Abstract
Atrial fibrillation (AF) is of public health importance and profoundly increases morbidity, mortality and health-related expenditures. Morbidities include the increased risks of cardiovascular outcomes such as heart failure and stroke, and the deleterious effects on quality of life, functional status and cognition. The clinical epidemiology of AF, its risk factors and outcomes, have been extensively investigated. Genetic advances over the last decade have facilitated the identification of mutations and common polymorphisms associated with AF. Metabolomics, proteomics and other “omics” technologies have only recently been applied to the study of AF, and have not yet been systematically investigated. Systems biology approaches, while still in their infancy, offer the promise of providing novel insights into pathways influencing AF risk. In the present review, we address the current state of the epidemiology and genomics of AF. We seek to emphasize how epidemiology and “omic” advances will contribute towards a systems biology approach that will help to unravel the pathogenesis, risk stratification, and novel targets for AF therapies. Our purpose is to articulate questions and challenges that hinge on integrating novel scientific advances in the epidemiology and genomics of AF. As a reference we have provided a glossary in the inset box.
Epidemiology
AF – increasing burden and burgeoning costs
In the US and Western Europe, the aging of the population and accompanying rise in the prevalence of AF have magnified its toll on morbidity and health care costs. The estimated US prevalence of 2.7 to 6.1 million is expected to increase to 5.6 to 12.1 million by the middle of the current century (Figure 1).1,2 Cumulative lifetime risk estimates demonstrate that AF is largely a disease of aging. In US and European community-based cohort studies the lifetime risk of AF is 22–26% in men and 22–23% in women by age 80 years (Figure 2).3,4 AF risk doubles with each progressive decade of age; <1% in individuals 50–59 years are affected, whereas about 10% of those 80–84 years and 11–18% of those ≥85 years have AF.1,2,5
Figure 1.
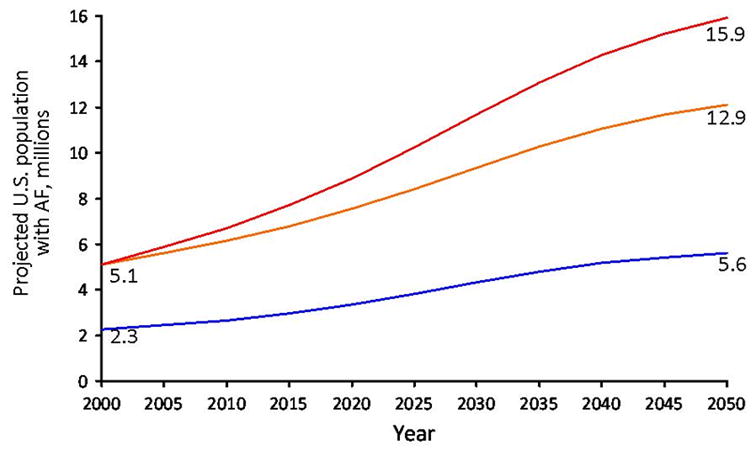
The estimated U.S. prevalence of atrial fibrillation (AF) in the year 2050 ranges from 5.6 million to as high as 15.9 million individuals. The blue line shows estimates derived from the AnTicoagulation and Risk Factors in Atrial Fibrillation Study, comprised of cross-sectional data from a large California health maintenance organization.1 The orange line shows data estimating the number of individuals with AF in the US through 2050. The investigators identified cases of ECG-confirmed incident AF in Olmsted County Minnesota, from 1980–2000, and employed U.S. Census Bureau data to estimate that the number of individuals with AF will reach 12.1 million by 2050. However, an increase in age-adjusted incidence of AF in Olmsted County was identified from 1980–2000. The red line shows the projected number of individuals with AF in the U.S. should this increase in age-adjusted incidence of AF persist. Figure used with permission and modified from Miyasaka et al.2
Figure 2.
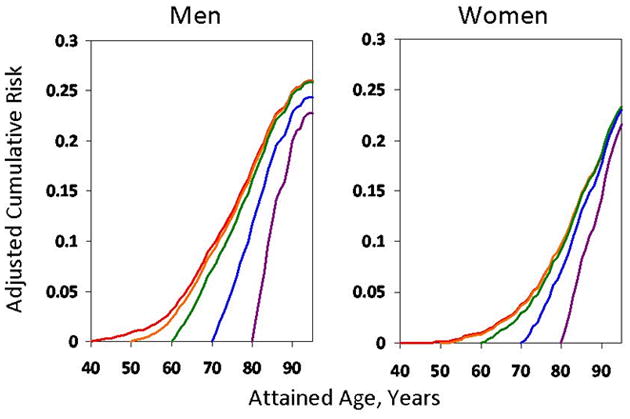
Lifetime risk for developing atrial fibrillation (AF) from the Framingham Heart Study. Men and women without AF at age 40 were determined to have a 26% and 23% likelihood of developing incident AF by age 80. Reproduced from Lloyd-Jones et al.3
The increasing prevalence of AF has resulted in greater health care utilization and costs. From 1985 through 1999, hospitalizations for AF increased nearly 3-fold.6 A recent analysis of medical costs associated with AF employed commercial and Medical databases representing approximately 38 million individuals in the U.S. The study employed propensity-matching techniques to determine individuals with AF had 73% higher medical costs compared to matched controls. The incremental cost was $8075 per individual with AF in the U.S., resulting in a total national incremental cost ranging from $6.0 to $26.0 billion in 2008 values.7 Data from Western Europe similarly have demonstrated escalating resources consumed by AF hospitalizations.8–12
The increased health care costs reflect sequelae of AF, including heart failure, dementia, stroke, and mortality. Clinical trial registries have established a direct increase in the prevalence of AF with each successive New York Heart Association class.13 In population-based studies the incidence of heart failure among individuals with AF ranged from 3314 to 4415 per 1000 person-years. Concomitant heart failure with AF is associated with a particularly adverse prognosis. In Framingham Heart Study participants with AF, the onset of heart failure was associated with a tripling in mortality compared to those with AF alone.14 Similarly, in an adjusted meta-analysis of observational heart failure studies, AF was associated with increased mortality (odds ratio [OR], 1.14; 95% CI, 1.03 to 1.26; P<0.05).16
In longitudinal, community-based analyses, AF is associated with an increased risk of silent and clinically-evident stroke.17 With nonrheumatic AF the risk of stroke is increased about 5 fold. Compared to strokes without AF, strokes occurring in the setting of AF have greater severity, tend to be more disabling, and have an increased risk of mortality compared to strokes of other etiologies (30-day mortality OR, 1.84; 95% CI, 1.04 to 3.27).18 In addition, AF is associated with a 2-fold increased risk of silent cerebral infarction (OR, 2.16; 95% CI, 1.07 to 4.40).19 Medicare registries and clinical trials have demonstrated the protective effect of warfarin on reducing stroke events.20,21 Furthermore, subtherapeutic anticoagulation results in 5-times the odds of an ischemic stroke.22 Multiple alternatives to warfarin, i.e. factor IIa and Xa antagonists, have been successfully developed for AF stroke prevention.23 Ongoing challenges include long-term studies assessing adherence, cost effectiveness, quality of life and access to such agents.
Individuals with AF have a 1.7-fold odds of impaired cognitive function (95% CI, 1.2 to 2.5) and a 2-fold increased odds for dementia even after adjustment for anticoagulation status (OR, 2.3 95% CI, 1.4 to 3.7).24 Similarly, there is evidence that decreased time in the therapeutic range, while on anticoagulation therapy, is associated with impaired cognition.25 In Olmsted County, the cumulative rate of dementia in individuals with AF exceeded 10% at 5-year follow-up and contributed to an almost 3-fold increased mortality risk (hazard ratio, 2.98; 95% CI, 2.5 to 3.3).26
Increased symptoms, diminished functional performance, and decreased quality of life (QOL) are receiving more attention as metrics for gauging AF treatments. Symptoms have been incorporated into AF guidelines as therapeutic targets, yet there remains a lack of agreement on standardized symptom ascertainment. The significant adverse association of AF with impairments in physical, mental, cognitive and social QOL domains has been documented.27 However, the relation between QOL metrics in individuals with AF and clinical end-points such as heart failure, stroke, and mortality has had limited investigation.28
An analysis in the Canadian Trial of Atrial Fibrillation showed improvement in QOL with AF treatment. The study was limited by 12 months of follow-up, was small in size, and was primarily comprised of individuals with preserved cardiac function.29 In longer follow-up, the Atrial Fibrillation Follow-up Investigation of Rhythm Management (AFFIRM) study identified no difference in QOL outcomes in the rate or rhythm control strategy arms.30
AF epidemiology: future directions
There remains a paucity of epidemiologic data on AF from many parts of the world including Eastern Europe, Africa and South America. Recent epidemiologic investigations in China and Japan reported that the AF prevalence was less than in the US and European countries.31,32 Further, the effects of industrialization and urbanization on AF incidence have not been ascertained. Marshaling resources in areas with social and economic challenges will remain an obstacle towards estimating the global burden of AF.
AF ascertainment, monitoring, and classification in ethnic and racial minorities also have been limited. Much of the AF epidemiology described here was developed in cohorts of European descent. Classification schemes and treatment guidelines have been developed from studies limited by racial and ethnic homogeneity. The longitudinal risk factors, outcomes, and quantification of the burden of AF in ethnic/racial minorities need further elucidation.
Fundamental questions remain concerning AF subsets and the life course of AF including the long-term assessment of the occurrence, recurrence and progression of AF from onset till death. Guidelines and classification schemes describe AF as first detected33 (first diagnosed34), paroxysmal, persistent or permanent.33 However, the classifications have clinical and epidemiologic limitations, in that an individual’s classification may only be evident retrospectively, and the durations embedded in the classification system are somewhat arbitrary. Likewise, it is uncertain how AF in the setting of acute illness, decompensation, or trauma relates to risk of subsequent AF. In addition, lone AF is a well-recognized clinical entity, and its environmental milieu and triggers merit continued investigation.
QOL and symptoms metrics have only recently been introduced,35 but have not become a standardized component for clinical trial, epidemiologic study, or clinical practice. How QOL measures are affected by comorbidity and burden of AF requires continued investigation. The intersections of QOL, aging, and AF have similarly not been well explored, and nor has the relation of AF to diminished functional status and frailty.
Clinical risk factors of AF
Standard AF risk factors may be summarized succinctly as older age, male sex, smoking, obesity, hypertension, diabetes, myocardial infarction, heart failure, valvular heart disease, and cardiac surgery.36,36–39 Figure 3 depicts the population attributable risk for AF risk factors in a longitudinal community-based study.36 The biracial Atherosclerosis Risk in Communities (ARIC) recently reported that 56.5% of the population attributable risk was explained by having at least one borderline or elevated risk factor with high blood pressure being the most important risk factor.40 Although a prominent component of the population attributable AF risk remains unknown, the ARIC data suggest important opportunities for AF prevention through risk factor modification.
Figure 3.
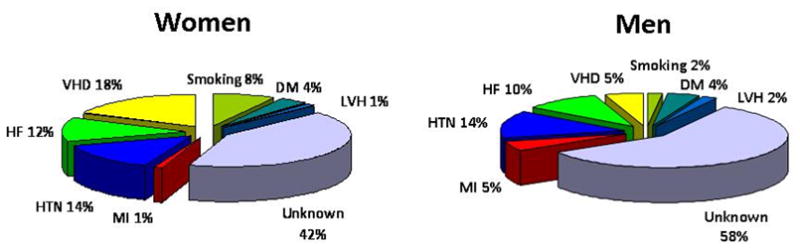
The population attributable risk of atrial fibrillation in men and women determined from a community-based longitudinal study.36 For both men and women a substantial portion of atrial fibrillation risk remains unexplained. MI, myocardial infarction; HTN, hypertension; HF, heart failure; VHD, valvular heart disease; DM, diabetes mellitus; LVH, electrocardiographic left ventricular hypertrophy.
