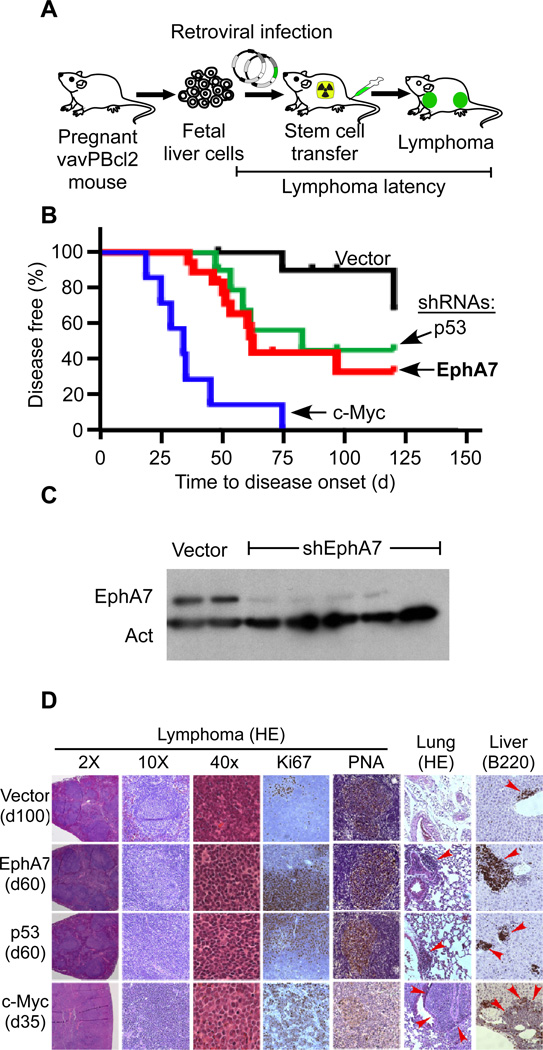
Mosaic mouse models of cancer enable the rapid functional assessment of genetic interactions in tumorigenesis (Heyer et al., 2010). The vavPBcl2 model closely recapitulates the genetics and morphology of human FL (Egle et al., 2004). In order to directly test the role of EphA7 in FL we transduced vavPBcl2 transgenic hematopoietic progenitors (HSCs) with retroviral shRNA vectors and transplanted these genetically engineered cells into irradiated recipients (Wendel et al., 2004) (Figure 3A). Ninety percent of controls remained tumor free for > 100d (vector n=11). c-MYC and p53 have established roles in FL transformation (Nanjangud et al., 2007). Accordingly, we find that enforced c-Myc expression (p < 0.01, n = 7) and also p53 knockdown (p < 0.01, n = 9) accelerated lymphomagenesis in vivo. Knockdown of EphA7 had a similar effect on tumor latency as p53 inactivation (p < 0.01, n = 18) (Figure 3B). Immunoblots on purified vavBcl2 lymphoma cells readily confirmed efficient loss of EphA7 expression in vivo, by contrast EphA7 was abundantly expressed in murine splenocytes and HSCs (Figure 3C, Suppl. Fig. 3A). A second shRNA against EphA7 was similarly enriched during lymphomagenesis in vivo (Suppl. Fig. 3B). Hence, EphA7 knockdown promotes lymphoma development in the vavPBcl2 transgenic model of FL.
Figure 3. EphA7 opposes tumor development in a murine model of FL.
A, A mosaic model of FL based on vavPBcl2 transgenic mice; B, Tumor latencies for animals receiving vavPBcl2 transgenic HSCs transduced with empty vector (black, n = 11), or shRNAs against EphA7 (red, n = 18) and p53 (green, n = 9) or over expressing c-Myc (blue, n = 7). C, Immunoblot on FACS purified vavPBcl2 lymphoma cells expressing vector or an shRNA against EphA7 and probed as indicated. D, Microscopic pathology and immunohistochemistry of vavPBcl2 lymphomas expressing the indicated constructs, red arrows indicate infiltrating tumor cells. See also Figure S3.
A detailed pathological analysis of the vavPBcl2 tumors confirmed key features of human follicular lymphomas. In particular, the lymphomas retained the typical follicular structures, showed expression of PNA, a marker indicating a germinal center B-cell phenotype, and had low Ki-67 indicating slow proliferation like human FLs (Figure 3D, Suppl. Fig. 3C–E, Suppl. Table 5). As expected these Bcl2 expressing tumors showed little or no apoptosis by TUNEL. The c-Myc expressing tumors were notably different and grew in a diffuse pattern that resembled transformed FLs or aggressive DLBCL. These tumors may represent the aggressive transformation that is also seen in human FL. All tumors expressed B-cell markers (B220, CD19), and had varying degrees of T-cell infiltration (Suppl. Table 5) (Egle et al., 2004). Further supporting the germinal center origin of the murine lymphomas, sequencing of the immunoglobulin JH4 intron confirmed somatic hypermutation (SHM) that is also seen in human FL (Suppl. Table 6) (Egle et al., 2004; Mandelbaum et al., 2010; McBride et al., 2008). PCR analysis of the immunoglobulin heavy chain locus confirmed clonal origin of tumors (Suppl. Fig. 3C–E) (Egle et al., 2004). Thus, the mosaic model based on the vavPBcl2 transgenic mice retains key features of human FL and reveals that EphA7 behaves as a tumor suppressor in follicular lymphomagenesis.
