Abstract
Summary: The survcomp package provides functions to assess and statistically compare the performance of survival/risk prediction models. It implements state-of-the-art statistics to (i) measure the performance of risk prediction models; (ii) combine these statistical estimates from multiple datasets using a meta-analytical framework; and (iii) statistically compare the performance of competitive models.
Availability: The R/Bioconductor package survcomp is provided open source under the Artistic-2.0 License with a user manual containing installation, operating instructions and use case scenarios on real datasets. survcomp requires R version 2.13.0 or higher. http://bioconductor.org/packages/release/bioc/html/survcomp.html
Contact: bhaibeka@jimmy.harvard.edu; mschroed@jimmy.harvard.edu
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Building risk prediction or survival models is an important area of research, especially in cancer where gene expression signatures are used to predict risk of metastasis, response to therapy and overall survival. However, assessing the relative performance of such models is complex due to the lack of standards regarding the best criterion to use in survival analysis (Table 1; Haibe-Kains et al., 2008). Although a number of model performance estimators have been described (Cox, 1972; Graf et al., 1999; Harrel et al., 1996; Heagerty et al., 2000; Royston and Sauerbrei, 2004; Verweij and Houwelingen, 1993), they are not widely used. This is partly because these are implemented in many different R packages that use heterogeneous interfaces, which makes it difficult for the non-specialist to easily use or compare the performance of these models. Another challenge in assessing performance of expression-based prediction models is a lack of power due to small sample size. Meta-analytical methods could leverage power from the numerous microarray datasets that are now publicly available by summarizing model performance estimated in multiple-independent studies. Moreover, because they enable efficient joint analysis of multiple datasets, such an analytical framework reduces the risk of artifactual discoveries that are due to bias or confounding factors that may be present in one dataset. This is particularly important when comparing competitive risk prediction models; often authors claim better performance of a new model without properly assessing whether a model significantly outperforms its competitors. To the best of our knowledge, there is no commercial or open-source tool to enable statistical comparison of risk prediction models in a meta-analytical framework.
Table 1.
Functions in survcomp to measure the performance of risk prediction models
| Name | Function in survcomp | References |
|---|---|---|
| Concordance index | concordance.index | Harrel et al. (1996) |
| D index | D.index | Royston and Sauerbrei (2004) |
| Hazard ratio | hazard.ratio | Cox (1972) |
| Brier score | sbrier.score2proba | Graf et al. (1999) |
| Cross-validated partial likelihood | cvpl | Verweij and Houwelingen (1993) |
| Time-dependent ROC curve | tdrocc | Heagerty et al. (2000) |
| Kaplan–Meier curve | km.cox.plot | Kaplan and Meier (1958) |
| Forestplot | forestplot.surv | Lewis and Clarke (2001) |
To address these issues, we developed a new Bioconductor package, survcomp, which implements several performance criteria for risk prediction models (Table 1), together with meta-analytical methods that enable combination of performance estimations from multiple-independent datasets [fixed- and random-effects models (Cochrane, 1954); ?combine.est], and statistical comparison of predictions between competitive models (?cindex.comp for the concordance index). The concordance index as described by Harrel et al. (1996) and implemented in survcomp may be sensitive to the study-specific censoring distribution, therefore we are working to implement a modified concordance index by Uno et al. (2011) that avoids this problem and which should be available in the next release of survcomp.
Although the performance criteria that are implemented in survcomp are mostly available in other R packages (except the D index which, to the best of our knowledge, is only in survcomp), our package provides a common interface to facilitate easy use of all these estimators. Moreover, with the exception of ipdmeta by Broeze et al. (2009) and survJamda by Yasrebi (2011), few R packages perform meta-analysis of survival data. survcomp provides a uniform interface to simplify the use of performance assessment and statistical comparison of risk prediction models, and provides new R functions to statistically compare these in a meta-analytical framework. It is worth noting that the aim of survcomp is to provide efficient computation of several performance estimates (Table 1) and not to implement a full validation framework. Proper validation using a fully independent test dataset is of utmost importance to avoid overoptimistic results (Jelizarow et al., 2010) and frameworks for cross-validation, multiple random splits or a single split into training/validation datasets are implemented in several existing R packages including peperr (Porzelius et al., 2009).