The increasing prevalence of many AF risk factors including the aging of the population, obesity, and diabetes coincide with the rising prevalence of AF. Age is a foremost risk factor for AF,41 from 2010 to 2050, the U.S. population ≥65 years of age is expected to increase from 13.0% to 20.2% and the population ≥85 years from 1.9% to 4.3%.42 Similarly, the prevalence of obesity, now up to one-third of the US population,43 may contribute to an increase in the prevalence of AF. Obesity has been associated with an increase in AF risk from 1.5 to 2.3-fold in community-based cohorts with follow-up ranging from 5 to 14 years.37,38,44 Obesity (HR, 1.54; 95% CI, 1.2 to 2.0) and severe obesity (HR 1.87, 95% CI 1.4 to 2.5) are associated with progression from paroxysmal to permanent AF over a median 5-year follow-up.45 Obstructive sleep apnea, rising in prevalence with obesity, also is associated with increased AF risk (OR, 2.19; 95% CI, 1.40 to 3.42).46
Miscellaneous risk factors for AF that have been reported include hyperthyroidism,47,48 alcohol use,49–51 and exercise.52–56 Of interest, for both alcohol and physical activity, a threshold effect has been reported wherein increased risk of AF was only observed in individuals with heavy alcohol consumption51 or frequent vigorous exercise.56
Indices of AF risk: biomarkers and intermediate phenotypes
Multiple biomarkers have been assessed for their association with AF. N-terminal pro-B-type natriuretic peptide (BNP) has been associated with a multivariable-adjusted, 4-fold increased risk of AF in 10 year median follow-up (HR 4.0, 95% CI, 3.2 to 5.0) when comparing the highest to lowest quartiles in a longitudinal, community-based study.57 Inclusion of BNP likewise significantly strengthened an AF risk score derived in a community-based cohort.58 Interleukin-6 has been elevated in participants with AF in diverse cohorts including individuals with chronic AF and coronary artery disease.59,60 Other biomarkers associated with incident or recurrent AF include osteoprotegerin,61 troponin,62,63 endothelin,63 and plasminogen activator inhibitor-1.64
P wave indices, electrocardiographic markers of intra- and interatrial conduction, have been proposed as non-invasive markers of electrical remodeling. The vectorcardiographic measurements of P wave duration, area, terminal force and dispersion have been used in AF risk prediction.65,66 Associated with age, sex, diabetes, body mass index and waist circumference,67 P wave indices serve as intermediate phenotypes of AF risk and reflect the delayed atrial conduction described in the setting of atrial inflammation, structural remodeling, and progressive atrial fibrosis.68–70 Left ventricular hypertrophy also has been reported to predict new-onset AF in prospective analysis.71 Echocardiographic predictors of AF have effect size estimates similar to ECG predictors and include increasing left atrial diameter and left ventricular wall thickness and decreasing left ventricular systolic function.72
Risk score development has received increasing attention in AF epidemiology. Risk scores serve multiple purposes, including providing benchmarks against which to test novel risk markers; assisting as tools for risk estimation and communication with patients; and identifying low or high risk individuals in decisions regarding clinical management or clinical trial enrollment. The Framingham Heart Study’s AF risk score identified significant predictors for AF risk including age, male sex, treatment for hypertension, significant cardiac murmur, history of heart failure, and increasing body mass index, blood pressure and PR interval.73 The Framingham model’s c statistic was 0.78 (95% CI 0.76 to 0.80).73 In subsequent models developed in the ARIC Study,71 as well as the Age, Gene/Environment Susceptibility-Reykjavik Study and the Cardiovascular Heart Study,74 similar predictive abilities were achieved. The ARIC Study AF risk score validated the Framingham model, achieving a c-statistic of 0.68 in the ARIC cohort, and developed a novel study-specific risk score, which adjusted for race and achieved a c-statistic of 0.78.71 C-reactive protein and natriuretic peptides significantly enhance reclassification for risk of AF.58
AF risk factors: future directions
Racial differences in AF remain poorly understood. Overall, blacks have a higher prevalence of multiple AF risk factors (obesity, diabetes, hypertension and heart failure),75 yet lower incidence of AF.1,76 In the ARIC Study, the cumulative risk of AF at age 80 years reached 21% in white men and 17% in white women, but was only 11% in black men and women.77 The contrasting burden of risk factors with decreased AF incidence has been termed a “racial paradox.”78 There is limited understanding of the biological and pathophysiologic differences modulating the racial differences in AF. An analysis of the Heart and Soul Study identified racial differences in left atrial dimensions.79 Racial variation in P wave indices has been identified in both the ARIC study and National Health and Nutrition Examination Survey (NHANES III).65,80 Some of the racial paradox of AF may be mediated by genetics. In a biracial meta-analysis of the Cardiovascular Health Study and ARIC study using ancestry informative genetic markers, European ancestry was identified as a risk factor for AF (estimated hazard ratio per 10% increase in European ancestry of 1.17, 95% CI 1.07 to1.29).81
Biomarkers represent important opportunities for their role in AF risk assessment, and criteria to evaluate new markers have been articulated.82 Numerous candidate biomarkers representing inflammatory, hemostatic, oxidative stress, matrix remodeling, and neurohormonal pathways may be tested for their associations with AF risk. We advocate a rigorous methodology for examining the value of putative novel biomarkers (or subclinical testing measure) in risk prediction. Biomarker assessment requires metrics of discrimination, calibration and risk reclassification. Providing independent replication of the relation between AF and one or clusters of biomarker(s) will further strengthen the credibility and utility of biomarkers in assessing AF risk. Finally, putative biomarkers should add incremental information over and above established AF predictors in order to demonstrate their value.
Dietary risk factors in AF epidemiology have had limited exploration, likely from challenges from longitudinal tracking of accurate exposures, as recently reviewed.83 There have been contrasting findings on the protective effect of specific kinds of fish on AF prevention.84,85 Long-term, highly accurate monitoring of nutritional intake, or development of nutritional biomarkers, is essential for clarifying dietary contributions to AF risk. The clinical and genetic interactions augmenting such risk also have not been explored. Finally, environmental exposures such as carbon monoxide and air pollution and their associations with AF risk have not been examined in community- or population-based studies.
Genetics of AF
Familial AF
The familial nature of AF was reported as early as 1942.86 In the ensuing 50 years, there were infrequent reports of similar families with an apparent Mendelian inheritance of AF. In 1996, Brugada and colleagues described the first genetic locus for AF;87 however, the causative gene at the locus remains elusive. In 2003, Chen and colleagues described the first mutation in a Chinese family with early-onset AF. The causative mutation resulted in a gain of function in KCNQ1 (the alpha subunit of IKs).88 In subsequent years, multiple mutations have been identified in potassium88–93 and sodium94–96 channels, gap junction proteins,97 and signaling molecules.98 However, screening each of the identified genes has revealed few additional mutations in families with AF.99,100 Thus, unlike other familial monogenic cardiac disorders such as hypertrophic cardiomyopathy and long QT syndrome, wherein a finite set of genes are responsible for the vast majority of the cases, AF is more genetically heterogenous.99,100
Although familial AF has long been recognized, it has only recently been appreciated that AF occurring in the general population is heritable. In the Framingham Heart Study a parental history of AF was shown to nearly double AF risk in offspring.101 More recently, familial AF and premature familial AF improved discrimination and category-less reclassification of AF risk.102 Similarly, investigators from Iceland found that the risk of AF approached nearly 2-fold in first degree relatives of those with AF (risk ratio, 1.77; 95% CI, 1.67 to 1.88) and increased even further with an earlier age of AF onset.103 Finally, in a study of Danish twins, monozygotic twins were significantly more likely to be concordant for AF than dizygotic twins.104
Genome-wide association studies have identified 3 loci for AF
The application of genome-wide association studies (GWAS) to AF has the potential to transform our understanding of molecular mechanisms underlying the arrhythmia. Employing large cohorts, GWAS utilize single nucleotide polymorphisms, or SNPs, as markers of genetic variation between affected and unaffected individuals. High-density genotyping arrays now combine the power of association studies with systematic genome-wide research. In contrast to candidate gene studies GWAS arrays are unconstrained by pre-existing knowledge of which genes contribute to disease. To date, AF GWAS have been successful in identifying 3 novel genetic loci.105–109
The first locus, on chromosome 4q25, is upstream of the gene PITX2.107,109 PITX2 encodes for the paired-like homeodomain transcription factor 2 that is a plausible candidate gene for AF, albeit unrecognized as such prior to the first AF GWAS. Mommersteeg et al. demonstrated that PITX2c-deficient mice fail to form embryonic pulmonary myocardial cells and as a result lack pulmonary myocardial sleeves. The PITX2c isoform suppresses the default formation of a sinus node in the left atrium.110,111 In the last year, two additional studies have advanced our understanding of the role of PITX2c in AF. Wang et al. have demonstrated that PITX2c is expressed in post-natal mouse left atria, pulmonary veins, and right ventricles, and that PITX2c −/− predisposes mice to atrial arrhythmias.112 Further, microarray analysis revealed that in PITX2c −/− mice, gene expression in sinus node tissue was up-regulated. Up-regulated genes included SHOX2 and TBX3, in addition to multiple channel genes (including KCNQ1).112 Similarly, in human atrial tissue, Kirchhof et al. found that PITX2 expression levels were approximately two orders of magnitude higher in the left atrium compared to the right atrium or the ventricles.113 PITX2c heterozygote mice had shorter atrial action potential durations compared to the wild type, and were susceptible to AF induced by pacing, whereas no differences in cardiac morphology, including interstitial fibrosis and function, were observed.113 However, while PITX2 is an attractive candidate gene for AF at this locus, the precise mechanism through which PITX2, or other genes at this locus, result in AF remains unknown.
The second genetic locus for AF is located at the transcription factor ZFHX3 on chromosome 16q22. ZFHX3 (also known as ATBF1) encodes the transcription factor zinc finger homeobox protein 3, and is expressed primarily in the brain, but also has been isolated in myocardium.114 The mechanism through which genetic variation at this locus relates to AF is unclear.
The third locus for AF is intronic to the calcium-activated potassium channel KCNN3 on chromosome 1q21.106 The KCNN3 (or SK3) channel is part of a family of Ca2+-activated potassium channels gated by changes in intracellular Ca2+ concentration.115 KCNN1 (SK1), and KCNN2 (SK2) channels have predominantly atrial expression. In contrast, KCNN3 channels are equally distributed in atria and ventricles. In human and mouse cardiac repolarization models, KCNN3 channels have a prominent role during the late phase of the cardiac action potential.116–118 Li et al. observed action potential duration prolongation and an increased number of early after depolarizations in the atrial myocytes of KCNN2 −/− knockout mice.119 Atrial arrhythmias, mainly pacing-induced AF, were more frequent in both heterozygous and homozygous KCNN2 mice.119 Interestingly, the familial occurrence of AF remains a risk factor for AF even after the adjustment for the 3 genetic variants identified by GWAS.102
Figure 4 displays the 3 loci on a Manhattan plot, providing a means of illustrating their chromosomal position and statistical significance in meta-analyses. In addition, multiple genetic loci related to AF or established risk factors for AF have been identified throughout the genome. The multiple loci are illustrated in Figure 5, which is intended to indicate the number of loci, their proximity, and their relations.120 As discussed below, such a map may inform development of a genetic systems-based approach towards AF.
