Abstract
It is well known that spontaneously hypertensive rats (SHR) develop muscle pathologies with hypertension and heart failure, though the mechanism remains poorly understood. Woon et al. (2007) linked the circadian clock gene Bmal1 to hypertension and metabolic dysfunction in the SHR. Building on these findings, we compared the expression pattern of several core-clock genes in the gastrocnemius muscle of aged SHR (80 weeks; overt heart failure) compared to aged-matched control WKY strain. Heart failure was associated with marked effects on the expression of Bmal1, Clock and Rora in addition to several non-circadian genes important in regulating skeletal muscle phenotype including Mck, Ttn and Mef2c. We next performed circadian time-course collections at a young age (8 weeks; pre-hypertensive) and adult age (22 weeks; hypertensive) to determine if clock gene expression was disrupted in gastrocnemius, heart and liver tissues prior to or after the rats became hypertensive. We found that hypertensive/hypertrophic SHR showed a dampening of peak Bmal1 and Rev-erb expression in the liver, and the clock-controlled gene Pgc1α in the gastrocnemius. In addition, the core-clock gene Clock and the muscle-specific, clock-controlled gene Myod1, no longer maintained a circadian pattern of expression in gastrocnemius from the hypertensive SHR. These findings provide a framework to suggest a mechanism whereby chronic heart failure leads to skeletal muscle pathologies; prolonged dysregulation of the molecular clock in skeletal muscle results in altered Clock, Pgc1α and Myod1 expression which in turn leads to the mis-regulation of target genes important for mechanical and metabolic function of skeletal muscle.
Introduction
The role of the molecular clock as an underlying factor contributing to cardiovascular and skeletal muscle disease is a new but growing area of research. It is now recognized that most, if not all, cells in the body contain a self-sustaining molecular circadian clock [1]. In general, the synchronization of all the body's clocks is orchestrated by a central clock (SCN: suprachiasmatic nucleus) located in the hypothalamus acting through neurohumoral mechanisms [2]. The synchronization of circadian clocks has been experimentally shown to provide an adaptive advantage by enhancing an organisms ability to respond to daily changes in light, temperature and humidity [3]. Pathologies emerge when there is a misalignment between internal circadian rhythms and daily cycles in environmental cues such as light which can occur when the expression of the molecular clock becomes shifted or dampened [4].
Circadian rhythms, endogenously generated rhythms with ≈24 hour-period, are driven by an intrinsic molecular clock which works as a transcription/translation feedback system. In mammals, the proteins encoded by core-clock genes, Bmal1 and Clock, dimerize to drive transcription of Period (Per) and Cryptochrome (Cry) and the protein products of these genes down-regulate BMAL1 and CLOCK function. BMAL1∶CLOCK heterodimers directly regulate the expression of a group of genes referred to as primary clock-controlled genes (CCGs). Many other genes are expressed in a circadian manner and these genes are indirectly influenced by the core clock factors as well as environmental cues, such as hormones like melatonin and cortisol [5]–[13].
Woon and colleagues (2007) identified polymorphisms in the promoter region of Bmal1 (Brain and muscle Arnt-like protein-1) that were within the same congenic interval associated with hypertension in the spontaneously hypertensive rat (SHR) [14]. Furthermore, the same group reported a significant genetic association of Bmal1 polymorphisms and hypertension and type II diabetes in humans [14]. These findings implicate the regulation of Bmal1 expression and proper molecular clock function in the development of cardiometabolic pathologies. Consistent with these findings, Andrews and coworkers (2010) identified a common skeletal muscle pathology in two clock-compromised mouse strains that were characterized by diminished force capacity and reduced mitochondrial content and function [5].
It is well recognized that chronic heart failure (CHF) and cardiovascular disease are often associated with distinct skeletal muscle pathologies [15], [16]. This association has been shown in different rodent models of cardiovascular disease as well as in humans. These studies have reported the presence of a number of pathologies in skeletal muscle related to both function and metabolism. Specifically, structural and biochemical alterations have been demonstrated in patients with CHF, including fiber atrophy [17], fiber type transformation [18] reduced sensitivity to insulin, decrease in oxidative capacity [19] and abnormalities in mitochondrial structure [20]–[22]. The mechanism(s) underlying the development of these skeletal muscle pathologies associated with cardiovascular disease remain undefined.