EPHA7 expression is lost in 72% of follicular lymphomas
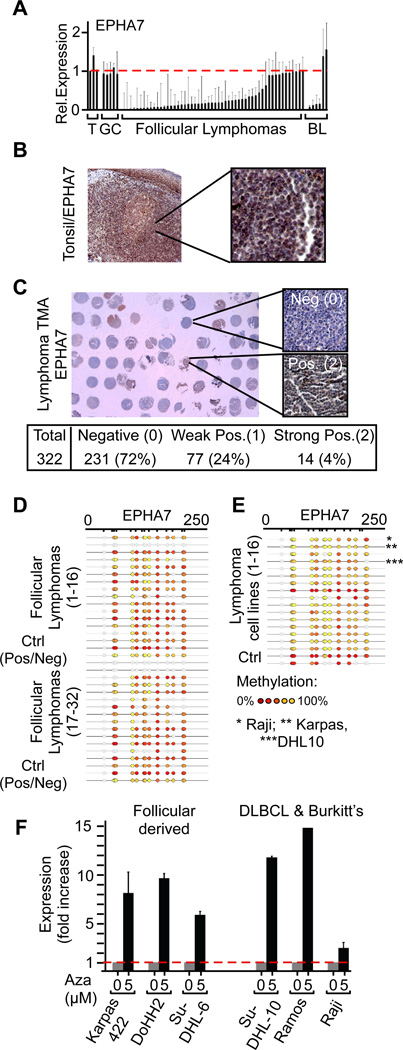
In our array-CGH analysis we found that EPHA7 is affected by frequent hemizygous 6q deletions. We wondered whether EPHA7 may also be subject to epigenetic silencing or mutational inactivation or if it acts as a haploinsufficient tumor suppressor. We initially noted a differential reduction of EPHA7 mRNA expression levels in lymphomas compared to germinal center (GC) B-cells, which are the normal counterpart and cell of origin of these tumors (Klein and Dalla-Favera, 2008). In detail, qRT-PCR showed decreased EPHA7 mRNA levels in purified lymphoma cells (FL: 41 of 50 (82%); BL: 4 of 6 (67%)) compared to GC B-cells or tonsils (p(B-cell vs. lymphoma) < 0.02) (Figure 4A). In our collection, reduced levels of EPHA7 were significantly associated with tumor grade, which is linked to clinical outcomes (Suppl. Fig. 4A). We easily detected cytpolasmic EPHA7 in normal tonsils by immunohistochemistry. By contrast, EPHA7 expression was completely absent in 231 of 332 (72%) of FLs samples on a tissue microarray (TMA) (Figure 4B and 4C, Suppl. Fig. 4B). Hence, EPHA7 protein expression closely resembled the results of our qRT-PCR analysis.
Figure 4. EPHA7 is differentially silenced in FLs and expressed in germinal center (GC) B-cells.
A, qRT-PCR results for EPHA7 in purified B-cells from reactive tonsils (T), GC B-cells (GC), purified B-cells from follicular lymphomas (FL), and Burkitt’s lymphomas (BL) (mean +/− standard deviation; p (tumor vs. normal < 0.05 for FL and BL); B, Immunohistochemical detection of the EPHA7 protein in a normal tonsil; C, Representative section of tissue microarrays (TMA) representing 322 human FLs and stained for EPHA7; D and E, Mass-array analysis of EPHA7 promoter methylation in 32 follicular lymphomas (D), and 16 human lymphoma lines (E) and positive / negative controls (Ctrl); the color scale indicates the degree of methylation (Red: 0%; yellow: 100%); f, qRT-PCR of EPHA7 mRNA levels in human lymphoma cell lines treated with 5’aza-2’-deoxycytidine (Aza); for all cell lines: p(untr. vs. Aza.) < 0.01). See also Figure S4.
Loss of EPHA7 expression in lymphomas was due to differential promoter methylation. Mass-array analysis on 32 primary FLs and 16 lymphoma cell lines revealed extensive CpG island methylation (Figure 4D and 4E, Suppl. Fig. 4C–E). We confirmed differential epigenetic silencing in normal GC B-cells (n = 9) and lymphomas (FL; n = 9 and DLBCL; n = 155; lymphoma lines; n = 24) using the HELP (HpaII tiny fragment enrichment by ligation-mediated PCR) assay for methylation detection (Suppl. Fig. 4D, 4E). Accordingly, in vitro treatment of human lymphoma cells with 5’Aza-2’-deoxycytidine caused re-expression of EPHA7 (Figure 4F). EPHA7 silencing occurs differentially in lymphomas and not in GC-B-cells. Therefore silencing cannot be attributed to cellular differentiation stage. Consistent with our observations in FL and in DLBCL, differential EPHA7 silencing has been reported in murine lymphomas arising in Tcl1 transgenic animals and in human B-lymphoblastic leukemia (B-ALL) (Dawson et al., 2007; Kuang et al., 2010). These findings may indicate a broader role for EPHA7 in lymphocyte malignancies. While somatic mutations affecting EPHA7 have been reported in lung cancer (Ding et al., 2008), we did not detect EPHA7 mutations in the 24 FL cases we analyzed (not shown). Hence, EPHA7 acts as a tumor suppressor in vivo and is targeted by genomic deletions and differential epigenetic silencing in human lymphomas.
EPHA7 blocks oncogenic signals in human lymphoma cells
The role of ephrin signaling in cancer is unclear and both oncogenic and tumor suppressive functions have been discussed (Noren et al., 2006; Pasquale, 2010). Ephrin receptors are tyrosine kinases that form dimers and are activated upon contact with ephrin expressing cells (Himanen et al., 2010; Seiradake et al., 2010). In this manner ephrin signaling is thought to mediate cell-cell signals in a variety of physiological contexts (Pasquale, 2010). Notably, alternate splice forms have been described for the murine and human EPHA7 genes that result in truncated proteins that lack kinase activity and can be shed from the cell surface (Dawson et al., 2007; Holmberg et al., 2000; Valenzuela et al., 1995). Alternate EPHA7 splicing has an important role in embryonic development, where expression of the short variant and its binding to the full-length EPHA7 receptor mediate a switch from cellular repulsion to adhesion during neural tube closure (Holmberg et al., 2000).