Figure 4.
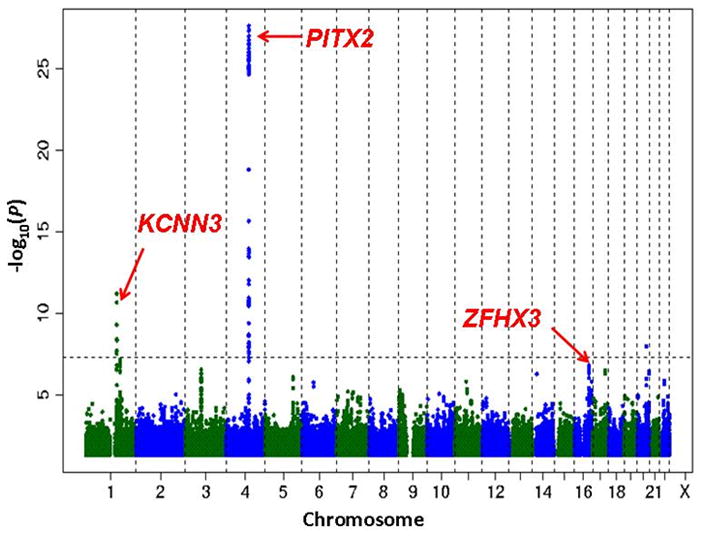
The figure displays the chromosomal positions for each of the three loci associated with atrial fibrillation (AF) in meta-analyses. Such a representation is designated a “Manhattan plot,” as it suggests the New York City skyline. While the x-axis displays the chromosomal position, the y-axis indicates the statistical significance for the association between the single-nucleotide polymorphism (SNP) and the phenotype of interest. The dotted line shows the threshold for statistical significance, adjusted for multiple testing to P<10−8. SNPs with P >0.05 are not displayed on the plot. The initial SNPs identified were on chromosome 4q25 located adjacent to PITX2 and had an effect size as high as 2-fold (Odds ratio [OR] 2.12; 95% confidence interval [CI], 1.77 to 2.54, P=6.3×10−18) in Icelandic individuals diagnosed with AF at ≤60 years.108 The second locus, associated with ZFHX3, identified 3 SNPs with relative risk ranging from 1.26–1.65.105 The third and most recently published SNP, identified on the 4q25 locus with OR 2.03, 95% CI, 1.79–2.30, P=2.5×10−28.106 Figure modified with permission from Ellinor PT, et al.106
Figure 5.
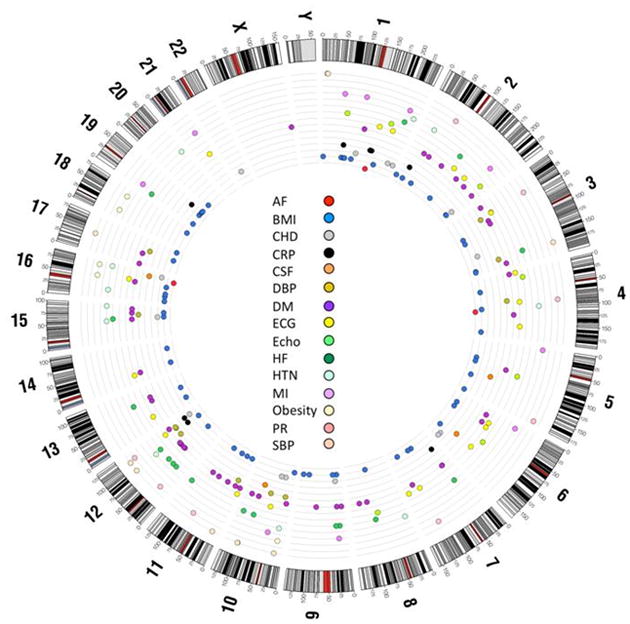
Circos120 plot representing the genetic variants found by genome-wide association study for atrial fibrillation (AF) and AF risk factors. The outer ring represents the chromosomes and the inner rings detail the location of different single nucleotide polymorphisms related to AF and AF risk factors. The colors represent each phenotype or AF risk factor. AF, atrial fibrillation; BMI, body mass index; CHD, coronary heart disease; CRP, C-reactive protein; CSF, cardiac structure and function; DBP, diastolic blood pressure; DM, diabetes mellitus; ECG, electrocardiographic traits; echo, echocardiographic traits; HF, heart failure; HTN, hypertension; MI, myocardial infarction; PR, PR interval; SBP, systolic blood pressure. See electronic supplement for methods.
Genome-wide association studies of AF: future directions
Whereas recent advances with GWAS have identified potential new pathways for AF, the reported genomic loci contribute minimally towards risk of AF in the population. So far, studies of familial AF have discovered rare, highly penetrant genetic variants of AF. In contrast candidate gene studies and GWAS have revealed more common genetic variants with smaller effect sizes in AF cohorts and the general population (Figure 6). However, a large proportion of the heritability of AF cannot be explained by genetic variants reported to date. It has been postulated that the missing heritability of most common diseases, such as AF, partially may be explained by low-frequency variants with intermediate effect sizes (Figure 6).121,122 Advances in sequencing technologies may make it possible to rapidly and reliably detect these low-frequency genetic variants. Whether exome sequencing and potentially whole genome sequencing studies will be able to uncover a substantial portion of the missing heritability of AF remains to be determined.
Figure 6.
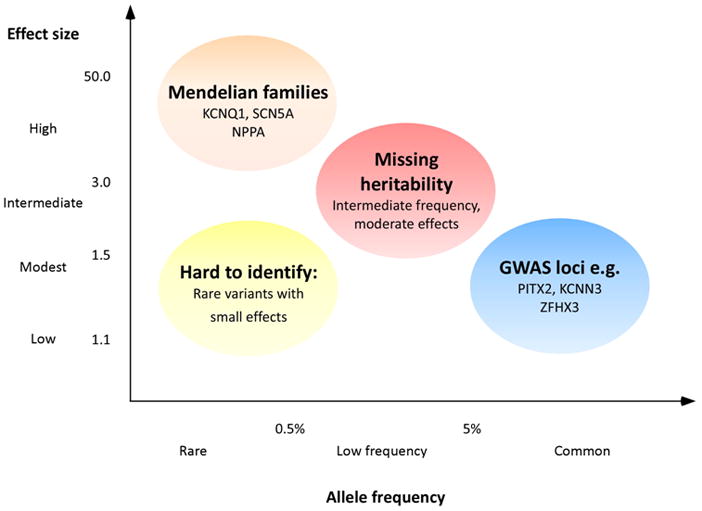
The figure summarizes the identified genetic associations of atrial fibrillation (AF) and future areas of genetic study. Investigators have identified Mendelian families with AF genes having large effect size but rare frequency. Both candidate gene studies and genome-wide association studies (GWAS) have identified variants and loci with low-to-modest effect sizes. Among the two study types, candidate gene studies have tended to report larger effect sizes. Such an overestimation of effect size has been described in a phrase borrowed from economics as the “winner’s curse.” In contrast GWAS have identified three distinct loci with low-to-modest effect sizes. Sequencing efforts may fill a substantive genomic gap by identifying unrecognized low-frequency variants with moderate effect sizes. As a prominent component of the population attributable risk (see Figure 3) remains unknown, further genetic study may contribute additional insights into the etiology of AF. Figure modified from Manolio et al.121
Lastly, the contributions of structural variation123 towards AF risk require investigation. Structural variation refers to the array of genomic microscopic and submicroscopic diversity within the human genome. Specific examples include copy-number variants, deletions, insertions, and other components introducing genomic heterogeneity.124 Unidentified structural variants may contribute towards the heritability of AF, augment AF risk, or indirectly mediate AF risk through other clinical pathways. Furthermore, exploration of structural variants may yield insights into gene-gene and gene-environment pathways for AF risk, which have had only limited investigation.
“Omics” in AF: current status and future directions
To fill the gap between genetic variants and the development of AF new advances are being made in the field of epigenomics, transcriptomics, proteomics, and metabolomics. Epigenetic phenomena, such as DNA methylation or histone modifications may alter gene expression, thereby influencing the potential impact of changes in the DNA sequence.125
Research in the expansive fields of “omics” is emerging rapidly. Although, such endeavors will elucidate novel pathways in the initiation and maintenance of AF, currently “omics” research is still in its infancy. Two recent advances demonstrate the utility of “omics” data in AF investigation, but also illustrate the challenges in interpretation. First, individuals with permanent AF were demonstrated to have up-regulation of transcripts involved in metabolic activities, specifically multiple glycolytic enzymes, thereby suggesting up-regulation of glucose metabolism during AF.126 Second, transcriptomic and proteomic analysis in individuals with persistent AF identified a rise in ketone bodies, suggesting ketone bodies may contribute towards negative feedback on glucose metabolism.127
Published transcriptomic and proteomic investigations need to be interpreted with some caution. The study samples were typically small and the findings have not had opportunity for replication. Analysis of atrial tissue is complicated by anatomic and physiologic variability.126 AF is a highly heterogeneous disease, with multiple contributing mechanisms, predisposing risk factors and conditions. Patient dissimilarities by age, sex, race, risk factors, AF patterns, medication use, and underlying comorbid conditions may contribute to variability in gene expression.
Regulatory loops involving microRNAs are intermediate regulators and modifiers of genes. While microRNAs may serve as broad regulatory mechanisms of an entire pathway, most of their targets and precise elements of their regulation have not been clarified. MicroRNAs have been linked to fibrotic and apoptotic pathways which may contribute to AF susceptibility, via electrical or structural remodeling.128 Several microRNAs have been targeted for their involvement in AF, based on their regulation or association with genes encoding cardiac ion channels or Ca2+-handling proteins. In both animal models and humans with AF, over 3-fold up-regulation of microRNAs specific to calcium channel subunits has been identified from atrial tissue.129 In a cardiac surgery series (n=62, half of whom had AF), left atrial samples from individuals with AF demonstrated significantly reduced levels of a microRNA associated with potassium channel regulation.130
Important challenges remain for the study of microRNAs in AF. There is redundancy in microRNAs targeting specific genes and likewise single microRNAs may have multiple genetic targets. How the multiplicity of effects contribute to AF and intermediate pathways for AF, such as cardiac hypertrophy and failure, renders the interpretation of microRNAs even more challenging.131 In addition, variable microRNA expression in specific patterns of AF (e.g. newly diagnosed, paroxysmal, persistent or permanent AF) or in the setting of clinical or genetic risk factors will require study. Prospective investigations such as those conducted by Lu, et al.129 may provide insights into microRNA expression across the spectrum of AF.
Systems biology: application towards AF
AF epidemiology and genetic investigations stand at an intellectual and scientific crossroad. Similar to many medical fields,132 much of the prior work in AF has applied a reductionist approach, utilizing an exposure-disease paradigm for understanding AF and its risk factors. Most prior genetic epidemiology similarly has employed a two-dimensional framework for examining genome-wide associations.
Accumulating evidence indicates that complex diseases are usually influenced by the interactions of multiple genes, which are incompletely captured by single-gene based approaches.133–135 Furthermore, biological systems are comprised of circuitries of interacting components, such as proteins, nucleic acids and other small molecules, which operate in concert to create complicated molecular networks.136–139 Added to such myriad factors are gene-gene and gene-environment interactions, followed in turn by longitudinal epidemiologic risk factor modification.
