Abstract
Summary: The large amount of data produced by proteomics experiments requires effective bioinformatics tools for the integration of data management and data analysis. Here we introduce a suite of tools developed at Vanderbilt University to support production proteomics. We present the Backup Utility Service tool for automated instrument file backup and the ScanSifter tool for data conversion. We also describe a queuing system to coordinate identification pipelines and the File Collector tool for batch copying analytical results. These tools are individually useful but collectively reinforce each other. They are particularly valuable for proteomics core facilities or research institutions that need to manage multiple mass spectrometers. With minor changes, they could support other types of biomolecular resource facilities.
Availability and Implementation: Source code and executable versions are available under Apache 2.0 License at http://www.vicc.org/jimayersinstitute/data/
Contact: daniel.liebler@vanderbilt.edu
1 INTRODUCTION
Mass spectrometry (MS)-based proteomics offers a remarkably powerful technology for identification of proteins in complex biological samples. Bioinformatics tools are essential to this process (Nesvizhskii, 2010). Proteomics services are often provided through shared MS instrumentation with the support of external computing resources. Core facilities often struggle to manage the volume of data that these instruments can generate, resulting in irregular or non-existent backup plans and long delays separating sample receipt and information release for end-users. Existing freely available tools such as TPP (Keller et al., 2005), CPAS (Nelson et al., 2011) and CPFP (Trudgian et al., 2010) focus only on data analysis by providing a set of tools for peptide identification and validation. A Laboratory Information Management System may offer some capabilities for integration of instrument data backup and data analysis, but these services usually catalog samples and bench procedure more effectively than they manage bioinformatics workflows, with little support for proteomics studies. We addressed these challenges by developing a suite of tools to support production proteomics. These tools are in daily use at the Vanderbilt Mass Spectrometry Research Center and the Jim Ayers Institute for Precancer Detection and Diagnosis. They are now freely available with source code to commercial, government and academic users.
2 SOFTWARE OVERVIEW
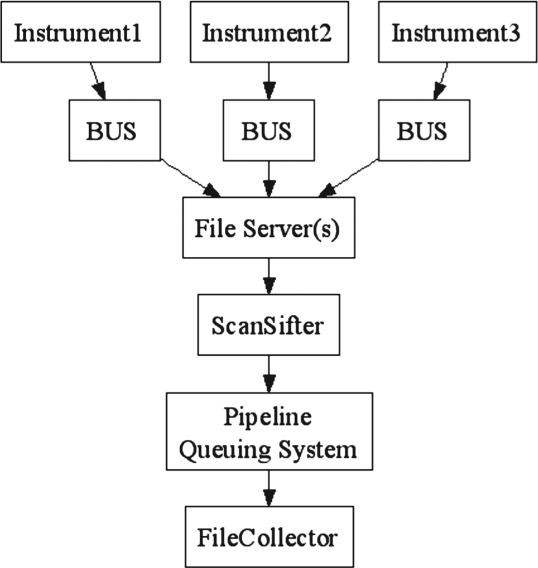
The workflow of these supporting tools is illustrated in Figure 1. The Backup Utility Service (BUS) tool offers automated backup of raw data to file servers. The ScanSifter tool reads these proprietary format files and converts them to open format files, which are submitted to a queuing system and identified by database search engines such as SEQUEST (Eng et al., 1994) and MyriMatch (Tabb et al., 2007). The identification results can be retrieved using the File Collector tool, enabling batch copying search results to local computer or network drives. We also provide an instrument file naming utility to quickly generate filenames in a standard consistent fashion as opposed to manually entering each filename into the instrument control software. Each tool can be used separately to fulfill its function. Combining these tools offers an integrated solution for production proteomics.
Fig. 1.
The workflow of supporting tools for production proteomics.
2.1 Backup utility service (BUS)
The BUS tool automates file backup from instruments to sets of file servers. It operates as a configurable, scheduled Windows service, seeking file system changes and copying updated files on a regular basis. A graphical user interface facilitates the configuration process. BUS is a Windows application written in C# and should be installed in each computer that controls a mass spectrometer. It can be used in either a standalone desktop or web service-coordinated capacity. In the latter case, the metadata of archived files are stored in an Oracle database or a free Oracle database Express Edition; a script to initiate the Oracle database is provided in the distribution package. This database can then assist the ScanSifter for data conversion to forestall additional file copy steps.