We illustrate the functionality of survcomp in a case study that investigates prognosis in breast cancer. In most studies, this is a difficult task due to the (generally) small sample size. Yet despite a large number of published studies, data heterogeneity, both in terms of data sources and microarray technologies, have limited the effectiveness of their joint analysis. Here we apply survcomp functions to statistically compare the prognostic value of a widely used clinical model, a single proliferation gene and two published gene signatures. The results suggest that the multi-gene signatures are not always superior to standard clinical models or to a simple single gene model.
2 CASE STUDY
The prognostic ability of gene expression of AURKA, a single proliferation-related gene, was compared with the Nottingham Prognostic Index (NPI, Galea et al., 1992) clinical model for prognosis, and to risk prediction scores from two published multigene prognostic signatures; GGI (Sotiriou et al., 2006) and GENIUS (Haibe-Kains et al., 2010). The NPI, GGI and GENIUS risk scores were calculated using the Bioconductor package genefu. Each score was computed in five publicly available datasets described in the Supplementary Material.
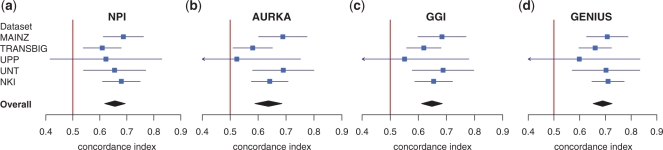
To compare the prognostic ability of these four different risk prediction models, we estimated the concordance index for each model in each dataset separately and used the function combine.est to compute the corresponding overall meta-estimate using the well-established random-effects model approach (Cochrane, 1954). As can be seen in Figure 1, although the performance varies between datasets, all models yielded highly significant overall prognostic value (high-risk predictions represent patient with poor survival, concordance index >0.5, one-sided P<0.001, see Supplementary Material for R code). AURKA, the single proliferation gene, was the worst predictor of survival (concordance index of 0.64), whereas GENIUS, the risk prediction model taking into account the breast cancer molecular subtypes, was the best (concordance index of 0.69). The continuous risk prediction of NPI, the traditional clinical model which combines nodal status, tumor size and histological grade, yielded a relatively high concordance index (concordance index of 0.66).
Fig. 1.
Forestplots representing the prognostic value of (a) NPI, (b) AURKA, (c) GGI and (d) GENIUS estimated by the concordance index in five independent breast cancer datasets [none of these datasets were used to train the classifiers (a)–(c), duplicated patients were removed what results in a combined dataset of 722 patients]. The blue square and horizontal line represent the concordance index and its 95% confidence interval which is clipped at 0.4 and 0.9 (represented by an arrow). The black rhombus is the overall meta-estimate from the combined five datasets. The greater the concordance index, the more prognostic the risk prediction model. The vertical red bar represents the concordance index of random risk predictions.
To identify the best risk prediction model(s), we statistically compared their concordance indices using the function cindex.comp.meta (Table 2). Concurring with Haibe-Kains et al. (2010) we observed that GENIUS outperforms AURKA, GGI and NPI (uncorrected P<0.10; Table 2). However, when P-values are corrected for multiple testing (Holm's method), no P-value remains significant suggesting that a larger meta-analysis is required to definitively claim the superiority of one classifier over another. This also suggests that prognostic clinical models such as NPI are still extremely competitive compared with more complex gene signatures. Repeating this analysis using performance criteria in survcomp other than the concordance index, including the D index or hazard ratio (Table 1), leads to similar conclusions (examples are provided in the package user manual and documentation).
Table 2.