The paradigm of systems biology provides novel and important opportunities for combining the classic epidemiology of exposure-disease association, genetic associations, “omics” and molecular investigations. A systems biology approach132 departs from the linear investigation of exposure-disease or gene-phenotype and seeks to investigate a network of interconnected hierarchies from the cellular to the population level. Informed by the preceding stage of investigation, systems biology networks will facilitate examination of the relations between genome, epigenome, transcriptome, proteome, metabolome and clinical (“environmental”) risk factors and their connection to the ‘phenome’ of interest. Systems biology models the complex interactions between the multiple epidemiologic, genomic, and ‘omic arenas, combining and distilling hierarchies to generate non-linear understanding of disease pathogenesis.140,141 Figure 7 illustrates selected components which may be integrated in a systems biology approach towards AF. Ultimately, we anticipate systems biology will provide a mechanistic basis for discerning phenotypic differences among individuals with AF through development of the “interactome” of network-based approaches integrating “omic” and environmental factors.141 By identifying and characterizing the components of a biological system, systems biology can examine the interaction and hierarchical association between the distinct components.142,143
Figure 7.
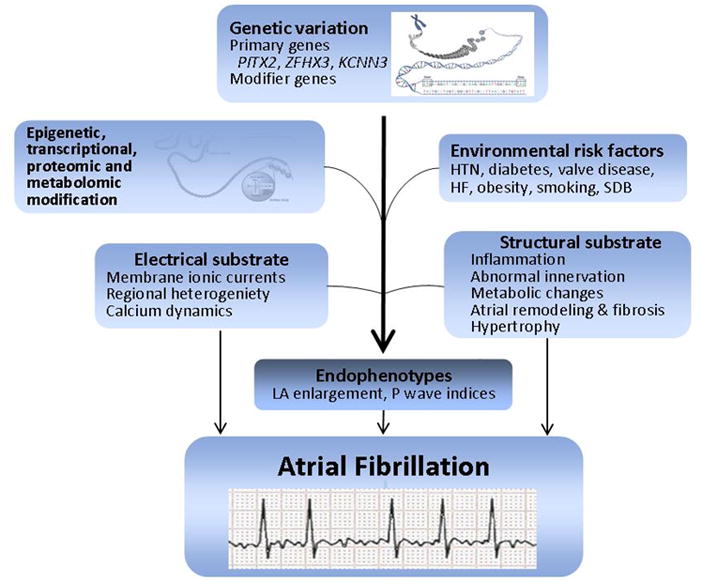
The figure shows the informational flow which comprises the systems biology approach. Genetic loci provide targets for epigenomic, transcriptomic, proteomic and metabolomic investigations. Epidemiologic investigations identify risk factors. In turn, systems biology translates epidemiologic and genomic pathways, providing insight into electrical and structural substrates. Systems biology in turn may yield novel hypotheses for risk factors, target intermediate endophenotypes for genetic dissection, or elucidate novel up-stream targets for therapeutic intervention. HTN, hypertension; HF, heart failure; SDB, sleep disordered breathing; LA, left atrial.
Systems biology has not had application to AF research and its potential has not yet been realized. As an iterative process, the systems biology approach will require repeated experimentation and refinement. No large studies of AF have conducted the comprehensive cell-to-cohort integration of systems networks which we envision. Future systems biology efforts will be most effective if they incorporate work across disciplines including cellular and clinical electrophysiology, in vitro and animal science, genomics, epidemiology, bioinformatics, statistics, etc. Finally, we are a full generation from a systems biology which integrates longitudinal gene-environment and risk exposures in developing networks for modeling AF risk. More realistically, we anticipate that in the future systems biology will inform basic and epidemiological investigations, reminding us that AF’s complex expression is not limited to isolated exposures or genetic loci.
Conclusion
AF has profound clinical and public health burdens, which have grown over the last several decades. Epidemiologic approaches have determined the major clinical risk factors but there are large areas of uncertainty. Robust biomarker assessment, racial and global variation, and dietary and environmental exposures are some persistent areas that require epidemiologic investigation and clarification. A better understanding of AF risk factors and risk stratification will hopefully accelerate efforts at prevention.
Advances by GWAS have identified new genomic loci associated with AF risk, but explain very little of the population attributable risk for AF. “Omics” studies, still in early development and confined to small experimental studies and clinical samples, provide new avenues for identifying cellular and developmental pathways for AF. We advocate the importance of developing a systems biology approach, which will integrate experimental and clinical data across multiple disciplines and “omics” studies. The Table summarizes gaps and research opportunities discussed in our review that are critical to address to accelerate discovery. We envision that systems biology, informed by population-based epidemiology, will identify phenotypic traits, integrate genetic loci, and utilize “omics” platforms to provide new insights into the pathogenesis, prevention, risk prediction, and therapeutic strategies for AF.
Table.
Knowledge Gaps and Areas Essential for Advancing Understanding of the Pathogenesis, Risk, Prevention and Treatment of Atrial Fibrillation.
| Research Domain | Important Knowledge Gaps | Areas of Potential Discovery and Scientific Advancement |
|---|---|---|
| Epidemiology |
|
|
| Genetics |
|
|
| “Omics” |
|
|
| Systems Biology |
|
|
AF, atrial fibrillation.
Acknowledgments
The authors would like to acknowledge and thank Dr. Joseph Losalzo, MD, PhD, for review of this article.
Funding: Dr. Magnani is supported by American Heart Association Award 09FTF219028. Dr. Rienstra is supported by a grant from the Netherlands Organization for Scientific Research (Rubicon grant 825.09.020). Dr. Sinner is supported by the German Heart Foundation. Dr. McManus is supported by grants from the NIH (R37HL69874-08, U01HL105268). This work was supported by grants from the NIH to Drs. Benjamin and Ellinor (HL092577), Dr. Benjamin (RO1AG028321, RC1-HL01056, 1R01HL102214) and Dr. Ellinor (5R21DA027021, 1RO1HL104156, 1K24HL105780) and 6R01-NS17950, N01-HC25195.
Glossary
- Allele
An allele is one or more versions of a gene. Different alleles may result in phenotypic differences. An individual gets one allele from his/her mother, and one from the father. In the context of common genetics, alleles typically refer to the two possible bases of a single nucleotide polymorphism, or SNP.
- Bioinformatics
The application of computer science to the analysis and integration of biological data. Bioinformatics has had an increasing role in genetics, genomics and other areas of “omics” investigation, and systems biology.
- Computational biology
A field of science that utilizes computer science techniques, applied mathematics and statistics to investigate and simulate problems motivated by biology.
- Calibration
Risk prediction models in which the predicted risk of events approximates the actual observed risk of events are considered well-calibrated. Calibration of prediction models is typically measured by dividing the distribution of predicted risk into different categories (quantiles) such as deciles, and comparing the expected number of events to the observed number of events, as in the Hosmer-Lemeshow chi-squared calibration test statistic.1 A significant difference between observed and expected numbers of events suggests a poorly calibrated model.
- Discrimination
Discrimination refers to the ability of a model to distinguish between two or more outcomes. In the case of risk prediction models, the degree to which a model assigns a higher predicted risk to those who develop events than to those who do not develop events may be quantified using the c-statistic. The c-statistic ranges from 0 to 1. Values of 0.5 represent a model that discriminates no better than chance alone, whereas higher values represent models with better discrimination.
- Endophenotype
Phenotypes that serve as proxies for another phenotype of investigative interest may be referred to as endophenotypes. Endophenotypes typically are heritable, are associated with condition or illness of interest, and may exist as an intermediate manifestation prior to the development of a manifest phenotype.
- Epigenomics
The field of genetics investigating pathways that regulate DNA modifications and expression, such as methylation or histone remodeling.
- Functional genomics
An investigative discipline focused on the phenotypic manifestations of genetic variation, and dynamic interactions between the genetic code and other factors. Examples include transcription and translation and the interaction of the genetic code with other genes (gene-gene interaction) or environmental factors (gene-environment interaction).
- Genome-wide association study (GWAS)
A common analytical approach to examine the relations between single nucleotide polymorphisms (SNPs) or other DNA sequence variants with a phenotype of interest. GWAS typically examine associations hundreds of thousands to millions of sequence variants and a trait. Due to the probability of false positive results with such multiple testing, GWAS generally employ conservative significance thresholds (e.g., P<5×10−8) and require replication of findings in order to report statistically robust associations (See Figure 4 and 6).
- Heritability
In the context of genetics, heritability refers to a measure typically quantified as the proportion of variation in a phenotype that can be attributed to genetic variation. Heritability therefore can range from 0 to 1.
- Integrated discrimination improvement
A statistical test which, when used for the assessment of novel biomarkers or risk factors, assesses the probability of altering risk classification.
- Interaction
Biological Interaction refers to the interrelation between two molecules or biological elements. Statistical interaction (also referred to as effect modification) describes the additional measurable effect on an outcome when considering two or more variables jointly, beyond that which occurs when measuring each variable separately. Interaction is central in genetics, proteomics and other “omics”, highlighting the association between genes, pathways of expression, and the environmental factors which result or influence such expression.
- Linkage analysis
A gene mapping technique that examines the degree to which genetic markers are transmitted in a pedigree in order to identify a locus associated with a disease. Genes close to a marker that is transmitted among affected individuals are more likely to be associated with disease; recombination is less likely to occur at shorter genetic distances.
- Metabolomics
The field of study that investigates the composition and effects of small molecules (metabolites) that remain as intermediary and end products of cellular processes.
- microRNA
MicroRNAs are short RNA sequences (22 base pairs on average) that are complementary to messenger RNA sequences. By binding to messenger RNAs, microRNAs are able to regulate gene function by up- or down-regulating messenger RNA translation.
- Monogenic
Disorders caused by defects in a single gene. Although a phenotype may have variable expression due to incomplete penetrance, the effect size of a monogenic defect is usually substantive enough to explain the disease occurrence.
- Mutation
A change in the genetic code of the DNA. Typically, a single base is modified, but other forms such as modifications of longer DNA stretches, insertions, deletions or duplications of genetic code are possible. Conceptually, mutations are rare events, typically with large effect sizes (see Figure 5), in contrast to polymorphisms, which are common.
- Network
A network in the context of systems biology contains the extensive relations between its constituent components (see Supplemental figures).
- “Omics”
Omics is a term that encompasses the study of genomic, epigenomic, transcriptomic, metabolomic, proteomic and phenomic data, and their interactions to understand complex processes involved in biological systems.
- Phenotype
A disease, characteristic or quantifiable biological measure that may be studied.
- Polygenic
Refers to diseases and conditions caused by multiple DNA alterations. Unlike mongenic conditions, the effect size of each single causal DNA sequence variant may be small. In current studies, polygenic conditions etiologies are implicated when single nucleotide polymorphisms (SNPs) at multiple genetic loci are associated with a condition (Figure 5).
- Population attributable risk
The extent to which a single exposure may be identified as the cause of an outcome in a defined population. Alternatively, the reduction in disease incidence if a single exposure could be completely eliminated from a population (Figure 3).
- Proteomics
The scientific field investigating proteins, their structure, functions and pathophysiologic implications.
- Risk reclassification
In a risk prediction model, risk reclassification refers to the degree to which predicted risk is correctly increased or decreased beyond a referent set of predictors, after considering potentially new predictor variables.2
- Single nucleotide polymorphism (SNP)
A common (≥5%) or low frequency2 (≥0.5% to <5%) type of DNA sequence variant comprised of a single nucleotide substitution. SNPs are usually biallelic (see Figure 6).