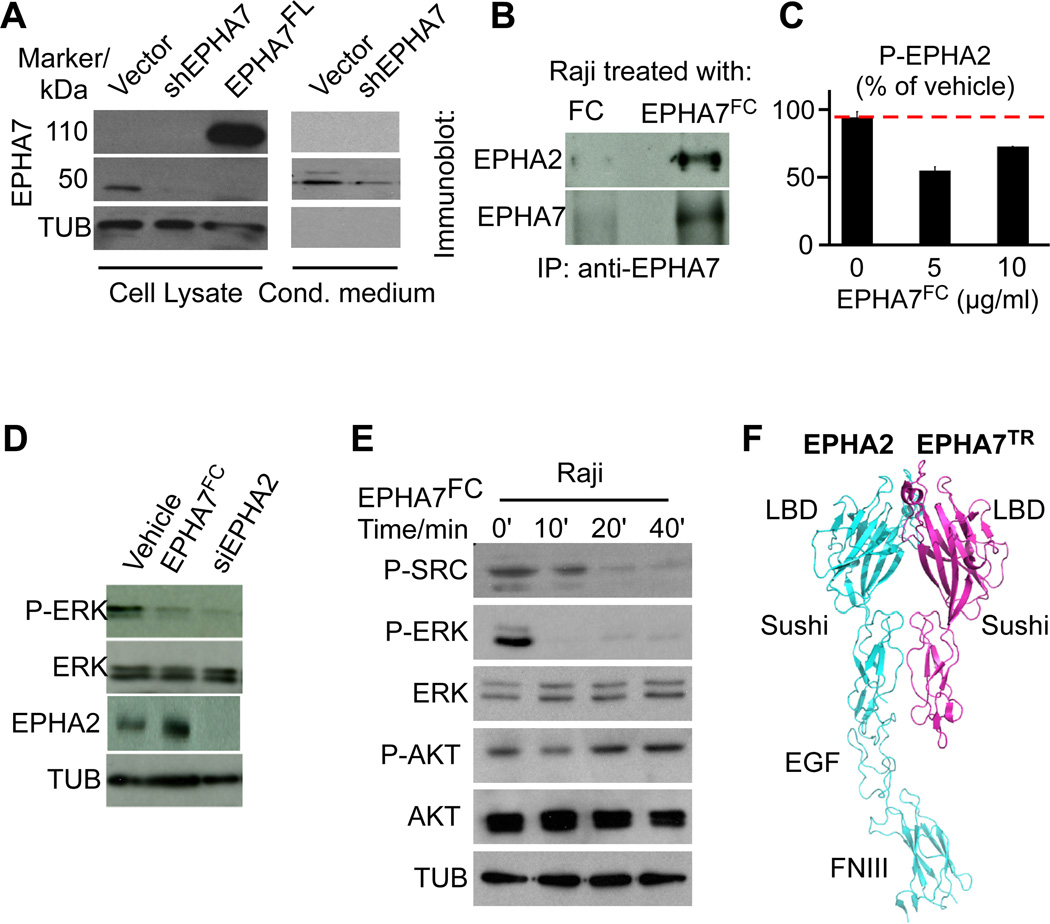
Normal B-lymphocytes express only a truncated EPHA7 protein (EPHA7TR) and not the full-length receptor (Figure 5A). Similarly, treatment of human SU-DHL-10 lymphoma cells with the de-methylating agent 5’aza-2’-deoxycytidine results in re-expression of only a truncated EPHA7TR protein that lacks the intracellular domains (Suppl. Fig. 4F – 4I). Consistent with potential shedding from the cell surface, we detect EphA7TR not only in lymphocyte lysates, but also in conditioned media from lymphocyte cultures, and also in the serum of volunteers (Figure 5A, Suppl. Fig. 5A). Lymphomas and derived cell lines do not express EPHA7 (Figure 4). However, Raji and DoHH2 cells express another EPHA receptor, the homologous EPHA2 receptor. Immunoprecipitation shows that an FC-tagged EPHA7 ectodomain protein (from hereon called EPHA7FC) can bind to the EPHA2 receptor (Figure 5B; Suppl. Figure 5B, 5C). In vitro treatment with EphA7FC further decreases EPHA2 phosphorylation by ELISA, and both, EPHA7FC treatment and EPHA2 knockdown by RNA interference lead to inhibition of ERK phosphorylation in lymphoma cell lines (Figure 5C, 5D, Suppl. Figure 5D, 5E). Conversely, knockdown of EPHA7 using additional EphA7 shRNAs causes ERK activation in B-lymphocytes, and this activation is reversed with purified EPHA7FC protein (Supp. Figure 5F–I). Using a phospho-protein array we identified additional signaling effects of EPHA7FC in lymphoma cells. Briefly, the array confirmed ERK inhibition and showed that EPHA7FC blocks several SRC family kinases including FYN, YES, FAK, SRC, also STAT3 and others (Suppl. Figure 5J, 5K). We confirmed the effects on ERK, STAT3 and SRC phosphorylation by immunoblot and also noted some cell line specific differences (Figure 5E, Suppl. Figure 5L, 5M). Hence, EPHA7FC acts, at least in part, as a dominant, soluble inhibitor of EPHA2 signaling in lymphoma cells.
Figure 5. EPHA7FC binds to EPHA2 and blocks oncogenic signals in lymphoma cells.
A, Lysates and conditioned media from FL5-12/Bcl2 cells expressing vector, an shRNA against EphA7 (shEphA7), or full length EphA7 (EphA7FL) probed with an antibody against EPHA7. B, Immunoprecipitation of lysates from Raji cells treated with EPHA7FC (FC-tagged ectodomain of EPHA7) or FC, IP with anti-EPHA7 and probed against EPHA7 and EPHA2; C, ELISA assay for EPHA2 phosphorylation on Raji cells treated with EPHA7FC or vehicle (FC); D, Immunoblot on lysates of Raji cells treated with vehicle, EPHA7FC (5µg) or an siRNA against EPHA2; E, Immunoblot on Raji cells treated with 5µg/ml EPHA7FC for the indicated times; F, Model of the EPHA2 – EPHA7TR interaction based on the known structure of EPHA2 and its homology with EPHA7 (LBD, ligand binding domain, EGF, EGF-like domain, FNIII, fibronectin domain). See also Figure S5.