The purpose of this study was to determine if changes occurred in the expression of core clock factors and CCGs in peripheral tissues either prior to or following the development of cardiovascular disease and insulin resistance. Our working hypothesis was that clock gene expression would change after the development of hypertension in muscle and non-muscle tissues. In this study we found that circadian gene expression was altered across all tissues studied (gastrocnemius, liver, heart) in the young pre-hypertensive SHR. Analysis of gene expression at the adult hypertensive/hypertrophic stage (22 week old), found that the disruption/dysregulation of clock gene expression persisted primarily in skeletal muscle of SHR compared to WKY rats. In addition, there were changes in expression of the core clock gene Clock and two clock-controlled genes in skeletal muscle, Myod1 and Pgc1α of adult SHR. Further examination revealed, that the expression of molecular clock factors were significantly altered in the skeletal muscle of SHR at end stage heart failure. Collectively, these results suggest that genetic difference(s) in the SHR vs. WKY are associated with altered expression of the core clock factors and downstream clock controlled genes in peripheral tissues prior to the development of hypertension. We also propose that prolonged alterations of the molecular clock factors in skeletal muscle may lead to a loss of regulation of downstream targets, Myod1 and Pgc1α with subsequent effects on skeletal muscle functional and metabolic pathologies described in the SHR.
Results
Dysregulation of core-clock genes is evident in the gastrocnemius of 8 week old SHR rats
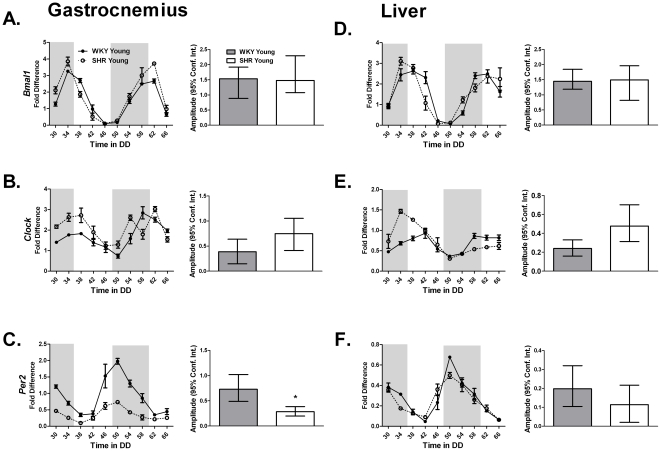
Preliminary experiments in 80 week old, overt heart failure rats demonstrated a diminished expression of several core clock genes as well as several non-circadian genes important in regulating skeletal muscle phenotype and function (Figures S1 and S2) in the gastrocnemius of SHR compared to age-matched WKY rats. While it was impossible to perform a circadian time course collection on the heart failure rats because of high mortality, we were able to perform a time course on 8 week old and 22 week old SHR to determine if the onset of hypertension was associated with disruption of the circadian clock. Tissues were collected from rats every 4 hours for 40 hours. Table 1 provides the results of the statistical analysis with the circadian parameters of the mRNA data for the core-clock genes Bmal1, Clock and Per2 in the gastrocnemius and liver of the young WKY and SHR rats using JTK_CYCLE [23]. The core molecular clock factors, Bmal1, Clock and Per2 exhibited circadian expression in both gastrocnemius and liver tissues ( Figure 1A–F ). Peak expression of Per2 was reduced in the gastrocnemius ( Figure 1C ) of the young SHR. Peak Per2 expression was unaffected in the liver and ( Figure 1F ) heart (Figure S3C) of the young SHR.
Table 1. The circadian parameters of the core-clock genes Bmal1, Clock, and Per2 in the gastrocnemius and liver of young WKY and SHR calculated using JTK_CYCLE analysis.
| Circadian Statistics Core Clock Genes | ||||
| Strain | Gene | JTK _CYCLE p value | Circadian JTK_CYCLE | JTK_CYCLE Amplitude |
| Gastrocnemius | ||||
| WKY Young | Bmal1 | 1.21E-11 | Yes | 1.54 |
| SHR Young | Bmal1 | 5.73E-08 | Yes | 1.48 |
| WKY Young | Clock | 1.29E-05 | Yes | 0.39 |
| SHR Young | Clock | 3.74E-04 | Yes | 0.75 |
| WKY Young | Per2 | 3.13E-09 | Yes | 0.73 |
| SHR Young | Per2 | 6.20E-09 | Yes | 0.28 |
| Liver | ||||
| WKY Young | Bmal1 | 2.20E-09 | Yes | 1.45 |
| SHR Young | Bmal1 | 2.26E-06 | Yes | 1.49 |
| WKY Young | Clock | 2.43E-07 | Yes | 0.24 |
| SHR Young | Clock | 5.95E-03 | Yes | 0.48 |
| WKY Young | Per2 | 8.65E-09 | Yes | 0.2 |
| SHR Young | Per2 | 2.00E-05 | Yes | 0.11 |
Figure 1. The expression of (A,D) Bmal1, (B,E) Clock, and (C,F) Per2 from the gastrocnemius and the liver of young WKY and SHR was determined by quantitative PCR.