- Systems biology
The endeavor to move beyond the boundaries of individual fields, and integrate diverse disciplines such as epidemiology, genetics, proteomics, and metabolomics. The objective of such an approach is to understand disease processes as comprehensively as possible in order to map pathways from genetic and cellular expression to their phenotypic variation.
- Transcription
The genetic process in which genetic code of the DNA is copied in the form of a messenger RNA. The messenger RNA will undergo various processing and regulation steps which ultimately result in protein expression.
- Transcriptomics
The field of study investigating transcribed nucleic acid molecules such as mRNA, rRNA, tRNA, and others.
Reference List
- 1.Hosmer DW, Hosmer T, Le CS, Lemeshow S. A comparison of goodness-of-fit tests for the logistic regression model. Stat Med. 1997;16:965–80. doi: 10.1002/(sici)1097-0258(19970515)16:9<965::aid-sim509>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, Menon S, Hanumanthu G, Gupta M, Upendran S, Gupta S, Mahesh M, Jacob B, Mathew P, Chatterjee P, Arun KS, Sharma S, Chandrika KN, Deshpande N, Palvankar K, Raghavnath R, Krishnakanth R, Karathia H, Rekha B, Nayak R, Vishnupriya G, Kumar HG, Nagini M, Kumar GS, Jose R, Deepthi P, Mohan SS, Gandhi TK, Harsha HC, Deshpande KS, Sarker M, Prasad TS, Pandey A. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Disclosures: None.
A useful genomics glossary is found at http://www.genome.gov/Glossary/index.cfm
Reference List
- 1.Go AS, Hylek EM, Phillips KA, Chang Y, Henault LE, Selby JV, Singer DE. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the AnTicoagulation and Risk Factors in Atrial Fibrillation (ATRIA) Study. JAMA. 2001;285:2370–5. doi: 10.1001/jama.285.18.2370. [DOI] [PubMed] [Google Scholar]
- 2.Miyasaka Y, Barnes ME, Gersh BJ, Cha SS, Bailey KR, Abhayaratna WP, Seward JB, Tsang TS. Secular trends in incidence of atrial fibrillation in Olmsted County, Minnesota, 1980 to 2000, and implications on the projections for future prevalence. Circulation. 2006;114:119–25. doi: 10.1161/CIRCULATIONAHA.105.595140. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Jones DM, Wang TJ, Leip EP, Larson MG, Levy D, Vasan RS, D’Agostino RB, Massaro JM, Beiser A, Wolf PA, Benjamin EJ. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation. 2004;110:1042–6. doi: 10.1161/01.CIR.0000140263.20897.42. [DOI] [PubMed] [Google Scholar]
- 4.Heeringa J, Van Der Kuip DA, Hofman A, Kors JA, van Herpen G, Stricker BH, Stijnen T, Lip GY, Witteman JC. Prevalence, incidence and lifetime risk of atrial fibrillation: the Rotterdam study. Eur Heart J. 2006;27:949–53. doi: 10.1093/eurheartj/ehi825. [DOI] [PubMed] [Google Scholar]
- 5.Kannel WB, Wolf PA, Benjamin EJ, Levy D. Prevalence, incidence, prognosis, and predisposing conditions for atrial fibrillation: population-based estimates. Am J Cardiol. 1998;82:2N–9N. doi: 10.1016/s0002-9149(98)00583-9. [DOI] [PubMed] [Google Scholar]
- 6.Wattigney WA, Mensah GA, Croft JB. Increasing trends in hospitalization for atrial fibrillation in the United States, 1985 through 1999: implications for primary prevention. Circulation. 2003;108:711–6. doi: 10.1161/01.CIR.0000083722.42033.0A. [DOI] [PubMed] [Google Scholar]
- 7.Kim MH, Johnston SS, Chu BC, Dalal MR, Schulman KL. Estimation of total incremental health care costs in patients with atrial fibrillation in the United States. Circ Cardiovasc Qual Outcomes. 2011;4:313–20. doi: 10.1161/CIRCOUTCOMES.110.958165. [DOI] [PubMed] [Google Scholar]
- 8.LE Heuzey JY, Paziaud O, Piot O, Said MA, Copie X, Lavergne T, Guize L. Cost of care distribution in atrial fibrillation patients: the COCAF study. Am Heart J. 2004;147:121–6. doi: 10.1016/s0002-8703(03)00524-6. [DOI] [PubMed] [Google Scholar]
- 9.Stewart S, Murphy N, Walker A, McGuire A, McMurray JJ. Cost of an emerging epidemic: an economic analysis of atrial fibrillation in the UK. Heart. 2004;90:286–92. doi: 10.1136/hrt.2002.008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frost L, Vestergaard P, Mosekilde L, Mortensen LS. Trends in incidence and mortality in the hospital diagnosis of atrial fibrillation or flutter in Denmark, 1980–1999. Int J Cardiol. 2005;103:78–84. doi: 10.1016/j.ijcard.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 11.Friberg J, Buch P, Scharling H, Gadsbphioll N, Jensen GB. Rising rates of hospital admissions for atrial fibrillation. Epidemiology. 2003;14:666–72. doi: 10.1097/01.ede.0000091649.26364.c0. [DOI] [PubMed] [Google Scholar]
- 12.Stewart S, MacIntyre K, MacLeod MM, Bailey AE, Capewell S, McMurray JJ. Trends in hospital activity, morbidity and case fatality related to atrial fibrillation in Scotland, 1986–1996. Eur Heart J. 2001;22:693–701. doi: 10.1053/euhj.2000.2511. [DOI] [PubMed] [Google Scholar]
- 13.Maisel WH, Stevenson LW. Atrial fibrillation in heart failure: epidemiology, pathophysiology, and rationale for therapy. Am J Cardiol. 2003;91:2D–8D. doi: 10.1016/s0002-9149(02)03373-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang TJ, Larson MG, Levy D, Vasan RS, Leip EP, Wolf PA, D’Agostino RB, Murabito JM, Kannel WB, Benjamin EJ. Temporal relations of atrial fibrillation and congestive heart failure and their joint influence on mortality: the Framingham Heart Study. Circulation. 2003;107:2920–5. doi: 10.1161/01.CIR.0000072767.89944.6E. [DOI] [PubMed] [Google Scholar]
- 15.Miyasaka Y, Barnes ME, Gersh BJ, Cha SS, Bailey KR, Abhayaratna W, Seward JB, Iwasaka T, Tsang TS. Incidence and mortality risk of congestive heart failure in atrial fibrillation patients: a community-based study over two decades. Eur Heart J. 2006;27:936–41. doi: 10.1093/eurheartj/ehi694. [DOI] [PubMed] [Google Scholar]
- 16.Mamas MA, Caldwell JC, Chacko S, Garratt CJ, Fath-Ordoubadi F, Neyses L. A meta-analysis of the prognostic significance of atrial fibrillation in chronic heart failure. Eur J Heart Fail. 2009;11:676–83. doi: 10.1093/eurjhf/hfp085. [DOI] [PubMed] [Google Scholar]
- 17.Ezekowitz MD, James KE, Nazarian SM, Davenport J, Broderick JP, Gupta SR, Thadani V, Meyer ML, Bridgers SL. Silent cerebral infarction in patients with nonrheumatic atrial fibrillation. The Veterans Affairs Stroke Prevention in Nonrheumatic Atrial Fibrillation Investigators. Circulation. 1995;92:2178–82. doi: 10.1161/01.cir.92.8.2178. [DOI] [PubMed] [Google Scholar]
- 18.Wolf PA, Dawber TR, Thomas HE, Jr, Kannel WB. Epidemiologic assessment of chronic atrial fibrillation and risk of stroke: the Framingham study. Neurology. 1978;28:973–7. doi: 10.1212/wnl.28.10.973. [DOI] [PubMed] [Google Scholar]
- 19.Das RR, Seshadri S, Beiser AS, Kelly-Hayes M, Au R, Himali JJ, Kase CS, Benjamin EJ, Polak JF, O’Donnell CJ, Yoshita M, D’Agostino RB, Sr, DeCarli C, Wolf PA. Prevalence and correlates of silent cerebral infarcts in the Framingham offspring study. Stroke. 2008;39:2929–35. doi: 10.1161/STROKEAHA.108.516575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gage BF, Waterman AD, Shannon W, Boechler M, Rich MW, Radford MJ. Validation of clinical classification schemes for predicting stroke: results from the National Registry of Atrial Fibrillation. JAMA. 2001;285:2864–70. doi: 10.1001/jama.285.22.2864. [DOI] [PubMed] [Google Scholar]
- 21.Birman-Deych E, Radford MJ, Nilasena DS, Gage BF. Use and effectiveness of warfarin in Medicare beneficiaries with atrial fibrillation. Stroke. 2006;37:1070–4. doi: 10.1161/01.STR.0000208294.46968.a4. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds MW, Fahrbach K, Hauch O, Wygant G, Estok R, Cella C, Nalysnyk L. Warfarin anticoagulation and outcomes in patients with atrial fibrillation: a systematic review and metaanalysis. Chest. 2004;126:1938–45. doi: 10.1378/chest.126.6.1938. [DOI] [PubMed] [Google Scholar]
- 23.Ezekowitz MD, Aikens TH, Brown A, Ellis Z. The evolving field of stroke prevention in patients with atrial fibrillation. Stroke. 2010;41:S17–S20. doi: 10.1161/STROKEAHA.110.598201. [DOI] [PubMed] [Google Scholar]
- 24.Ott A, Breteler MM, de Bruyne MC, van Harskamp F, Grobbee DE, Hofman A. Atrial fibrillation and dementia in a population-based study. The Rotterdam Study. Stroke. 1997;28:316–21. doi: 10.1161/01.str.28.2.316. [DOI] [PubMed] [Google Scholar]
- 25.Flaker GC, Pogue J, Yusuf S, Pfeffer MA, Goldhaber SZ, Granger CB, Anand IS, Hart R, Connolly SJ. Cognitive function and anticoagulation control in patients with atrial fibrillation. Circ Cardiovasc Qual Outcomes. 2010;3:277–83. doi: 10.1161/CIRCOUTCOMES.109.884171. [DOI] [PubMed] [Google Scholar]
- 26.Miyasaka Y, Barnes ME, Petersen RC, Cha SS, Bailey KR, Gersh BJ, Casaclang-Verzosa G, Abhayaratna WP, Seward JB, Iwasaka T, Tsang TS. Risk of dementia in stroke-free patients diagnosed with atrial fibrillation: data from a community-based cohort. Eur Heart J. 2007;28:1962–7. doi: 10.1093/eurheartj/ehm012. [DOI] [PubMed] [Google Scholar]
- 27.Dorian P, Jung W, Newman D, Paquette M, Wood K, Ayers GM, Camm J, Akhtar M, Luderitz B. The impairment of health-related quality of life in patients with intermittent atrial fibrillation: implications for the assessment of investigational therapy. J Am Coll Cardiol. 2000;36:1303–9. doi: 10.1016/s0735-1097(00)00886-x. [DOI] [PubMed] [Google Scholar]