Next, we examined the possibility that EPHA7 might interact with additional EPHA receptors or ephrin ligands. Gel shift revealed that purified EPHA7FC could interact with EPHA2 and also EPHA3 but not EPHA4 (Suppl. Fig. 5N). However, in FL samples we only detected mRNA expression of EPHA2 and not EPHA3, and we confirmed EPHA2 protein expression by staining the TMA (Suppl. Fig. 5O and data not shown). Ephrins are the physiological ligands for Eph receptors and gel-shift showed that purified EphA7FC can interact with ephrins A1, A4 and A5, but not with ephrins A2 and A3 (Suppl. Fig. 5P). We detected significant expression of ephrins A1 and A3 in human FL samples (Suppl. Fig. 5Q and data not shown). To discern the contribution of ephrin binding to the signaling and anti-proliferative effects of EPHA7 in lymphoma, we mutated the ephrin binding domain of EPHA7 (T105Q) (Suppl. Fig. 5R). The analogous mutation in EPHA3 has been shown to selectively disrupt ephrin binding to the receptor (Smith et al., 2004). The EPHA7 mutant (T105Q) retained the ability to block ERK activation and had anti-proliferative effects that were similar to the wild type EPHA7 (Suppl. Figs. 5S, 5T). We cannot exclude that EPHA7 may also bind additional surface molecules. Based on these data, the known structure of EPHA2, and its homology with the EPHA7 sequence (51%) and domain structure we built a structural model (Himanen et al., 2010; Seiradake et al., 2010). The model suggests that the EPHA2 receptor and EPHA7TR may interact through several sites in their Sushi and ligand binding domains, resulting in a heterodimer that cannot undergo auto-phosphorylation and activation (Figure 5F). In analogy to the function of EPHA7TR in nervous system development (Holmberg et al., 2000), we propose that EPHA7 acts as a decoy receptor that blocks EPHA2 activation and oncogenic signals including ERK and SRC in lymphoma cells.
EPHA7 has anti-tumor activity against xenografted human lymphomas
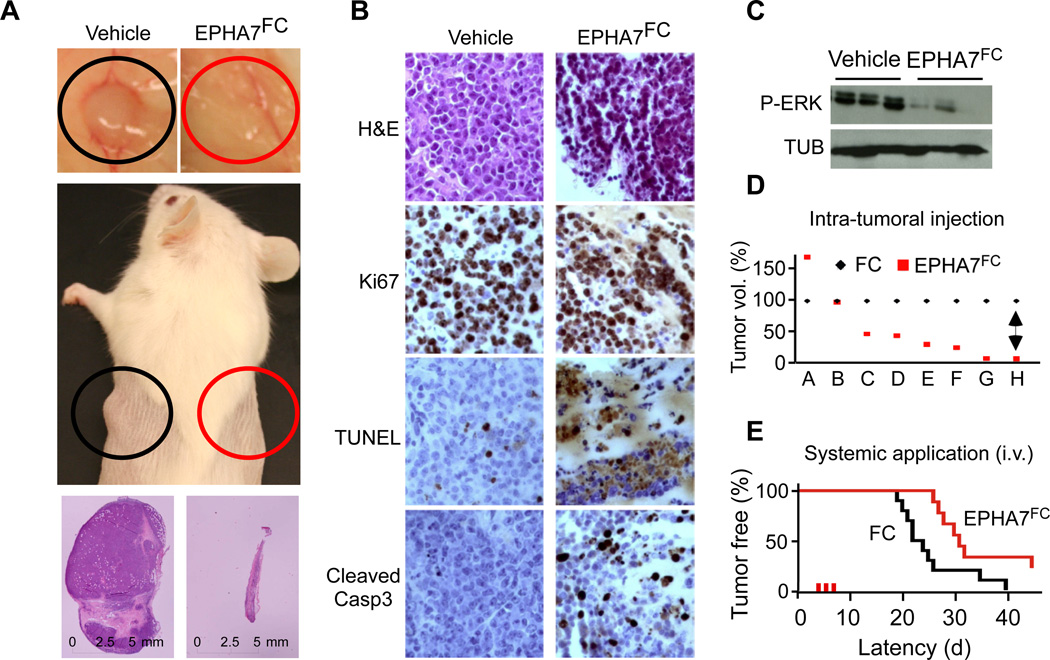
Given that EPHA7TR can block oncogenic signaling molecules and act as a tumor suppressor in a murine model, we wondered whether the EPHA7TR protein had therapeutic activity against xenografted human lymphomas. First, we examined the effects of restoring EPHA7 by either retroviral expression or by direct application of the purified EPHA7FC protein on human lymphoma cells in vitro. In these experiments EPHA7 had powerful anti-proliferative effects against Raji, DHL-10, DoHH2 and Karpas 422 cells in vitro (Suppl. Figure 6A–E). In vivo we observed the most striking effects upon local EPHA7 injection. The purified EPHA7FC protein (20µg for 3 days), but not vehicle (Fc), caused dramatic tumor regressions (n = 12; p(FC vs. EPHA7)<0.04) (Figure 6A, Suppl. Figure 6F and 6G). Residual EPHA7FC treated lymphoma xenografts showed extensive apoptosis, disrupted architecture, and reduced ERK phosphorylation (Figure 6B–D). We also observed that systemic administration of EPHA7FC via tail-vein injection (i.v. 20µg for 3 days) caused a significant delay in the development of xenografted Raji lymphomas (EPHA7FC: n = 5; Vehicle/Fc: n = 5; p < 0.05) (Figure 6E).
Figure 6. Exogenous administration of purified EPHA7 suppresses human lymphoma xenografts.
A, Xenografted Raji lymphomas grown in the flank of NOD/SCID mice and treated three times on alternate days by intra-tumoral administration of 20 µg EPHA7FC (red circle) or vehicle (FC; black circle); B, Microscopic pathology on EPHA7FC treated and mock treated Raji lymphomas stained as indicated; C, Immunoblot on lysates of tumors treated with EPHA7FC or vehicle in vivo; D, Matched pair analysis of tumor volumes of eight (A–H) treated (red) and control (black) Raji lymphomas; E, Intravenous (i.v.) administration of vehicle (FC, black) or EPHA7FC (20µg, for 3 days, red) delays tumor development from 1×106 injected Raji lymphoma cells. See also Figure S6.