Samples were collected every 4 hours for 40 hours. Collections were performed under red light. The dark and light bars on the graph represent extrapolated subjective day and night as defined by ZT according to the prior L∶D cycle before release into DD. A plot of amplitude with 95% confidence intervals as error bars is include next to each circadian gene expression plot.
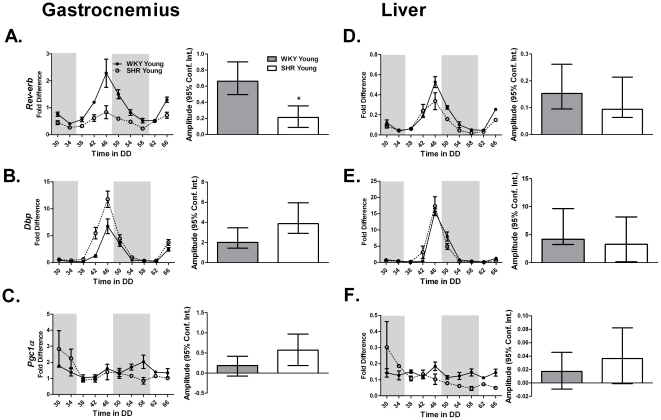
The core-clock genes Bmal1 and Clock heterodimerize and transcriptionally regulate a group of direct clock-controlled genes which are thought to be necessary for the maintenance of normal cell physiology. Rora and Rev-erbα are also considered components of the molecular clock. We examined the expression patterns of the Rev-erbα and downstream clock-controlled genes Dbp and Pgc1α. Rev-erbα expression was diminished in the heart and gastrocnemius with the most dramatic effect observed in the gastrocnemius ( Figure 2A and D ; Figure S4A). We found that Pgc1α did not cycle in either the gastrocnemius or the liver at this age ( Table 2 , Figure 2C and F ).
Figure 2. Rev-erbα is a core clock gene and Dbp, and Pgc1α are clock-controlled genes.
The expression of (A,D) Rev-erb, (B,E) Dbp, and (C,F) Pgc1α in the gastrocnemius and liver of young WKY and SHR was measured by quantitative PCR. Samples were collected every 4 hours for 40 hours. Collections were performed under red light. The dark and light bars on the graph represent extrapolated subjective day and night as defined by ZT according to the prior L∶D cycle before release into DD. A plot of amplitude with 95% confidence intervals as error bars is include next to each circadian gene expression plot.
Table 2. The circadian parameters of the core-clock gene Rev-erb and the clock-controlled genes Dbp and Pgc1α in the gastrocnemius and liver of young WKY and SHR calculated using JTK_CYCLE analysis.
| Circadian Statistics | ||||
| Strain | Gene | JTK _CYCLE p value | Circadian JTK_CYCLE | JTK_CYCLE Amplitude |
| Gastrocnemius | ||||
| WKY Young | Rev erb | 3.74E-11 | Yes | 0.66 |
| SHR Young | Rev erb | 1.55E-06 | Yes | 0.21 |
| WKY Young | Dbp | 2.28E-08 | Yes | 2.01 |
| SHR Young | Dbp | 2.70E-08 | Yes | 3.88 |
| WKY Young | Pgc1α | 7.40E-02 | No | 0.19 |
| SHR Young | Pgc1α | 1.24E-01 | No | 0.57 |
| Liver | ||||
| WKY Young | Rev erb | 3.74E-11 | Yes | 0.15 |
| SHR Young | Rev erb | 1.04E-07 | Yes | 0.09 |
| WKY Young | Dbp | 1.04E-07 | Yes | 4.19 |
| SHR Young | Dbp | 5.19E-06 | Yes | 3.28 |
| WKY Young | Pgc1α | 1 | No | 0.02 |
| SHR Young | Pgc1α | 1 | No | 0.04 |
Dysregulation of the molecular clock is increased in the 22 week old SHR
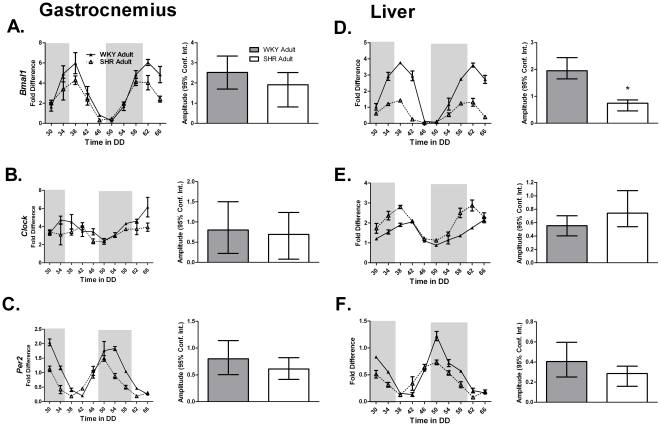
A circadian time-course collection was then performed with adult (22 week old) SHR and WKY rats to ascertain the expression of molecular clock factors associated with the progression of hypertension and insulin resistance. As seen with the young animals, JTK_CYCLE analysis confirmed core clock gene cycling in the gastrocnemius, liver, and heart of the WKY rats ( Table 3 ; Table S1). We found that Bmal1 expression was damped in the liver of the adult animals ( Figure 3D ). Clock expression also showed tissue specific changes in that it no longer cycled in the gastrocnemius ( Table 3 , Figure 3B ) but continued to do so in the liver ( Table 3 , Figure 3E ) and the heart of hypertensive SHR ( Table 1 , Figure S3E).