- 28.Darbar D, Roden DM. Symptomatic burden as an endpoint to evaluate interventions in patients with atrial fibrillation. Heart Rhythm. 2005;2:544–9. doi: 10.1016/j.hrthm.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 29.Dorian P, Paquette M, Newman D, Green M, Connolly SJ, Talajic M, Roy D. Quality of life improves with treatment in the Canadian Trial of Atrial Fibrillation. Am Heart J. 2002;143:984–90. doi: 10.1067/mhj.2002.122518. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins LS, Brodsky M, Schron E, Chung M, Rocco T, Jr, Lader E, Constantine M, Sheppard R, Holmes D, Mateski D, Floden L, Prasun M, Greene HL, Shemanski L. Quality of life in atrial fibrillation: the Atrial Fibrillation Follow-up Investigation of Rhythm Management (AFFIRM) study. Am Heart J. 2005;149:112–20. doi: 10.1016/j.ahj.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 31.Iguchi Y, Kimura K, Aoki J, Kobayashi K, Terasawa Y, Sakai K, Shibazaki K. Prevalence of atrial fibrillation in community-dwelling Japanese aged 40 years or older in Japan: analysis of 41,436 non-employee residents in Kurashiki-city. Circ J. 2008;72:909–13. doi: 10.1253/circj.72.909. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z, Hu D. An epidemiological study on the prevalence of atrial fibrillation in the Chinese population of mainland China. J Epidemiol. 2008;18:209–16. doi: 10.2188/jea.JE2008021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuster V, Ryden LE, Cannom DS, Crijns HJ, Curtis AB, Ellenbogen KA, Halperin JL, LE Heuzey JY, Kay GN, Lowe JE, Olsson SB, Prystowsky EN, Tamargo JL, Wann S, Smith SC, Jr, Jacobs AK, Adams CD, Anderson JL, Antman EM, Halperin JL, Hunt SA, Nishimura R, Ornato JP, Page RL, Riegel B, Priori SG, Blanc JJ, Budaj A, Camm AJ, Dean V, Deckers JW, Despres C, Dickstein K, Lekakis J, McGregor K, Metra M, Morais J, Osterspey A, Tamargo JL, Zamorano JL. ACC/AHA/ESC 2006 Guidelines for the Management of Patients with Atrial Fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Revise the 2001 Guidelines for the Management of Patients With Atrial Fibrillation): developed in collaboration with the European Heart Rhythm Association and the Heart Rhythm Society. Circulation. 2006;114:e257–e354. doi: 10.1161/CIRCULATIONAHA.106.177292. [DOI] [PubMed] [Google Scholar]
- 34.Camm AJ, Kirchhof P, Lip GY, Schotten U, Savelieva I, Ernst S, Van Gelder IC, Al-Attar N, Hindricks G, Prendergast B, Heidbuchel H, Alfieri O, Angelini A, Atar D, Colonna P, De CR, De SJ, Goette A, Gorenek B, Heldal M, Hohloser SH, Kolh P, LE Heuzey JY, Ponikowski P, Rutten FH. Guidelines for the management of atrial fibrillation: the Task Force for the Management of Atrial Fibrillation of the European Society of Cardiology (ESC) Eur Heart J. 2010;31:2369–429. doi: 10.1093/eurheartj/ehq278. [DOI] [PubMed] [Google Scholar]
- 35.Spertus J, Dorian P, Bubien R, Lewis S, Godejohn D, Reynolds MR, Lakkireddy DR, Wimmer AP, Bhandari A, Burk C. Development and validation of the Atrial Fibrillation Effect on QualiTy-of-Life (AFEQT) Questionnaire in patients with atrial fibrillation. Circ Arrhythm Electrophysiol. 2011;4:15–25. doi: 10.1161/CIRCEP.110.958033. [DOI] [PubMed] [Google Scholar]
- 36.Benjamin EJ, Levy D, Vaziri SM, D’Agostino RB, Belanger AJ, Wolf PA. Independent risk factors for atrial fibrillation in a population-based cohort. The Framingham Heart Study. JAMA. 1994;271:840–4. [PubMed] [Google Scholar]
- 37.Wang TJ, Parise H, Levy D, D’Agostino RB, Sr, Wolf PA, Vasan RS, Benjamin EJ. Obesity and the risk of new-onset atrial fibrillation. JAMA. 2004;292:2471–7. doi: 10.1001/jama.292.20.2471. [DOI] [PubMed] [Google Scholar]
- 38.Dublin S, French B, Glazer NL, Wiggins KL, Lumley T, Psaty BM, Smith NL, Heckbert SR. Risk of new-onset atrial fibrillation in relation to body mass index. Arch Intern Med. 2006;166:2322–8. doi: 10.1001/archinte.166.21.2322. [DOI] [PubMed] [Google Scholar]
- 39.Thomas MC, Dublin S, Kaplan RC, Glazer NL, Lumley T, Longstreth WT, Jr, Smith NL, Psaty BM, Siscovick DS, Heckbert SR. Blood pressure control and risk of incident atrial fibrillation. Am J Hypertens. 2008;21:1111–6. doi: 10.1038/ajh.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huxley RR, Lopez FL, Folsom AR, Agarwal SK, Loehr LR, Soliman EZ, Maclehose R, Konety S, Alonso A. Absolute and Attributable Risks of Atrial Fibrillation in Relation to Optimal and Borderline Risk Factors: The Atherosclerosis Risk in Communities (ARIC) Study. Circulation. 2011;123:1501–8. doi: 10.1161/CIRCULATIONAHA.110.009035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feinberg WM, Blackshear JL, Laupacis A, Kronmal R, Hart RG. Prevalence, age distribution, and gender of patients with atrial fibrillation. Analysis and implications. Arch Intern Med. 1995;155:469–73. [PubMed] [Google Scholar]
- 42. [Accessed on April 4, 2011]; http://www.census.gov/population/www/projections/summarytables.html.
- 43.Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006;295:1549–55. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 44.Frost L, Hune LJ, Vestergaard P. Overweight and obesity as risk factors for atrial fibrillation or flutter: the Danish Diet, Cancer, and Health Study. Am J Med. 2005;118:489–95. doi: 10.1016/j.amjmed.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 45.Tsang TS, Barnes ME, Miyasaka Y, Cha SS, Bailey KR, Verzosa GC, Seward JB, Gersh BJ. Obesity as a risk factor for the progression of paroxysmal to permanent atrial fibrillation: a longitudinal cohort study of 21 years. Eur Heart J. 2008;9:2227–33. doi: 10.1093/eurheartj/ehn324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gami AS, Pressman G, Caples SM, Kanagala R, Gard JJ, Davison DE, Malouf JF, Ammash NM, Friedman PA, Somers VK. Association of atrial fibrillation and obstructive sleep apnea. Circulation. 2004;110:364–7. doi: 10.1161/01.CIR.0000136587.68725.8E. [DOI] [PubMed] [Google Scholar]
- 47.Cappola AR, Fried LP, Arnold AM, Danese MD, Kuller LH, Burke GL, Tracy RP, Ladenson PW. Thyroid status, cardiovascular risk, and mortality in older adults. JAMA. 2006;295:1033–41. doi: 10.1001/jama.295.9.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawin CT, Geller A, Wolf PA, Belanger AJ, Baker E, Bacharach P, Wilson PW, Benjamin EJ, D’Agostino RB. Low serum thyrotropin concentrations as a risk factor for atrial fibrillation in older persons. N Engl J Med. 1994;331:1249–52. doi: 10.1056/NEJM199411103311901. [DOI] [PubMed] [Google Scholar]
- 49.Frost L, Vestergaard P. Alcohol and risk of atrial fibrillation or flutter: a cohort study. Arch Intern Med. 2004;164:1993–8. doi: 10.1001/archinte.164.18.1993. [DOI] [PubMed] [Google Scholar]
- 50.Conen D, Tedrow UB, Cook NR, Moorthy MV, Buring JE, Albert CM. Alcohol consumption and risk of incident atrial fibrillation in women. JAMA. 2008;300:2489–96. doi: 10.1001/jama.2008.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kodama S, Saito K, Tanaka S, Horikawa C, Saito A, Heianza Y, Anasako Y, Nishigaki Y, Yachi Y, Iida KT, Ohashi Y, Yamada N, Sone H. Alcohol consumption and risk of atrial fibrillation: a meta-analysis. J Am Coll Cardiol. 2011;57:427–36. doi: 10.1016/j.jacc.2010.08.641. [DOI] [PubMed] [Google Scholar]
- 52.Aizer A, Gaziano JM, Cook NR, Manson JE, Buring JE, Albert CM. Relation of vigorous exercise to risk of atrial fibrillation. Am J Cardiol. 2009;103:1572–7. doi: 10.1016/j.amjcard.2009.01.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frost L, Frost P, Vestergaard P. Work related physical activity and risk of a hospital discharge diagnosis of atrial fibrillation or flutter: the Danish Diet, Cancer, and Health Study. Occup Environ Med. 2005;62:49–53. doi: 10.1136/oem.2004.014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molina L, Mont L, Marrugat J, Berruezo A, Brugada J, Bruguera J, Rebato C, Elosua R. Long-term endurance sport practice increases the incidence of lone atrial fibrillation in men: a follow-up study. Europace. 2008;10:618–23. doi: 10.1093/europace/eun071. [DOI] [PubMed] [Google Scholar]
- 55.Mont L, Sambola A, Brugada J, Vacca M, Marrugat J, Elosua R, Pare C, Azqueta M, Sanz G. Long-lasting sport practice and lone atrial fibrillation. Eur Heart J. 2002;23:477–82. doi: 10.1053/euhj.2001.2802. [DOI] [PubMed] [Google Scholar]
- 56.Mozaffarian D, Furberg CD, Psaty BM, Siscovick D. Physical Activity and Incidence of Atrial Fibrillation in Older Adults. The Cardiovascular Health Study. Circulation. 2008;118:800–7. doi: 10.1161/CIRCULATIONAHA.108.785626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patton KK, Ellinor PT, Heckbert SR, Christenson RH, DeFilippi C, Gottdiener JS, Kronmal RA. N-terminal pro-B-type natriuretic peptide is a major predictor of the development of atrial fibrillation: the Cardiovascular Health Study. Circulation. 2009;120:1768–74. doi: 10.1161/CIRCULATIONAHA.109.873265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schnabel RB, Larson MG, Yamamoto JF, Sullivan LM, Pencina MJ, Meigs JB, Tofler GH, Selhub J, Jacques PF, Wolf PA, Magnani JW, Ellinor PT, Wang TJ, Levy D, Vasan RS, Benjamin EJ. Relations of biomarkers of distinct pathophysiological pathways and atrial fibrillation incidence in the community. Circulation. 2010;121:200–7. doi: 10.1161/CIRCULATIONAHA.109.882241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marcus GM, Whooley MA, Glidden DV, Pawlikowska L, Zaroff JG, Olgin JE. Interleukin-6 and atrial fibrillation in patients with coronary artery disease: data from the Heart and Soul Study. Am Heart J. 2008;155:303–9. doi: 10.1016/j.ahj.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roldan V, Marin F, Blann AD, Garcia A, Marco P, Sogorb F, Lip GY. Interleukin-6, endothelial activation and thrombogenesis in chronic atrial fibrillation. Eur Heart J. 2003;24:1373–80. doi: 10.1016/s0195-668x(03)00239-2. [DOI] [PubMed] [Google Scholar]
- 61.Schnabel RB, Larson MG, Yamamoto JF, Kathiresan S, Rong J, Levy D, Keaney JF, Jr, Wang TJ, Vasan RS, Benjamin EJ. Relation of multiple inflammatory biomarkers to incident atrial fibrillation. Am J Cardiol. 2009;104:92–6. doi: 10.1016/j.amjcard.2009.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bugnicourt JM, Rogez V, Guillaumont MP, Rogez JC, Canaple S, Godefroy O. Troponin levels help predict new-onset atrial fibrillation in ischaemic stroke patients: a retrospective study. Eur Neurol. 2010;63:24–8. doi: 10.1159/000258679. [DOI] [PubMed] [Google Scholar]
- 63.Latini R, Masson S, Pirelli S, Barlera S, Pulitano G, Carbonieri E, Gulizia M, Vago T, Favero C, Zdunek D, Struck J, Staszewsky L, Maggioni AP, Franzosi MG, Disertori M. Circulating cardiovascular biomarkers in recurrent atrial fibrillation: data from the GISSI-atrial fibrillation trial. J Intern Med. 2011;269:160–71. doi: 10.1111/j.1365-2796.2010.02287.x. [DOI] [PubMed] [Google Scholar]