Statistical performance comparison for the risk prediction models used in our case study
| Models | NPI | AURKA | GGI | GENIUS |
|---|---|---|---|---|
| NPI | 0.25 | 0.37 | 0.91 | |
| AURKA | 0.75 | 0.70 | 0.98 | |
| GGI | 0.63 | 0.30 | 0.97 | |
| GENIUS | 0.09 | 0.02 | 0.033 |
P-values are computed using a one-sided paired Student's t-test to test whether risk prediction models in rows are better than the ones in columns.
3 CONCLUSION
The R/Bioconductor package survcomp provides a uniform interface to an extensive set of performance assessment and statistical comparison methods for survival/risk prediction. It allows scientists to easily implement large comparative studies integrating multiple independent datasets while providing statistical tools to identify the best model(s) as supported by the data under study.
Funding: National Human Genome Research Institute (1P50 HG004233, to M.S.S.); Fulbright Commission for Educational Exchange to (B.H.-K.); US National Institutes of Health (R01 LM010129-01, to B.H.-K. and J.Q.); Claudia Adams Barr Program in Innovative Basic Cancer Research (A.C.C. and J.Q.); Career Development grant (to A.C.C.) from DFCI Breast Cancer SPORE: CA 08939.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Broeze K.A., et al. Individual patient data meta-analysis of diagnostic and prognostic studies in obstetrics, gynaecology and reproductive medicine. BMC Med. Res. Methodol. 2009;9:22. doi: 10.1186/1471-2288-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane W.G. The combination of estimates from different experiments. Biometrixs. 1954;10:101–129. [Google Scholar]
- Cox D.R. Regression models and life tables. J. R Stat. Soc. Ser. B. 1972;34:187–220. [Google Scholar]
- Galea M.H., et al. The Nottingham prognostic index in primary breast cancer. Breast Cancer Res. Treat. 1992;22:207–219. doi: 10.1007/BF01840834. [DOI] [PubMed] [Google Scholar]
- Graf E., et al. Assessment and comparison of prognostic classification schemes for survival data. Stat. Med. 1999;18:2529–2545. doi: 10.1002/(sici)1097-0258(19990915/30)18:17/18<2529::aid-sim274>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Haibe-Kains B., et al. A comparative study of survival models for breast cancer prognostication based on microarray data: does a single gene beat them all? Bioinformatics. 2008;24:2200–2208. doi: 10.1093/bioinformatics/btn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B., et al. A fuzzy gene expression-based computational approach improves breast cancer prognostication. Genome Biol. 2010;11:R18. doi: 10.1186/gb-2010-11-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrel F.E., Jr., et al. Tutorial in biostatistics: multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing error. Stat. Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Heagerty P.J., et al. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- Jelizarow M., et al. Over-optimism in bioinformatics: an illustration. Bioinformatics. 2010;26:1990–1998. doi: 10.1093/bioinformatics/btq323. [DOI] [PubMed] [Google Scholar]
- Kaplan E.L., Meier P. Nonparametric estimation from incomplete observations. J. Amer. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- Lewis S., Clarke M. Forest plots: trying to see the wood and the trees. Br. Med. J. 2001;322:1479–1480. doi: 10.1136/bmj.322.7300.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porzelius C., et al. Parallelized prediction error estimation for evaluation of high-dimensional models. Bioinformatics. 2009;25:827–829. doi: 10.1093/bioinformatics/btp062. [DOI] [PubMed] [Google Scholar]
- Royston P., Sauerbrei W. A new measure of prognostic separation in survival data. Stat. Med. 2004;23:723–748. doi: 10.1002/sim.1621. [DOI] [PubMed] [Google Scholar]
- Sotiriou C., et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Uno H., et al. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med. 2011;30:1105–1117. doi: 10.1002/sim.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij P.J.M., van Houwelingen H.C. Cross-validation in survival analysis. Stat. Med. 1993;12:2305–2314. doi: 10.1002/sim.4780122407. [DOI] [PubMed] [Google Scholar]
- Yasrebi H. SurvJamda: an R package to predict patients' survival and risk assessment using joint analysis of microarray gene expression data. Bioinformatics. 2011;27:1168–1169. doi: 10.1093/bioinformatics/btr103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.