Targeted tumor suppressor therapy with an anti-CD20-EPHA7 fusion protein
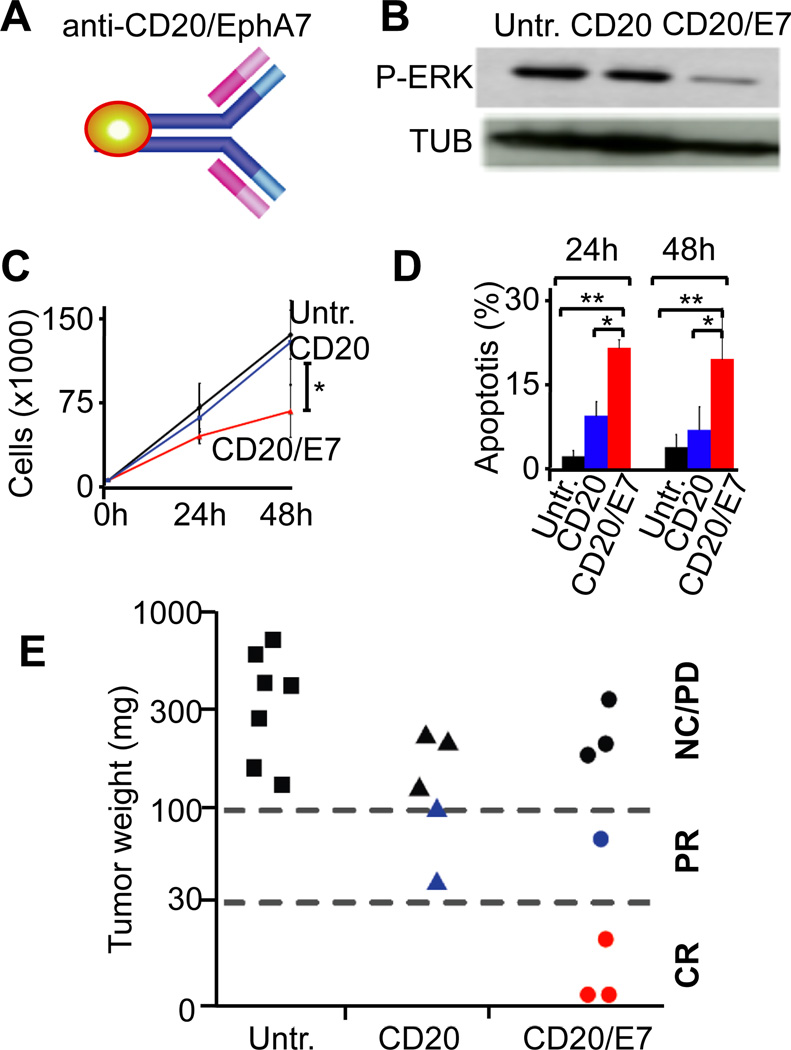
Next, we wondered whether fusing EPHA7 to the Fc-terminus of an anti-CD20 antibody (rituximab) could further enhance its therapeutic potential and selectively deliver the EPHA7 tumor suppressor to the lymphoma cells in vivo (Figure 7A, Suppl. Figure 7A). Initial characterization of the fusion antibody (anti-CD20-EPHA7TR) revealed that it retained properties of both proteins. Namely, the fusions recognized CD20+ Raji lymphoma cells in vitro (Suppl. Figure 7B), and the protein retained the ability to block EPHA2 and ERK phosphorylation in vitro (Figure 7B, Suppl. Figure 7C). Moreover, the fusion antibody was more efficient than anti-CD20 alone in slowing cell proliferation and in killing Raji or FL-derived DoHH2 cells in vitro (Figure 7C and 7D, Suppl. Figure 7D and 7E). In vivo treatment with either anti-CD20 or the fusion antibody (1µg i.v. for 5days) was well tolerated. However, in Raji xenografts that had reached a size of > 1cm3 at the time of treatment only the fusion antibody was able to produce complete responses (ex vivo tumor weight 0 – 30 mg) in 3 of 7 animals. Residual tumors examined immediately following the last antibody administration confirmed reduced ERK activation, and effects on apoptosis and proliferation by TUNEL and Ki-67 stain as the most immediate effects of therapy (Suppl. Figure 7F). At higher doses (20µg) rituximab is curative against xenografts (nor shown). However, when tested at the same low dose (1µg for 5 days) the anti-CD20 antibody was less effective than the fusion protein and produced only partial responses or slowed progression compared to vehicle (p = 0.039, Fisher’s exact for all three groups) (Figure 7E).
Figure 7. Targeted delivery of EPHA7 to xenografted lymphomas using an anti-CD20-EPHA7 fusion antibody.
A, Schematic of the anti-CD20/EPHA7 fusion antibody; B, Immunoblot on Raji cells that were untreated (Untr.), treated with anti-CD20 (CD20), or anti-CD20-EPHA7TR fusion (CD20/E7); C, Proliferation of Raji cells treated as indicated (*denotes p(CD20 vs. CD20/E7 < 0.05); D, Apoptosis of Raji cells 30 treated as indicated at 24h and 48h; * and ** denote significance (p < 0.05).; E, Mice bearing Raji xenografts (> 1cm3) left untreated (Untr.) or given 1µg of anti-CD20 (CD20) or anti-CD20-EPHA7TR (CD20/E7) for 5 days and collected 2 days after last treatment. Tumors (in matrigel) weighed ex vivo and classified as complete response (CR): 0 – 30 mg, partial responses (PR): 30–100 mg; no change (NC)/progressive disease (PD): > 100 mg. See also Figure S7.
Finally, we examined the potential toxicity of administrating either the purified EphA7FC protein or the fusion antibody. Briefly, animals treated with twice the therapeutic dose revealed no overt toxicity at 24h or 7d after treatment. Necropsy revealed no macro- or microscopic organ damage, and serum chemistry showed only marginally elevated glucose levels but was otherwise unremarkable. Differential blood counts showed a decrease in mean corpuscular hemoglobin in the absence of frank anemia (Suppl. Fig. 7G). Hence, the EPHA7 protein has well-tolerated in vivo tumor suppressive properties that can be directed to xenografted human lymphomas using an anti-CD20 antibody.