Table 3. The circadian parameters of the core-clock genes Bmal1, Clock, and Per2 in the gastrocnemius and liver of adult WKY and SHR calculated using JTK_CYCLE analysis.
| Circadian Statistics Core Clock Genes | ||||
| Strain | Gene | JTK _CYCLE p value | Circadian JTK_CYCLE | JTK_CYCLE Amplitude |
| Gastrocnemius | ||||
| WKY Adult | Bmal1 | 7.40E-10 | Yes | 2.52 |
| SHR Adult | Bmal1 | 1.20E-06 | Yes | 1.91 |
| WKY Adult | Clock | 1.68E-03 | Yes | 0.80 |
| SHR Adult | Clock | 7.82E-02 | No | 0.69 |
| WKY Adult | Per2 | 1.24E-13 | Yes | 0.80 |
| SHR Adult | Per2 | 3.92E-09 | Yes | 0.61 |
| Liver | ||||
| WKY Adult | Bmal1 | 9.87E-12 | Yes | 1.95 |
| SHR Adult | Bmal1 | 4.42E-09 | Yes | 0.75 |
| WKY Adult | Clock | 2.47E-05 | Yes | 0.55 |
| SHR Adult | Clock | 2.20E-09 | Yes | 0.74 |
| WKY Adult | Per2 | 3.13E-09 | Yes | 0.40 |
| SHR Adult | Per2 | 3.11E-08 | Yes | 0.29 |
Figure 3. The expression of (A,D) Bmal1, (B,E) Clock, and (C,F) Per2 from the gastrocnemius and the liver of adult WKY and SHR was determined by quantitative PCR.
Samples were collected every 4 hours for 40 hours. Collections were performed under red light. The dark and light bars on the graph represent extrapolated subjective day and night as defined by ZT according to the prior L∶D cycle before release into DD. A plot of amplitude with 95% confidence intervals as error bars is include next to each circadian gene expression plot.
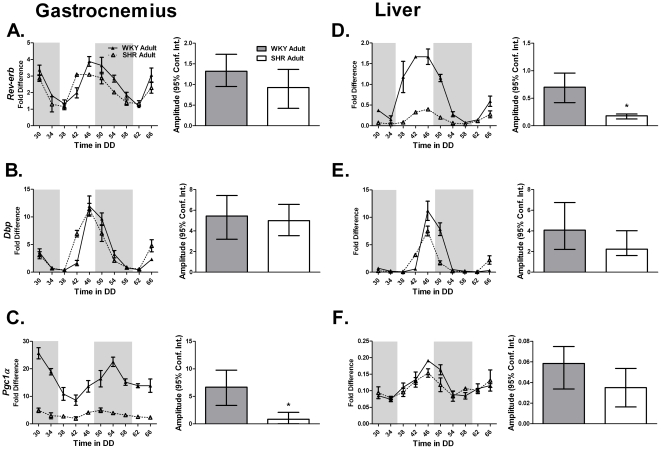
Rev-erbα is a negative transcriptional regulator of Bmal1 expression. Peak Rev-erbα expression was unchanged in skeletal muscle and heart ( Figure 4A , Table 4 ; Figure S4D, Table S2). Paradoxically, Rev-erbα expression was damped coincident with decreased Bmal1 expression in the liver of adult SHR ( Figure 4D ). However, expression of Pgc1α was damped in the gastrocnemius but not the liver of the 22 week old SHR throughout the circadian time-course ( Figure 4C and F ).
Figure 4. The expression of (A,D) Rev-erbα, (B) Dbp, and (C) Pgc1α in the gastrocnemius and liver of adult WKY and SHR was measured by quantitative PCR.
Samples were collected every 4 hours for 40 hours. Collections were performed under red light. The dark and light bars on the graph represent extrapolated subjective day and night as defined by ZT according to the prior L∶D cycle before release into DD. A plot of amplitude with 95% confidence intervals as error bars is include next to each circadian gene expression plot.