- 64.Pretorius M, Donahue BS, Yu C, Greelish JP, Roden DM, Brown NJ. Plasminogen activator inhibitor-1 as a predictor of postoperative atrial fibrillation after cardiopulmonary bypass. Circulation. 2007;116:I1–I7. doi: 10.1161/CIRCULATIONAHA.106.677906. [DOI] [PubMed] [Google Scholar]
- 65.Soliman EZ, Prineas RJ, Case LD, Zhang ZM, Goff DC., Jr Ethnic distribution of ECG predictors of atrial fibrillation and its impact on understanding the ethnic distribution of ischemic stroke in the Atherosclerosis Risk in Communities (ARIC) study. Stroke. 2009;40:1204–11. doi: 10.1161/STROKEAHA.108.534735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magnani JW, Johnson VM, Sullivan LM, Gorodeski EZ, Schnabel RB, Lubitz SA, Levy D, Ellinor PT, Benjamin EJ. P Wave Duration and Risk of Longitudinal Atrial Fibrillation Risk in Persons >/=60 Years Old (from the Framingham Heart Study) Am J Cardiol. 2011;107:917–21. doi: 10.1016/j.amjcard.2010.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Magnani JW, Lopez FL, Soliman EZ, Maclehose RF, Crow RS, Alonso A. P Wave Indices, Obesity, and the Metabolic Syndrome: The Atherosclerosis Risk in Communities Study. Obesity (Silver Spring) 2011 doi: 10.1038/oby.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frustaci A, Chimenti C, Bellocci F, Morgante E, Russo MA, Maseri A. Histological substrate of atrial biopsies in patients with lone atrial fibrillation. Circulation. 1997;96:1180–4. doi: 10.1161/01.cir.96.4.1180. [DOI] [PubMed] [Google Scholar]
- 69.Nattel S, Burstein B, Dobrev D. Atrial remodeling and atrial fibrillation: mechanisms and implications. Circ Arrhythm Electrophysiol. 2008;1:62–73. doi: 10.1161/CIRCEP.107.754564. [DOI] [PubMed] [Google Scholar]
- 70.Ohtani K, Yutani C, Nagata S, Koretsune Y, Hori M, Kamada T. High prevalence of atrial fibrosis in patients with dilated cardiomyopathy. J Am Coll Cardiol. 1995;25:1162–9. doi: 10.1016/0735-1097(94)00529-y. [DOI] [PubMed] [Google Scholar]
- 71.Chamberlain AM, Agarwal SK, Folsom AR, Soliman EZ, Chambless LE, Crow R, Ambrose M, Alonso A. A clinical risk score for atrial fibrillation in a biracial prospective cohort (from the Atherosclerosis Risk in Communities [ARIC] study) Am J Cardiol. 2011;107:85–91. doi: 10.1016/j.amjcard.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vaziri SM, Larson MG, Benjamin EJ, Levy D. Echocardiographic predictors of nonrheumatic atrial fibrillation. The Framingham Heart Study. Circulation. 1994;89:724–30. doi: 10.1161/01.cir.89.2.724. [DOI] [PubMed] [Google Scholar]
- 73.Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D’Agostino RB, Sr, Newton-Cheh C, Yamamoto JF, Magnani JW, Tadros TM, Kannel WB, Wang TJ, Ellinor PT, Wolf PA, Vasan RS, Benjamin EJ. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–45. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schnabel RB, Aspelund T, Li G, Sullivan LM, Suchy-Dicey A, Harris TB, Pencina MJ, D’Agostino RB, Sr, Levy D, Kannel WB, Wang TJ, Kronmal RA, Wolf PA, Burke GL, Launer LJ, Vasan RS, Psaty BM, Benjamin EJ, Gudnason V, Heckbert SR. Validation of an atrial fibrillation risk algorithm in whites and African Americans. Arch Intern Med. 2010;170:1909–17. doi: 10.1001/archinternmed.2010.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith SC, Jr, Clark LT, Cooper RS, Daniels SR, Kumanyika SK, Ofili E, Quinones MA, Sanchez EJ, Saunders E, Tiukinhoy SD. Discovering the full spectrum of cardiovascular disease: Minority Health Summit 2003: report of the Obesity, Metabolic Syndrome, and Hypertension Writing Group. Circulation. 2005;111:e134–e139. doi: 10.1161/01.CIR.0000157743.54710.04. [DOI] [PubMed] [Google Scholar]
- 76.Psaty BM, Manolio TA, Kuller LH, Kronmal RA, Cushman M, Fried LP, White R, Furberg CD, Rautaharju PM. Incidence of and risk factors for atrial fibrillation in older adults. Circulation. 1997;96:2455–61. doi: 10.1161/01.cir.96.7.2455. [DOI] [PubMed] [Google Scholar]
- 77.Alonso A, Agarwal SK, Soliman EZ, Ambrose M, Chamberlain AM, Prineas RJ, Folsom AR. Incidence of atrial fibrillation in whites and African-Americans: the Atherosclerosis Risk in Communities (ARIC) study. Am Heart J. 2009;158:111–7. doi: 10.1016/j.ahj.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soliman EZ, Goff DC., Jr Paradox of racial distribution of atrial fibrillation. J Natl Med Assoc. 2008;100:447–8. doi: 10.1016/s0027-9684(15)31282-7. [DOI] [PubMed] [Google Scholar]
- 79.Marcus GM, Olgin JE, Whooley M, Vittinghoff E, Stone KL, Mehra R, Hulley SB, Schiller NB. Racial differences in atrial fibrillation prevalence and left atrial size. Am J Med. 2010;123:375–7. doi: 10.1016/j.amjmed.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Magnani JW, Gorodeski EZ, Johnson VM, Sullivan LM, Hamburg NM, Benjamin EJ, Ellinor PT. P wave duration is associated with cardiovascular and all-cause mortality outcomes: the National Health and Nutrition Examination Survey. Heart Rhythm. 2011;8:93–100. doi: 10.1016/j.hrthm.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marcus GM, Alonso A, Peralta CA, Lettre G, Vittinghoff E, Lubitz SA, Fox ER, Levitzky YS, Mehra R, Kerr KF, Deo R, Sotoodehnia N, Akylbekova M, Ellinor PT, Paltoo DN, Soliman EZ, Benjamin EJ, Heckbert SR. European ancestry as a risk factor for atrial fibrillation in African Americans. Circulation. 2010;122:2009–15. doi: 10.1161/CIRCULATIONAHA.110.958306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hlatky MA, Greenland P, Arnett DK, Ballantyne CM, Criqui MH, Elkind MS, Go AS, Harrell FE, Jr, Hong Y, Howard BV, Howard VJ, Hsue PY, Kramer CM, McConnell JP, Normand SL, O’Donnell CJ, Smith SC, Jr, Wilson PW. Criteria for evaluation of novel markers of cardiovascular risk: a scientific statement from the American Heart Association. Circulation. 2009;119:2408–16. doi: 10.1161/CIRCULATIONAHA.109.192278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gronroos NN, Alonso A. Diet and risk of atrial fibrillation - epidemiologic and clinical evidence - Circ J. 2010;74:2029–38. doi: 10.1253/circj.cj-10-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mozaffarian D, Psaty BM, Rimm EB, Lemaitre RN, Burke GL, Lyles MF, Lefkowitz D, Siscovick DS. Fish intake and risk of incident atrial fibrillation. Circulation. 2004;110:368–73. doi: 10.1161/01.CIR.0000138154.00779.A5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen J, Johnson VM, Sullivan LM, Jacques PF, Magnani JW, Lubitz SA, Pandey S, Levy D, Vasan RS, Quatromoni PA, Junyent M, Ordovas JM, Benjamin EJ. Dietary factors and incident atrial fibrillation: the Framingham Heart Study. Am J Clin Nutr. 2011;93:261–6. doi: 10.3945/ajcn.110.001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolff L. Familial auricular fibrillation. N Engl J Med. 1943;229(10):396–398. [Google Scholar]
- 87.Brugada R, Tapscott T, Czernuszewicz GZ, Marian AJ, Iglesias A, Mont L, Brugada J, Girona J, Domingo A, Bachinski LL, Roberts R. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med. 1997;336:905–11. doi: 10.1056/NEJM199703273361302. [DOI] [PubMed] [Google Scholar]
- 88.Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, Jin HW, Sun H, Su XY, Zhuang QN, Yang YQ, Li YB, Liu Y, Xu HJ, Li XF, Ma N, Mou CP, Chen Z, Barhanin J, Huang W. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–4. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 89.Yang Y, Xia M, Jin Q, Bendahhou S, Shi J, Chen Y, Liang B, Lin J, Liu Y, Liu B, Zhou Q, Zhang D, Wang R, Ma N, Su X, Niu K, Pei Y, Xu W, Chen Z, Wan H, Cui J, Barhanin J, Chen Y. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genet. 2004;75:899–905. doi: 10.1086/425342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xia M, Jin Q, Bendahhou S, He Y, Larroque MM, Chen Y, Zhou Q, Yang Y, Liu Y, Liu B, Zhu Q, Zhou Y, Lin J, Liang B, Li L, Dong X, Pan Z, Wang R, Wan H, Qiu W, Xu W, Eurlings P, Barhanin J, Chen Y. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–9. doi: 10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 91.Otway R, Vandenberg JI, Guo G, Varghese A, Castro ML, Liu J, Zhao J, Bursill JA, Wyse KR, Crotty H, Baddeley O, Walker B, Kuchar D, Thorburn C, Fatkin D. Stretch-sensitive KCNQ1 mutation A link between genetic and environmental factors in the pathogenesis of atrial fibrillation? J Am Coll Cardiol. 2007;49:578–86. doi: 10.1016/j.jacc.2006.09.044. [DOI] [PubMed] [Google Scholar]
- 92.Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, Sattiraju S, Ballew JD, Jahangir A, Terzic A. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–91. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 93.Hong K, Bjerregaard P, Gussak I, Brugada R. Short QT syndrome and atrial fibrillation caused by mutation in KCNH2. J Cardiovasc Electrophysiol. 2005;16:394–6. doi: 10.1046/j.1540-8167.2005.40621.x. [DOI] [PubMed] [Google Scholar]
- 94.Chen LY, Ballew JD, Herron KJ, Rodeheffer RJ, Olson TM. A common polymorphism in SCN5A is associated with lone atrial fibrillation. Clin Pharmacol Ther. 2007;81:35–41. doi: 10.1038/sj.clpt.6100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, George AL, Jr, Roden DM. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–35. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Makiyama T, Akao M, Shizuta S, Doi T, Nishiyama K, Oka Y, Ohno S, Nishio Y, Tsuji K, Itoh H, Kimura T, Kita T, Horie M. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J Am Coll Cardiol. 2008;52:1326–34. doi: 10.1016/j.jacc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 97.Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, Liu X, Veinot JP, Tang AS, Stewart AF, Tesson F, Klein GJ, Yee R, Skanes AC, Guiraudon GM, Ebihara L, Bai D. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med. 2006;354:2677–88. doi: 10.1056/NEJMoa052800. [DOI] [PubMed] [Google Scholar]
