Abstract
Summary: The Visual DSD (DNA Strand Displacement) tool allows rapid prototyping and analysis of computational devices implemented using DNA strand displacement, in a convenient web-based graphical interface. It is an implementation of the DSD programming language and compiler described by Lakin et al. (2011) with additional features such as support for polymers of unbounded length. It also supports stochastic and deterministic simulation, construction of continuous-time Markov chains and various export formats which allow models to be analysed using third-party tools.
Availability: Visual DSD is available as a web-based Silverlight application for most major browsers on Windows and Mac OS X at http://research.microsoft.com/dna. It can be installed locally for offline use. Command-line versions for Windows, Mac OS X and Linux are also available from the web page.
Contact: aphillip@microsoft.com
Supplementary Information:Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Novel techniques for designing and manipulating synthetic DNA are making it possible to construct increasingly sophisticated biological computation devices. One such technique is DNA strand displacement (Seelig et al., 2006; Zhang and Seelig, 2011) in which hybridization between complementary DNA strands is the sole computational mechanism. Designing novel biomolecular computing devices has practical benefits as well as offering fundamental insights into understanding life as biomolecular information processing. As these devices increase in complexity, it becomes intractable to calculate the low-level interactions between the DNA species by hand. Thus, one of the main barriers to continued progress in this nascent field is the lack of automation and abstraction in the design process. Visual DSD implements a domain-specific language whose syntax is tailored to allow concise encodings of the species in a DSD system. The DSD programming language is described in (Lakin et al., 2011; Phillips and Cardelli, 2009). The system automatically computes all possible reactions between the species, providing the benefits of a reaction-based approach without the difficulty of manually constructing the reaction network. The user can choose an appropriate level of abstraction in the compilation process, from low-level detailed views to higher level simplified views. The design can then be visualized and analyzed to eliminate any unwanted behaviour. Visual DSD provides suitable analysis tools both for large-scale designs involving many thousands of formal and intermediate species, such as DNA logic gates (Qian and Winfree, 2011), and for systems involving low species populations, such as DNA implementations of stack machines (Qian et al., 2010).
2 METHODS AND IMPLEMENTATION
2.1 Encoding systems
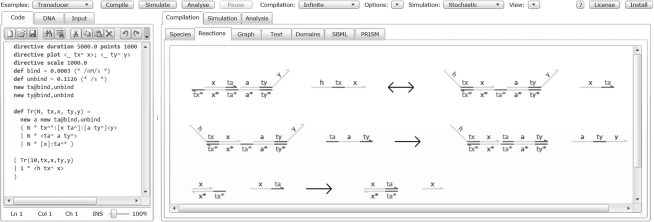
Program code is entered using the syntax from Lakin et al. (2011), as shown on the left-hand side of Figure 1. Our domain-specific language for DNA strand displacement includes support for parameterized modules which can be instantiated multiple times in a program. This encourages good software engineering practices such as code reuse and modular design. Settings which are specific to a particular system, such as the default reaction rates and the populations to plot during a simulation, are specified within the program code. More general settings, such as whether to use a stochastic or deterministic simulation, are selected through the graphical user interface. Input programs are checked for type safety and well-formedness of the DNA species before being passed to the compiler, though some of these checks can be disabled to allow rapid development of programs. The tool includes a variety of example programs and an integrated tutorial. A manual is available on the web page that contains a formal definition of the syntax.
Fig. 1.
Screenshot of the Visual DSD tool, with the code entry box on the left and the output tabs on the right. Along the top of the screen there are options to select example programs, adjust the semantics and control the simulator. The example shown implements a simple transducer gate. The Compilation tab on the right-hand side displays output from the compiler, in this case a visualization of all the individual reactions. The Simulation tab shows time-course plots and data tables from stochastic and deterministic simulations, and the Analysis tab shows various representations of the continuous-time Markov chain.
2.2 Compilation, simulation and analysis
Visual DSD implements the compilation scheme outlined in Lakin et al. (2011), with an extension to support reactions which form DNA polymers. The user interface offers a selection of semantic rules, which formalize the possible interactions between the DNA species with varying levels of detail. This provides a great deal of flexibility. Visual DSD includes a stochastic simulator that uses the Gillespie algorithm (Gillespie, 1977) to generate trajectories when the entire chemical reaction network has been pre-computed. There is also a deterministic simulator that uses the Runge-Kutta-Fehlberg method (Fehlberg, 1969) to solve an ordinary differential equation model. For systems where the chemical reaction network is very large (or infinite, in the case of polymer systems), there is a just-in-time compiler which allows the reaction network to be computed incrementally as needed during a stochastic simulation (Paulevé et al., 2010). This can produce significant performance gains in certain situations. The tool can also construct and visualize the continuous-time Markov chain of a (sufficiently small) system, which is particularly useful for low-level debugging of individual circuit components.