Table 4. The circadian parameters of the core-clock gene Rev-erb and the clock-controlled genes Dbp and Pgc1α in the gastrocnemius and liver of adult WKY and SHR calculated using JTK_CYCLE analysis.
| Circadian Statistics | ||||
| Strain | Gene | JTK _CYCLE p value | Circadian JTK_CYCLE | JTK_CYCLE Amplitude |
| Gastrocnemius | ||||
| WKY Adult | Rev erb | 2.66E-09 | Yes | 1.32 |
| SHR Adult | Rev erb | 5.87E-06 | Yes | 0.93 |
| WKY Adult | Dbp | 5.22E-13 | Yes | 5.45 |
| SHR Adult | Dbp | 6.20E-09 | Yes | 5.00 |
| WKY Adult | Pgc1α | 5.87E-06 | Yes | 6.67 |
| SHR Adult | Pgc1α | 3.76E-05 | Yes | 0.86 |
| Liver | ||||
| WKY Adult | Rev erb | 2.00E-05 | Yes | 0.70 |
| SHR Adult | Rev erb | 4.78E-12 | Yes | 0.18 |
| WKY Adult | Dbp | 5.07E-10 | Yes | 4.07 |
| SHR Adult | Dbp | 1.20E-06 | Yes | 2.24 |
| WKY Adult | Pgc1α | 5.19E-06 | Yes | 0.06 |
| SHR Adult | Pgc1α | 2.66E-03 | Yes | 0.04 |
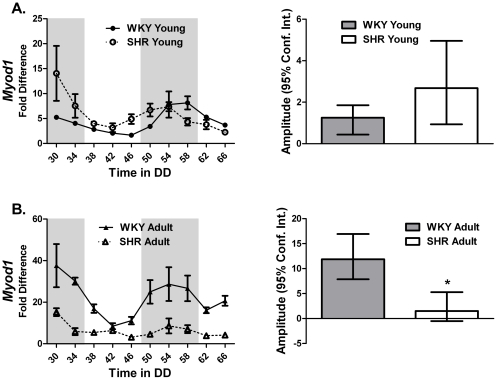
Myod1 is dysregulated in the muscle of 22 week old SHR
Myod1 is a well-described master regulator of the skeletal muscle transcriptional program [24]. JTK_CYCLE results confirm previously reported circadian cycling of Myod1 ( Table 5 ). Disruption of Myod1 expression was severe in the adult 22 week old SHR animals. We found that expression of Myod1 was no longer circadian and levels were damped throughout the circadian cycle ( Figure 5B ). Although little is known about the function of Myod1 in adult skeletal muscle, the observation that Myod1 oscillates is consistent with an expanded role in the daily maintenance of skeletal muscle tissue.
Table 5. The circadian parameters of the skeletal muscle specific clock-controlled gene Myod1 in the gastrocnemius of young and adult WKY and SHR calculated using JTK_CYCLE analysis.
| Circadian Statistics | ||||
| Strain | Gene | JTK _CYCLE p value | Circadian JTK_CYCLE | JTK_CYCLE Amplitude |
| Gastrocnemius | ||||
| WKY Young | Myod1 | 2.87E-04 | Yes | 1.25 |
| SHR Young | Myod1 | 2.54E-06 | Yes | 2.68 |
| WKY Adult | Myod1 | 8.79E-04 | Yes | 11.86 |
| SHR Adult | Myod1 | 5.65E-02 | No | 1.52 |
Figure 5. Myod1 is a skeletal muscle specific clock-controlled gene.
The expression of Myod1 in the gastrocnemius of (A) young and (B) adult WKY and SHR was determined by quantitative PCR. Samples were collected every 4 hours for 40 hours. Collections were performed under red light. The dark and light bars on the graph represent extrapolated subjective day and night as defined by ZT according to the prior L∶D cycle before release into DD. Note that Myod1 does not cycle in the gastrocnemius from adult SHR rats. A plot of amplitude with 95% confidence intervals as error bars is include next to each circadian gene expression plot.