- 98.Hodgson-Zingman DM, Karst ML, Zingman LV, Heublein DM, Darbar D, Herron KJ, Ballew JD, de Andrade M, Burnett JC, Jr, Olson TM. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N Engl J Med. 2008;359:158–65. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ellinor PT, Moore RK, Patton KK, Ruskin JN, Pollak MR, MacRae CA. Mutations in the long QT gene, KCNQ1, are an uncommon cause of atrial fibrillation. Heart. 2004;90:1487–8. doi: 10.1136/hrt.2003.027227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ellinor PT, Petrov-Kondratov VI, Zakharova E, Nam EG, MacRae CA. Potassium channel gene mutations rarely cause atrial fibrillation. BMC Med Genet. 2006;7:70. doi: 10.1186/1471-2350-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fox CS, Parise H, D’Agostino RB, Sr, Lloyd-Jones DM, Vasan RS, Wang TJ, Levy D, Wolf PA, Benjamin EJ. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291:2851–5. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 102.Lubitz SA, Yin X, Fontes JD, Magnani JW, Rienstra M, Pai M, Villalon ML, Vasan RS, Pencina MJ, Levy D, Larson MG, Ellinor PT, Benjamin EJ. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA. 2010;304:2263–9. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arnar DO, Thorvaldsson S, Manolio TA, Thorgeirsson G, Kristjansson K, Hakonarson H, Stefansson K. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J. 2006;27:708–12. doi: 10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- 104.Christophersen IE, Ravn LS, Budtz-Joergensen E, Skytthe A, Haunsoe S, Svendsen JH, Christensen K. Familial aggregation of atrial fibrillation: a study in Danish twins. Circ Arrhythm Electrophysiol. 2009;2:378–83. doi: 10.1161/CIRCEP.108.786665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van NC, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, Dehghan A, Lubitz SA, D’Agostino RB, Sr, Lumley T, Ehret GB, Heeringa J, Aspelund T, Newton-Cheh C, Larson MG, Marciante KD, Soliman EZ, Rivadeneira F, Wang TJ, Eiriksdottir G, Levy D, Psaty BM, Li M, Chamberlain AM, Hofman A, Vasan RS, Harris TB, Rotter JI, Kao WH, Agarwal SK, Stricker BH, Wang K, Launer LJ, Smith NL, Chakravarti A, Uitterlinden AG, Wolf PA, Sotoodehnia N, Kottgen A, van Duijn CM, Meitinger T, Mueller M, Perz S, Steinbeck G, Wichmann HE, Lunetta KL, Heckbert SR, Gudnason V, Alonso A, Kaab S, Ellinor PT, Witteman JC. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–81. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA, Fox E, Darbar D, Smith NL, Smith JD, Schnabel RB, Soliman EZ, Rice KM, Van Wagoner DR, Beckmann BM, van NC, Wang K, Ehret GB, Rotter JI, Hazen SL, Steinbeck G, Smith AV, Launer LJ, Harris TB, Makino S, Nelis M, Milan DJ, Perz S, Esko T, Kottgen A, Moebus S, Newton-Cheh C, Li M, Mohlenkamp S, Wang TJ, Kao WH, Vasan RS, Nothen MM, MacRae CA, Stricker BH, Hofman A, Uitterlinden AG, Levy D, Boerwinkle E, Metspalu A, Topol EJ, Chakravarti A, Gudnason V, Psaty BM, Roden DM, Meitinger T, Wichmann HE, Witteman JC, Barnard J, Arking DE, Benjamin EJ, Heckbert SR, Kaab S. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–4. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–7. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 108.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbaumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjornsdottir S, Valdimarsson EM, Lochen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–8. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaab S, Darbar D, van NC, Dupuis J, Pfeufer A, Newton-Cheh C, Schnabel R, Makino S, Sinner MF, Kannankeril PJ, Beckmann BM, Choudry S, Donahue BS, Heeringa J, Perz S, Lunetta KL, Larson MG, Levy D, MacRae CA, Ruskin JN, Wacker A, Schomig A, Wichmann HE, Steinbeck G, Meitinger T, Uitterlinden AG, Witteman JC, Roden DM, Benjamin EJ, Ellinor PT. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009;30:813–9. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mommersteeg MT, Hoogaars WM, Prall OW, Gier-de Vries C, Wiese C, Clout DE, Papaioannou VE, Brown NA, Harvey RP, Moorman AF, Christoffels VM. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–62. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 111.Mommersteeg MT, Brown NA, Prall OW, Gier-de Vries C, Harvey RP, Moorman AF, Christoffels VM. Pitx2c and Nkx2–5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–9. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 112.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A. 2010;107:9753–8. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, Rotering H, Fortmueller L, Laakmann S, Verheule S, Schotten U, Fabritz L, Brown NA. PITX2c is Expressed in the Adult Left Atrium, and Reducing Pitx2c Expression Promotes Atrial Fibrillation Inducibility and Complex Changes in Gene Expression. Circ Cardiovasc Genet. 2011 doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 114.Ido A, Miura Y, Watanabe M, Sakai M, Inoue Y, Miki T, Hashimoto T, Morinaga T, Nishi S, Tamaoki T. Cloning of the cDNA encoding the mouse ATBF1 transcription factor. Gene. 1996;168:227–31. doi: 10.1016/0378-1119(95)00740-7. [DOI] [PubMed] [Google Scholar]
- 115.Kohler M, Hirschberg B, Bond CT, Kinzie JM, Marrion NV, Maylie J, Adelman JP. Small-conductance, calcium-activated potassium channels from mammalian brain. Science. 1996;273:1709–14. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 116.Tuteja D, Xu D, Timofeyev V, Lu L, Sharma D, Zhang Z, Xu Y, Nie L, Vazquez AE, Young JN, Glatter KA, Chiamvimonvat N. Differential expression of small-conductance Ca2+-activated K+ channels SK1, SK2, and SK3 in mouse atrial and ventricular myocytes. Am J Physiol Heart Circ Physiol. 2005;289:H2714–H2723. doi: 10.1152/ajpheart.00534.2005. [DOI] [PubMed] [Google Scholar]
- 117.Tuteja D, Rafizadeh S, Timofeyev V, Wang S, Zhang Z, Li N, Mateo RK, Singapuri A, Young JN, Knowlton AA, Chiamvimonvat N. Cardiac small conductance Ca2+-activated K+ channel subunits form heteromultimers via the coiled-coil domains in the C termini of the channels. Circ Res. 2010;107:851–9. doi: 10.1161/CIRCRESAHA.109.215269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu Y, Tuteja D, Zhang Z, Xu D, Zhang Y, Rodriguez J, Nie L, Tuxson HR, Young JN, Glatter KA, Vazquez AE, Yamoah EN, Chiamvimonvat N. Molecular identification and functional roles of a Ca(2+)-activated K+ channel in human and mouse hearts. J Biol Chem. 2003;278:49085–94. doi: 10.1074/jbc.M307508200. [DOI] [PubMed] [Google Scholar]
- 119.Li N, Timofeyev V, Tuteja D, Xu D, Lu L, Zhang Q, Zhang Z, Singapuri A, Albert TR, Rajagopal AV, Bond CT, Periasamy M, Adelman J, Chiamvimonvat N. Ablation of a Ca2+-activated K+ channel (SK2 channel) results in action potential prolongation in atrial myocytes and atrial fibrillation. J Physiol. 2009;587:1087–100. doi: 10.1113/jphysiol.2008.167718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sinner MF, Ellinor PT, Meitinger T, Benjamin EJ, Kaab S. Genome-wide association studies of atrial fibrillation: past, present, and future. Cardiovasc Res. 2011;89:701–9. doi: 10.1093/cvr/cvr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39:S7–15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 125.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010;3:567–73. doi: 10.1161/CIRCGENETICS.110.958744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kaab S, Hinterseer M, Kartmann H, Kreuzer E, Dugas M, Steinbeck G, Nabauer M. Reprogramming of the human atrial transcriptome in permanent atrial fibrillation: expression of a ventricular-like genomic signature. Circ Res. 2005;96:1022–9. doi: 10.1161/01.RES.0000165480.82737.33. [DOI] [PubMed] [Google Scholar]
- 127.Mayr M, Yusuf S, Weir G, Chung YL, Mayr U, Yin X, Ladroue C, Madhu B, Roberts N, De Souza A, Fredericks S, Stubbs M, Griffiths JR, Jahangiri M, Xu Q, Camm AJ. Combined metabolomic and proteomic analysis of human atrial fibrillation. J Am Coll Cardiol. 2008;51:585–94. doi: 10.1016/j.jacc.2007.09.055. [DOI] [PubMed] [Google Scholar]
- 128.Wang Z, Lu Y, Yang B. MicroRNAs and atrial fibrillation: new fundamentals. Cardiovasc Res. 2011;89:710–21. doi: 10.1093/cvr/cvq350. [DOI] [PubMed] [Google Scholar]
- 129.Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, Zhang Y, Shan H, Luo X, Bai Y, Sun L, Song W, Xu C, Wang Z, Yang B. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122:2378–87. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 130.Girmatsion Z, Biliczki P, Bonauer A, Wimmer-Greinecker G, Scherer M, Moritz A, Bukowska A, Goette A, Nattel S, Hohnloser SH, Ehrlich JR. Changes in microRNA-1 expression and IK1 up-regulation in human atrial fibrillation. Heart Rhythm. 2009;6:1802–9. doi: 10.1016/j.hrthm.2009.08.035. [DOI] [PubMed] [Google Scholar]
- 131.Luo X, Zhang H, Xiao J, Wang Z. Regulation of human cardiac ion channel genes by microRNAs: theoretical perspective and pathophysiological implications. Cell Physiol Biochem. 2010;25:571–86. doi: 10.1159/000315076. [DOI] [PubMed] [Google Scholar]
- 132.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Freimer NB, Sabatti C. Human genetics: variants in common diseases. Nature. 2007;445:828–30. doi: 10.1038/nature05568. [DOI] [PubMed] [Google Scholar]
- 135.Askland K, Read C, Moore J. Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum Genet. 2009;125:63–79. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- 136.Pereira-Leal JB, Enright AJ, Ouzounis CA. Detection of functional modules from protein interaction networks. Proteins. 2004;54:49–57. doi: 10.1002/prot.10505. [DOI] [PubMed] [Google Scholar]
- 137.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100:12123–8. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Snel B, Bork P, Huynen MA. The identification of functional modules from the genomic association of genes. Proc Natl Acad Sci U S A. 2002;99:5890–5. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–4. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 140.Lusis AJ, Weiss JN. Cardiovascular networks: systems-based approaches to cardiovascular disease. Circulation. 2010;121:157–70. doi: 10.1161/CIRCULATIONAHA.108.847699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–4. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 143.Kohl P, Crampin EJ, Quinn TA, Noble D. Systems biology: an approach. Clin Pharmacol Ther. 2010;88:25–33. doi: 10.1038/clpt.2010.92. [DOI] [PubMed] [Google Scholar]