2.3 Visualization and export
Visual DSD offers numerous visualizations of the DNA species in the system, using the graphical notation introduced in Lakin et al. (2011). These range from the structure of the individual DNA species through pictorial representations of individual reactions (as shown on the right-hand side of fig. 1) to a graph-based visualization of the entire chemical reaction network or continuous-time Markov chain. Simulation results are plotted as line graphs of species populations over time. Simple arithmetic functions on populations can also be plotted. The tool can also output nucleotide sequences as a starting point for construction of the systems.
Data and models can be exported from Visual DSD in a variety of formats. Chemical reaction networks can be exported to SBML (Hucka et al., 2003) and the time series data from simulations can be saved in CSV format for loading into a spreadsheet. Continuous-time Markov chains can be exported as a model for the PRISM probabilistic model checker (Hinton et al., 2006). The user can then express system properties as temporal logic formulae and verify them using stochastic model checking (Kwiatkowska et al., 2007). PRISM can also perform detailed quantitative analysis of reaction kinetics. Examples queries include the probability of reaching a given state within a particular time, or the probability of reaching an undesirable state due to interference between DNA strands.
3 DISCUSSION
Visual DSD has been used to model and analyse a wide range of DNA strand displacement devices, including logic gates (Qian and Winfree, 2011), neural network computation (Qian et al., 2011), fork and join gates (Cardelli, 2010), oscillators (Lakin et al., 2011) and a range of other devices including catalytic gates and schemes for simulating arbitrary chemical systems (Phillips and Cardelli, 2009). Note that secondary structures such as hairpin, pseudoknot and multiloop junctions are not currently supported; however, some of these structures, including hairpins, are currently under development for a future release. We have found that the tool is particularly beneficial when designing new systems from scratch, as the visualizations make it easy to debug programs. Support for reusable modules in program code makes it quick and easy to construct new, larger systems using pre-designed components. The tool embodies the scientific workflow and can increase efficiency and productivity of DNA device design and analysis by allowing users to simulate systems before attempting to build them in the lab, thereby saving time and lab resources. It also enables in silico investigation of the behaviour of systems that are beyond the current state of the art in fabrication, such as particularly large and/or complex systems. Furthermore, once a design has been formalized in the DSD language it becomes possible to perform formal verification of certain aspects of its behaviour, for example using a stochastic model checker or a theorem prover. We are continually developing the software and adding new features.
ACKNOWLEDGEMENTS
We thank Luca Cardelli for valuable feedback on the design of the Visual DSD language and tool.
Conflict of Interest: none declared.
REFERENCES
- Cardelli L. Two-domain DNA strand displacement. In: Cooper S.B., et al., editors. Proceedings of DCM 2010. Vol. 26. Open Publishing Association; 2010. pp. 47–61. EPTCS. [Google Scholar]
- Fehlberg E. Low-order classical Runge-Kutta formulas with stepsize control and their application to some heat transfer problems. NASA, USA: Technical Report NASA-TR-R-315; 1969. [Google Scholar]
- Gillespie D. Exact stochastic simulation of coupled chemical reactions. J. Phy. Chem. 1977;81:2340–2361. [Google Scholar]
- Hinton A., et al. PRISM: a tool for automatic verification of probabilistic systems. In: Hermanns H., Palsberg J., editors. Proceedings of TACAS 2006. Vol. 3920. Berlin/Heidelberg, Germany: Springer; 2006. pp. 441–444. LNCS. [Google Scholar]
- Hucka M., et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;9:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Kwiatkowska M., et al. Stochastic model checking. In: Bernardo M., Hillston J., editors. Proceedings of SFM 2007. Vol. 4486. Berlin/Heidelberg, Germany: Springer; 2007. pp. 220–270. LNCS. [Google Scholar]
- Lakin M.R., et al. Abstractions for DNA circuit design. J. R. Soc. Interface. 2011 doi: 10.1098/rsif.2011.0343. [Epub ahead of print, doi:10.1098/rsif.2011.0343, July 20, 2011] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulevé L., et al. A generic abstract machine for stochastic process calculi. In: Quaglia P., editor. Proceedings of CMSB 2010. New York, NY, USA: ACM; 2010. pp. 43–54. [Google Scholar]
- Phillips A., Cardelli L. A programming language for composable DNA circuits. J. R. Soc. Interface. 2009;6(4):S419–S436. doi: 10.1098/rsif.2009.0072.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- Qian L., et al. Efficient Turing-universal computation with DNA polymers. In: Sakakibara Y., Mi Y., editors. Proceedings of DNA 16. Vol. 6518. Berlin/Heidelberg, Germany: Springer; 2010. pp. 123–140. Lecture Notes in Computer Science. [Google Scholar]
- Qian L., et al. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- Seelig G., et al. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- Zhang D.Y., Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]