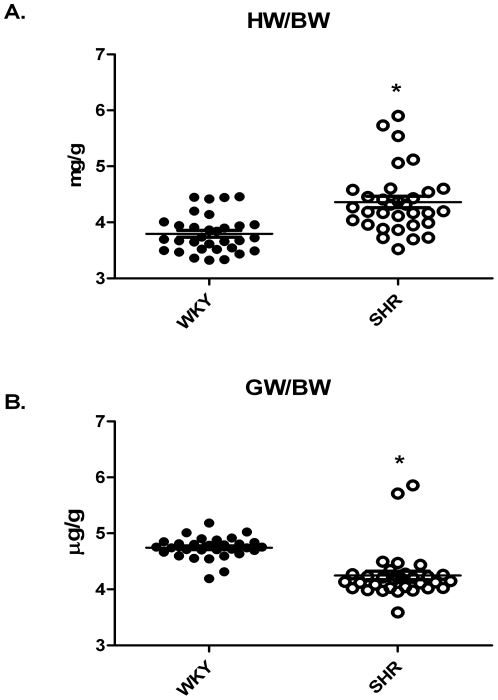
The presence of a hypertrophic heart was confirmed in that; heart weight normalized to body weight was significantly increased in the SHR ( Figure 6A ). Consistent with the observed loss of muscle mass in patients with cardiac hypertrophy/heart failure, the gastrocnemius muscle weight normalized to body weight of the SHR was significantly reduced compared to WKY ( Figure 6B ).
Figure 6. Heart weight normalized to body weight in the WKY and SHR demonstrating the presence of cardiac hypertrophy in the SHR (A).
Gastrocnemius wet weight normalized to body weight measured from the hypertensive WKY and SHR demonstrating a loss of muscle mass in the SHR (B).
Discussion
In this study, we examined a circadian time-course expression pattern of the molecular clock and clock-controlled genes in the gastrocnemius and liver in WKY and SHR rats at different disease stages to determine if polymorphisms upstream of the Bmal1 gene in the SHR were associated with effects on the molecular clock either before or after disease onset. Overall, we found that there were differences in the expression of the molecular clock factors both pre- and post-disease onset but the patterns in different tissues varied across ages. The observation that molecular clock factor expression was altered across all peripheral tissues studied (skeletal muscle, liver and heart) prior to disease onset suggests that an underlying problem with the molecular clock may contribute to the both the hypertension and metabolic disease in the SHR rat. When looking at clock gene expression after disease onset, we found more pronounced effects in skeletal muscle with a loss of Clock cycling and a dampening of the clock-controlled genes Pgc1α and Myod1. The damped expression of Myod1 and Pgc1α in the hypertensive and insulin resistant SHR was associated with a significant loss of muscle mass. Examination of single time point gene expression in tissues from heart failure SHR rats revealed significant down-regulation of several molecular clock genes in skeletal muscle as well as genes responsible for daily muscle maintenance and function. In summary, these findings confirm that molecular clock expression in the SHR is disrupted prior to the onset of cardiovascular and metabolic disease. The continued disruption in clock and clock-controlled genes in the skeletal muscle following disease onset suggests that the expression and/or stability of the molecular clock may contribute to the pathophysiology phenotype seen in this model.
Core-clock disruption precedes the development of hypertension
Our studies in the WKY and SHR demonstrate a pattern of core-clock disruption beginning in the pre-hypertensive SHR. Hypertension and metabolic disease are systemic diseases involving many organs beyond vasculature and heart. Subtle disruption of the molecular clock through changing peak amplitude (Per2, Rev-erbα) may be characteristic of an unstable clock mechanism in these tissues thus, making the tissues/organs more prone to disease triggers and increasing the probability of disease onset. Since the clock mechanism is not totally lost upon disease onset we favor a model in which there is a delicate time-dependent balance of expression of core-clock genes necessary to diminish the potential for developing pathology. An example of subtle changes in core-clock expression patterns contributing to pathology occurs in human breast cancer. Dysregulation or loss of PER1 and/or PER2 has been shown in human breast cancer tumor cells when compared with normal cells from adjacent tissue [25]. Lack of PER synchrony was even observed in the expression patterns of PER in separate cancer cell populations of the same cancer tissue. In addition, many clinical studies have associated circadian regulation/synchrony with cancer [25]–[27]. Studies in liver and adipose tissue of ob/ob mice have also shown that dysregulation of Per1, Per2 and Dbp occurs prior to the development of metabolic abnormalities suggesting a causal link [28]. These data under-score the need for future studies focusing on targeting the molecular clock to stabilize phase and amplitude and coordinate the circadian system.