Discussion
Our study illustrates the power of combining tumor genomic data with functional genetic screens and exemplifies the translation of genetic insights into therapies. The size and hemizygosity of chromosome 6q deletions in follicular lymphoma would typically preclude the identification of a target gene based on genomic analyses alone. By cross-referencing the genomic data with the results of an unbiased, deletion-targeted loss of function screen, we functionally identify the truncated EPHA7 receptor as a soluble tumor suppressor in FL. We describe converging lines of evidence including in vivo studies in a mouse model, and in human lymphoma cell lines that EPHA7 acts as a tumor suppressor in follicular lymphoma and is a promising candidate for translational development.
We provide insight into the molecular pathogenesis of follicular lymphoma (FL). FLs are characterized by the translocation t(14;18)(q32;q21) that causes increased expression of BCL2, which is considered an initiating event in the malignant transformation of germinal center B-cells (Bende et al., 2007). However, several facts indicate that elevated BCL2 is not sufficient for lymphoma development. For example, the t(14;18)(q32;q21) is often found in healthy individuals, we and others find a multitude of genomic aberration in diagnosis samples of FL, and also the long latency to lymphoma development in Bcl2 transgenic animals indicate that additional events are required for lymphomagenesis (Bende et al., 2007; Egle et al., 2004; Nanjangud et al., 2007). Deletions affecting chromosome 6q11-27 are particularly common in FL and can affect the outcome of treatment in lymphoma patients (Johnson et al., 2009; Nanjangud et al., 2007; Viardot et al., 2002). We find a complex pattern of large and hemizygous 6q deletions in FL and most likely this pattern suggests the presence of multiple and potentially cooperating tumor suppressor genes. However the absence of bi-allelic losses precludes the direct identification of a specific target based on the genomic deletion data and may indicate the presence of haploinsufficient tumor suppressors. We have used a genetic screening approach to functionally identify tumor suppressor genes in the 6q region. The screen uncovered a surprising role for the ephrin receptor A7 (EPHA7) as a candidate tumor suppressor in FL. We confirm its ability to oppose lymphomagenesis using a genetically and pathologically accurate mouse model of the disease. We also demonstrate that EPHA7 has dramatic antitumor effects on human lymphoma cells. Further, an analysis of several hundred primary FL specimens reveals differential silencing in that EPHA7 in tumors, and EPHA7 expression in germinal center B-lymphocytes, which are considered the cell of origin and normal counterpart of these lymphomas (Klein and Dalla-Favera, 2008). Hence, loss of EPHA7 expression does not merely reflect cellular differentiation or transformation, and instead EPHA7 inactivation directly contributes to lymphomagenesis. Consistent with our findings in FL, EPHA7 silencing has been observed in lymphomas arising in Tcl1 transgenic animals, in human marginal zone lymphomas (MZL), and in acute lymphoblastic leukemia (B-ALL) (Dawson et al., 2007; Kuang et al., 2010). These data may suggest a broader role for EPHA7 in lymphoid cancers. Deletions of chromosome 6q have also been described in other types of lymphomas. These lesions have been studied in detail in DLBCL, where tumor suppressive activities have been shown for BLIMP1 and TNFAIP3 (Calado et al., 2010; Compagno et al., 2009; Kato et al., 2009; Mandelbaum et al., 2010; Pasqualucci et al., 2006). Inactivation of these genes has been specifically associated with the activated B-cells (ABC) type of DLBCL, but not the germinal center type of DLBCL. Our screen confirms a cell survival function for TNFAIP3/A20, a negative regulator of NFkB signaling. BLIMP1 fell outside the common region of loss in FL and did not pass our screen, which does not probe effects on lymphocyte differentiation. The pattern of genomic loss suggests the presence of multiple oftentimes haploinsufficient tumor suppressors in this region and that these may cooperate in lymphomagenesis. We did not investigate these interactions, and decided to focus on the soluble tumor suppressor EPHA7 because of its potential for therapeutic application.
Ephrin receptors form a large family of receptor tyrosine kinases with established functions in cell-cell signaling, embryonic development and neuro- and angiogenesis (Pasquale, 2008). In epithelial cancers both oncogenic and tumor suppressive roles have been described for specific ephrin receptors and their ligands (Dawson et al., 2007; Macrae et al., 2005; Noren et al., 2006; Pasquale, 2010). Specifically, EPHA7 is frequently silenced in gastric, colon and prostate cancers and somatic mutations have been found in non-small cell lung cancer (Ding et al., 2008; Pasquale, 2010). We now identify EPHA7 as a target of recurrent deletions and differential epigenetic silencing in lymphoma and we demonstrate its ability to oppose lymphoma development in mice and kill human lymphoma cells. Notably, normal B-lymphocytes express only a short splice variant of EPHA7 (Dawson et al., 2007; Holmberg et al., 2000; Valenzuela et al., 1995). The expression of EPHA7TR is completely lost in over 70% of FLs, and is re-activated in human lymphoma cells following 5’azacytidine treatment. EPHA7TR is shed from normal B-cells and likely acts in an auto- and paracrine manner. Specifically, the soluble EPHA7 protein binds to another Eph-receptor (EPHA2) and blocks its activity and downstream signals including ERK and SRC kinases. Interestingly, this mechanism is reminiscent of EPHA7’s function in central nervous system development (Holmberg et al., 2000). Briefly, during neural tube closure the short splice form of EPHA7 binds to and blocks the signaling activity of the full-length EPHA7 receptor. In the brain, this interaction causes a switch from cellular repulsion to adhesion and permits closure of the neural tube. In lymphocytes we do not detect expression of the full-length EphA7 receptor, instead these cells express the homologous full-length EphA2 receptor and EphA7TR can bind and block the activity of that receptor. Accordingly, knockdown of EPHA2 in lymphoma cells produces the same effect as EPHA2 blockade with EPHA7TR. Disabling the ephrin binding site on EPHA7 does not affect its signaling and anti-tumor properties, however we cannot rule out that EPHA7 may also bind other surface receptors. We propose a mechanism whereby the soluble EPHA7 protein is a decoy receptor that acts, at least, in part by interfering with Eph-receptor dimerization, auto-phosphorylation and signaling activity.