2.2 ScanSifter
ScanSifter is a data conversion tool to transcode and filter mass spectra. It reads and writes a variety of formats via use of the ProteoWizard (Kessner et al., 2008) library. Currently, it supports spectral data in proprietary formats from Bruker and Thermo; adding other formats supported by ProteoWizard requires few changes. It exports data in mzML (Deutsch, 2008), mzXML (Pedrioli et al., 2004), mzData, MGF and DTA formats. It can also translate files from MGF or XML-based formats to the others.
ScanSifter implements several algorithms for recognizing mass spectra that can be filtered prior to export. By default, the software will output all mass spectra, but users can, for example, export only MS/MS/MS scans. Noise spectra can be filtered by imposing a minimum required peak count or total ion current (TIC). In special cases, a researcher may be interested in scans that correspond to a narrow range of precursor m/z values, and this filter is also implemented.
Two versions of ScanSifter are included in the distribution package: ScanSifter Web and ScanSifter Desktop. The former is installed in a Microsoft IIS server to provide web services that read input files from designated network directories. Alternatively, it can be configured to read a BUS database and find archived raw files automatically. The converted files are stored in file servers and subsequently processed by identification tools. ScanSifter Web can coordinate the use of shared file servers among multiple labs. If multiple ScanSifter jobs are submitted at the same time, they will be queued for processing.
ScanSifter Desktop is a standalone Windows application written in C#/.NET. Besides the filtering features described above, it provides three algorithms to enable the removal of duplicate spectra during data conversion. The first algorithm adapts the scoring approach for MyriMatch (Tabb et al., 2007) to recognize similar spectra. The second algorithm computes a normalized dot product for spectral comparison, as is typical in spectral library search. The third algorithm treats m/z values of two spectra as two groups, and calculates the distance between the distributions of these two groups through the Kolmogorov–Smirnov test. If duplicate spectra are observed during data conversion, only the spectrum with the highest TIC is retained and exported to the output file.
2.3 Identification pipeline and queuing system
A queuing system and web interfaces to run peptide identification pipelines are included in the distribution package and should be installed in a Linux server. The queuing system is written in Perl and coordinates with the TORQUE resource manager or Moab Cluster Manager. Web interfaces to run database search by SEQUEST (Eng et al., 1994) and MyriMatch (Tabb et al., 2007) are provided. Sequence tagging-based modification search by TagRecon (Dasari et al., 2010) is also enabled. A FASTA database maintenance tool is provided to upload new sequence databases and view the status of current databases. The queuing system can separate LC-MS/MS experiments to many tasks, maintaining a target number of running jobs for a cluster. Status queries and error logs are fielded by provided support tools.
2.4 File Collector
The File Collector tool enables batch copying search results and spectral files from file servers to local computer or network drives. It is a Windows application written in C#/.NET. Filters such as file type or file name are enabled. A configuration file is used to specify the source file locations. Network resources are not required to be mounted as drive letters; the system supports UNC path description.
3 SUMMARY
Handling data for multiple mass spectrometers requires support tools. We developed these tools to enable analysis of data from several instruments operating continuously in our laboratories. We hope that these systems for data transport and computing coordination will be broadly useful to others in their present form or with slight modifications. All applications are released under an open source license to accommodate both academic and commercial users. The queue management and data archival tools could easily be applied in other biotechnologies with minimal modification. As high-throughput instrumentation becomes ubiquitous, the challenges of managing these data flows will only increase. Software systems such as these will only grow in importance to shared instrument facilities.
Funding: National Cancer Institute through the Clinical Proteomic Technology Assessment for Cancer (CPTAC) program (U24CA126470 to D.C.L. and R01CA126218 to D.L.T.).
Conflict of Interest: none declared.
REFERENCES
- Dasari S., et al. TagRecon: high-throughput mutation identification through sequence tagging. J. Proteome Res. 2010;9:1716–1726. doi: 10.1021/pr900850m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch E. mzML: a single, unifying data format for mass spectrometer output. Proteomics. 2008;8:2776–2777. doi: 10.1002/pmic.200890049. [DOI] [PubMed] [Google Scholar]
- Eng J.K., et al. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Keller A., et al. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005;1:E1–E8. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D., et al. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson E., et al. LabKey Server: an open source platform for scientific data integration, analysis and collaboration. BMC Bioinformatics. 2011;12:71. doi: 10.1186/1471-2105-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A.I. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrioli P.G.A., et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- Tabb D.L., et al. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudgian D.C., et al. CPFP: a central proteomics facilities pipeline. Bioinformatics. 2010;26:1131–1132. doi: 10.1093/bioinformatics/btq081. [DOI] [PubMed] [Google Scholar]