Hypertension and insulin resistance is associated with pronounced changes of the core-clock in the gastrocnemius muscle
As the SHR progressed from pre-hypertensive to hypertensive and insulin resistant (22 Weeks, adult), the disruption of the molecular clock was more pronounced in the gastrocnemius compared to the liver. The tissue specificity of the clock disruption in skeletal muscle suggests 1) there is likely a gene – environment interaction leading to tissue specific disruption of the molecular clock; 2) clock gene disruption in skeletal muscle is associated with insulin resistance and 3) clock disruption of skeletal muscle likely contributes significantly to the whole body insulin resistance in this model. The SHR strain has been used extensively as a genetic model of essential hypertension. Skeletal muscle is a primary site of insulin resistance in essential hypertension [29], [30] It is interesting to note that SHR exhibit several skeletal muscle abnormalities and dysfunctions when compared to WKY counterparts including decreased fatigue resistance [31], insulin resistance [32], development of less contractile force [33], increased interstitial norepinephrine levels [34], altered sodium pump number and activity [35], elevated intracellular free calcium [36], fiber type transformation [37], and decreased capillary density [38]. Major alterations in skeletal muscle ultrastructure and biochemical properties have also been demonstrated in patients with chronic heart failure including fiber atrophy [17], transformation of fast-twitch type I to slow-twitch type II fibers [18], [39], decrease in oxidative enzyme capacity [19] and abnormal mitochondrial structure [20]–[22]. Recent studies in two clock-compromised mouse strains identified a common skeletal muscle pathology that was characterized by diminished force capacity and reduced mitochondrial content and function [5] both of which have been observed in SHR. Loss of Clock cycling in the hypertensive SHR provide insights that that modification of core-clock components may contribute to the functional and metabolic phenotype observed in SHR skeletal muscle. Metabolic dysfunction may also manifest through decreased expression of Rev-erb in the SHR gastrocnemius. The orphan nuclear receptors Rev-erb and Rora (RAR-related orphan receptor) link the feedback loops of the clock by repressing and activating Bmal1 gene transcription, respectively [40]–[43]. Rora and Rev-erb have also been shown to regulate lipid homeostasis in skeletal muscle [44], [45]. Changes in the expression of these genes could have a broad impact beyond circadian regulation, in that skeletal muscle relies heavily on fatty acids as a fuel source.
Circadian expression of Myod1 and Pgc1α is disrupted in the hypertensive gastrocnemius
Recent work from our lab has demonstrated that expression of Myod1 oscillates in a circadian pattern in skeletal muscle [10] and is a direct clock-controlled gene [5]. Cycling of Myod1, a direct target of the core-clock genes Bmal1 and Clock in skeletal muscle, ceased in the gastrocnemius muscle of the hypertensive and insulin resistant SHR. While speculative, the loss in gastrocnemius muscle weight in the hypertensive SHR may be connected to the disruption in Myod1 expression [46]. Another gene, Pgc1α, known to play a critical role in regulating the expression of metabolic and mitochondrial genes in skeletal muscle was damped in the gastrocnemius of the hypertensive rats throughout the circadian collection time-course. Data from Patti and colleagues is highly suggestive of a potential link between PGC1 expression and insulin resistance in diabetic and non-diabetic individuals with a family history of diabetes [47]. A reduction of the transcriptional co-activator Pgc1α in the gastrocnemius of the hypertensive SHR is indicative of a role for the molecular clock in the insulin resistance observed in these hypertensive animals. This work is in agreement with previous work from Andrews et al., (2010) demonstrating a down-regulation of both Pgc1α and Pgc1ß in mice with disruption of the core-clock genes ClockΔ19 or Bmal1− /−. These mice exhibit altered mitochondrial structure and function.
Multiple core-clock components are down regulated in skeletal muscle of heart failure SHR
Our data revealed a pattern of down-regulation of several core-clock genes, including Clock and Bmal1, and non-circadian skeletal muscle genes, such as Mef2 and Ttn, responsible for daily muscle maintenance in the gastrocnemius of the SHR during overt heart failure. Work from this lab recently reported that muscle from Bmal1−/− and ClockΔ19 mice exhibit disrupted myofilament architecture and exhibit reduced normalized maximal force [5] demonstrating a role for the molecular clock in maintaining structure and function at the cellular level in skeletal muscle. However, the skeletal muscle specific circadian gene Myod1 was unchanged in the heart failure animals (Figure S1A). This finding may more reflect a limitation of the experimental design rather than the biology. Given the high mortality of the heart failure animals, it was only possible to collect at a single time point. Tissue for heart failure experiments were collected during the daylight hours and this lack of change may be more a result of the time of day of collection. As seen in Figure 5B , Myod1 expression in the gastrocnemius from the WKY and hypertensive SHR shows that Myod1 expression levels were not different during the daylight hours. Our observation that Myod1 target genes Mck and Ttn, were down-regulated is suggestive that the loss of oscillation of Myod1 in the SHR is maintained in the heart failure rats.