Our findings concerning EPHA7 are directly relevant to lymphoma treatment. Tumor cells are highly sensitive to the restoration of tumor suppressor genes and even brief reactivation can produce powerful therapeutic effects (Feldser et al., 2010; Ventura et al., 2007; Xue et al., 2007). EPHA7 is especially interesting in this regard because it encodes an extrinsic tumor suppressor and can disrupt oncogenic signals emanating from other Eph-A receptors. Accordingly, we find that exogenous administration of EPHA7 produces powerful anti-tumor effects against xenografted human lymphoma cells. To further enhance the potential for clinical application we engineered a fusion antibody based on the anti-CD20 (Rituximab) antibody that is already in clinical use. Specifically, we fused EPHA7 to the anti-CD20 antibody, this allows us to deliver EPHA7’s tumor suppressive activity directly to the CD20 expressing lymphoma cells. The EPHA7-antibody fusion limits the potential for unwanted off-target effects and has single agent activity against lymphoma xenografts at very low concentrations. Potentially, EPHA7TR may be active against other cancers with 6q loss or amplifications of EPHA2. Further studies are needed also to define a minimum tumor suppressive peptide, and to compare its long-term efficacy to current anti-CD20-based therapies. In summary, our study reveals EPHA7 as a tumor suppressor in follicular lymphoma, and indicates a therapeutic strategy that exploits the tumor suppressor hypersensitivity of these tumors.
Experimental procedures
Tumor ascertainment
Sixty-four newly diagnosed FLs collected at MSKCC since 1984 and evaluated by 3 hematopathologists, classified according to WHO criteria, were selected based on diagnosis and presence of an abnormal karyotype. TMAs were constructed from a separate series of 322 follicular lymphomas treated at MSKCC (see Suppl. Methods for details).
Array-CGH
DNA from fresh frozen tissue or OCT-embedded tissue was analyzed on an Agilent 244K oligonucleotide array and compared to human male DNA obtained from Promega (Cat# G147A) as a reference. All genomic positions described refer to UCSC May 2004 human reference sequence (hg17/NCBI build 35). The modified CBS algorithm was used to identify segmental gains and losses along the autosomes (see Suppl. Methods for details).
Cell culture and pooled shRNA library screen
FL5-12 murine lymphocytes were stably transduced with Bcl2 (FL5-12/Bcl2). Briefly, the shRNA library was constructed by pooling 262 individually cloned shRNAs and shRNAs were identified by sequencing upon enrichment of GFP expressing cells (see Suppl. Methods for details).
Generation of mice
The vavPBcl2 mouse model of FL(Egle et al., 2004) and EµMyc lymphoma model were adapted to the transplantation approach using retrovirally transduced HSCs (Wendel et al., 2004). Data were analyzed in Kaplan-Meier format using the log-rank (Mantel-Cox) test for statistical significance (see Suppl. Methods for details).
Supplementary Material
Acknowledgments
We thank L. Pasqualucci, E. Papapetrou, S.L. Morrison, and M. Teitell for reagents; Saara Korpela for help in modeling the EphA2/EphA7 interaction; several MSK core facilities, including the Research Animal Facility, A. Viale of the Genomics Core, Elisa de Stanchina of the Anti-tumor Assessment Core, A. Lash of the Computational Biology Core, Katy Huberman of the Geoffrey Beene Translational Oncology Core Facility, Aleksey V. Kamenshchikov of the Antibody Facility, Irina Linkov of the Pathology Core. This work is supported by grants from the NCI (R01-CA142798-01), a P30 supplemental award from the NCI and the AIDS Malignancy Consortium (AMC) (HGW), the Leukemia Research Foundation (HGW), the Louis V. Gerstner Foundation (HGW), the WLBH Foundation (HGW), the Society of MSKCC (HGW), the Starr Cancer Consortium grant I4-A410 (HGW and JH), NIH MSTP grant GM07739 (J.B.), NIH-K08 CA127353 (R.S), the ASCO Cancer Foundation (JHS), LLS TRP (R.S.), LLS SCOR S 7032-04 (A.M.), NCI R01 CA104348 (A.M.) LLS scholar (A.M.), Burroughs Wellcome Clinical Translational Scientist (A.M.), Cancer Center Support Grant P30- CA008748 (FWG) and a SCOR grant from The Leukemia and Lymphoma Society (RSKC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barr PM, Lazarus HM. Follicular non-Hodgkin lymphoma: long-term results of stem-cell transplantation. Curr Opin Oncol. 2008;20:502–508. doi: 10.1097/CCO.0b013e32830b61ac. [DOI] [PubMed] [Google Scholar]
- Bende RJ, Smit LA, van Noesel CJ. Molecular pathways in follicular lymphoma. Leukemia. 2007;21:18–29. doi: 10.1038/sj.leu.2404426. [DOI] [PubMed] [Google Scholar]
- Bidere N, Ngo VN, Lee J, Collins C, Zheng L, Wan F, Davis RE, Lenz G, Anderson DE, Arnoult D, et al. Casein kinase 1alpha governs antigen-receptor-induced NF-kappaB activation and human lymphoma cell survival. Nature. 2009;458:92–96. doi: 10.1038/nature07613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado DP, Zhang B, Srinivasan L, Sasaki Y, Seagal J, Unitt C, Rodig S, Kutok J, Tarakhovsky A, Schmidt-Supprian M, et al. Constitutive canonical NF-kappaB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell. 2010;18:580–589. doi: 10.1016/j.ccr.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson DW, Hong JS, Shen RR, French SW, Troke JJ, Wu YZ, Chen SS, Gui D, Regelson M, Marahrens Y, et al. Global DNA methylation profiling reveals silencing of a secreted form of Epha7 in mouse and human germinal center B-cell lymphomas. Oncogene. 2007;26:4243–4252. doi: 10.1038/sj.onc.1210211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bath ML, O'Reilly L, Cory S. VavP-Bcl2 transgenic mice develop follicular lymphoma preceded by germinal center hyperplasia. Blood. 2004;103:2276–2283. doi: 10.1182/blood-2003-07-2469. [DOI] [PubMed] [Google Scholar]
- Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, Sanchez-Rivera FJ, Resnick R, Bronson R, Hemann MT, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468:572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidano G, Hauptschein RS, Parsa NZ, Offit K, Rao PH, Lenoir G, Knowles DM, Chaganti RS, Dalla-Favera R. Deletions involving two distinct regions of 6q in B-cell non-Hodgkin lymphoma. Blood. 1992;80:1781–1787. [PubMed] [Google Scholar]
- Hauptschein RS, Gamberi B, Rao PH, Frigeri F, Scotto L, Venkatraj VS, Gaidano G, Rutner T, Edwards YH, Chaganti RS, et al. Cloning and mapping of human chromosome 6q26–q27 deleted in B-cell non-Hodgkin lymphoma and multiple tumor types. Genomics. 1998;50:170–186. doi: 10.1006/geno.1998.5321. [DOI] [PubMed] [Google Scholar]
- Heyer J, Kwong LN, Lowe SW, Chin L. Non-germline genetically engineered mouse models for translational cancer research. Nat Rev Cancer. 2010;10:470–480. doi: 10.1038/nrc2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen JP, Yermekbayeva L, Janes PW, Walker JR, Xu K, Atapattu L, Rajashankar KR, Mensinga A, Lackmann M, Nikolov DB, et al. Architecture of Eph receptor clusters. Proc Natl Acad Sci U S A. 2010;107:10860–10865. doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, Clarke DL, Frisen J. Regulation of repulsion versus adhesion by different splice forms of an Eph receptor. Nature. 2000;408:203–206. doi: 10.1038/35041577. [DOI] [PubMed] [Google Scholar]
- Johnson NA, Savage KJ, Ludkovski O, Ben-Neriah S, Woods R, Steidl C, Dyer MJ, Siebert R, Kuruvilla J, Klasa R, et al. Lymphomas with concurrent BCL2 and MYC translocations: the critical factors associated with survival. Blood. 2009;114:2273–2279. doi: 10.1182/blood-2009-03-212191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Kuang SQ, Bai H, Fang ZH, Lopez G, Yang H, Tong W, Wang ZZ, Garcia-Manero G. Aberrant DNA methylation and epigenetic inactivation of Eph receptor tyrosine kinases and ephrin ligands in acute lymphoblastic leukemia. Blood. 2010;115:2412–2419. doi: 10.1182/blood-2009-05-222208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- Macrae M, Neve RM, Rodriguez-Viciana P, Haqq C, Yeh J, Chen C, Gray JW, McCormick F. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 2005;8:111–118. doi: 10.1016/j.ccr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Maloney DG. Rituximab for follicular lymphoma. Curr Hematol Rep. 2003;2:13–22. [PubMed] [Google Scholar]
- Mandelbaum J, Bhagat G, Tang H, Mo T, Brahmachary M, Shen Q, Chadburn A, Rajewsky K, Tarakhovsky A, Pasqualucci L, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. 2010;12:372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Schwickert TA, Chait BT, Nussenzweig MC. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008;205:2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- Nanjangud G, Rao PH, Teruya-Feldstein J, Donnelly G, Qin J, Mehra S, Jhanwar SC, Zelenetz AD, Chaganti RS. Molecular cytogenetic analysis of follicular lymphoma (FL) provides detailed characterization of chromosomal instability associated with the t(14;18)(q32;q21) positive and negative subsets and histologic progression. Cytogenet Genome Res. 2007;118:337–344. doi: 10.1159/000108318. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Foos G, Hauser CA, Pasquale EB. The EphB4 receptor suppresses breast cancer cell tumorigenicity through an Abl-Crk pathway. Nat Cell Biol. 2006;8:815–825. doi: 10.1038/ncb1438. [DOI] [PubMed] [Google Scholar]
- Novak U, Rinaldi A, Kwee I, Nandula SV, Rancoita PM, Compagno M, Cerri M, Rossi D, Murty VV, Zucca E, et al. The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood. 2009;113:4918–4921. doi: 10.1182/blood-2008-08-174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offit K, Parsa NZ, Gaidano G, Filippa DA, Louie D, Pan D, Jhanwar SC, Dalla-Favera R, Chaganti RS. 6q deletions define distinct clinicopathologic subsets of non-Hodgkin's lymphoma. Blood. 1993;82:2157–2162. [PubMed] [Google Scholar]
- Oricchio E, Wolfe AL, Schatz JH, Mavrakis KJ, Wendel HG. Mouse models of cancer as biological filters for complex genomic data. Dis Model Mech. 2010 doi: 10.1242/dmm.006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, Aster JC, Murty VV, Shipp MA, Dalla-Favera R. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relander T, Johnson NA, Farinha P, Connors JM, Sehn LH, Gascoyne RD. Prognostic factors in follicular lymphoma. J Clin Oncol. 2010;28:2902–2913. doi: 10.1200/JCO.2009.26.1693. [DOI] [PubMed] [Google Scholar]
- Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, Klapper W, Vater I, Giefing M, Gesk S, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiradake E, Harlos K, Sutton G, Aricescu AR, Jones EY. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat Struct Mol Biol. 2010;17:398–402. doi: 10.1038/nsmb.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FM, Vearing C, Lackmann M, Treutlein H, Himanen J, Chen K, Saul A, Nikolov D, Boyd AW. Dissecting the EphA3/Ephrin-A5 interactions using a novel functional mutagenesis screen. J Biol Chem. 2004;279:9522–9531. doi: 10.1074/jbc.M309326200. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Rojas E, Griffiths JA, Compton DL, Gisser M, Ip NY, Goldfarb M, Yancopoulos GD. Identification of full-length and truncated forms of Ehk-3, a novel member of the Eph receptor tyrosine kinase family. Oncogene. 1995;10:1573–1580. [PubMed] [Google Scholar]
- Velculescu VE. Defining the blueprint of the cancer genome. Carcinogenesis. 2008;29:1087–1091. doi: 10.1093/carcin/bgn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Viardot A, Moller P, Hogel J, Werner K, Mechtersheimer G, Ho AD, Ott G, Barth TF, Siebert R, Gesk S, et al. Clinicopathologic correlations of genomic gains and losses in follicular lymphoma. J Clin Oncol. 2002;20:4523–4530. doi: 10.1200/JCO.2002.12.006. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.