Patterns of dysregulation in the molecular clock including diminished amplitude, phase shifts, and period changes have been observed in many disease states like diabetes, cardiovascular disease and cancer [4], [48]–[52]. Dyssynchrony can be the result of genetic alterations or changes in environmental cues. As a whole, our data demonstrate a pattern of molecular clock dysregulation that begins prior to the development of hypertension and insulin resistance and may be linked to the genetic background of the SHR progressing independent of the development of hypertension or worsening because of the development of hypertension. In association with previous studies, these data are highly suggestive of a role for the molecular clock in skeletal muscle pathology associated with hypertension/hypertrophy in SHR. Whether these genetic variables are identical or overlap with those in hypertensive patients is unknown. Future studies will need to examine other models of hypertension in order to better understand the mechanism behind hypertension, heart failure and circadian gene regulation in skeletal muscle.
Methods
Animals
All animal procedures were conducted in accordance with institutional guidelines for the care and use of laboratory animals as approved by the University of Kentucky Institutional Animal Care and Use Committees. This work was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Kentucky, protocol number 00890M2005. WKY and SHR rats were obtained from Harlan Laboratories. Rats were housed in a temperature- and humidity-controlled room maintained on a 12 h light–12 h dark cycle with food and water ad libitum.
Tissue Collections (80 wks old)
Due to the high mortality of the heart failure rats (80 wks old) tissue samples were collected at a single time point, Tissue samples from old rats (80 wks old) were collected at the same time of day, during 8AM and 9AM. This corresponds to time between ZT1 and ZT2.
Circadian Collections
All rats were maintained in the 12 h L/D cycles, then released into DD. Starting 30 h after entry into DD skeletal muscle, heart and liver from three WKY and three SHR rats were collected every 4 h for 40 h under dim red light (<5 lux). The muscles, liver and heart were removed from each rat and frozen in liquid nitrogen.
RNA isolation
Total RNA was prepared from frozen tissue samples using TRIzol (Invitrogen) according to the manufacturer's directions. RNA samples were treated with TURBO DNase (Ambion, Austin, TX) to remove genomic DNA contamination. Isolated RNA was quantified by spectrophotometry (λ = 260 nm). First-strand cDNA synthesis from total RNA was performed with a mixture of oligo(dT) primer and random hexamers using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). All isolated RNA and cDNA samples were stored at −80°C until further analysis. Real-time quantitative PCR using TaqMan (Applied Biosystems) assays was used to examine the gene expression of several core-clock, clock-controlled, and skeletal muscle specific genes. The gene expression assays were as follows: Bmal1 Rn00562847_m1*; Per2 Rn01427704_m1*; Clock Rn00573120_m1*; Rora Rn01173769_m1*; Rev- erb Rn00595671_m1*; Myod Rn00591291_m1*; Pgc1α Rn00580241_m1*; Rpl26 Rn02127510_s1*; Mck Rn01644605_m1*; Dbp Rn00497539_m1*. RPL26 was used as an internal calibration control [5], [10], [11], [53]. The ΔΔCT method was used for the quantification of real-time PCR data in the circadian collections. Gene expression in each sample was shown as the relative value compared to the WKY soleus sample No. 1 from DD 30 hours group. This enabled us to compare not only inter-species differences but also inter-organ differences in the expression pattern/amplitude of each gene.
Statistics
A new version of JTK_CYCLE was used to identify and characterize cycling variables in our datasets [23]. JTK_CYCLE analyses were performed using the R statistical package, version 2.12.1 to look specifically for 24 hour rhythms. Briefly, the original version of JTK_CYCLE estimated the amplitude of the most probable period/lag combination by calculating the median sign-adjusted deviation from the median over the first complete cycle. The new version-2 of JTK_CYCLE provides a more precise estimate of amplitude by replacing the median with the “pseudo median” (Hodges-Lehmann estimator) and additionally reports the associated confidence interval calculated by the Wilcox test function in the standard R stats package. A clear explanation of the mathematical basis for these new features is available online [54]. Differences in heart weight/body weight ratios and gastrocnemius wet weight/body weight were calculated with Graphpad Prism software using a two-tailed student t test.
Supporting Information
Bmal1 , Clock , Per2 and Rora are core-clock genes. Expression of (A) Bmal1, (B) Clock, (C) Per2, and (D) Rora from the gastrocnemius of WKY and SHR in heart failure (80 weeks) was determined by quantitative PCR. (*p<0.05).
(TIF)
The expression of the skeletal muscle genes (A) Myod1 , (B) Mck , (C) Mef2c , and (D) Ttn from the gastrocnemius of WKY and SHR in heart failure (80 weeks) was determined by quantitative PCR. (*p<0.05).
(TIF)
The expression of (A,D) Bmal1 , (B,E) Clock , and (C,F) Per2 from the heart of young and adult WKY and SHR was determined by quantitative PCR. Samples were collected every 4 hours for 40 hours. Collections were performed under total red light. The dark and light bars on the graph represent extrapolated subjective day and night as defined by ZT according to the prior L∶D cycle before release into DD. A plot of amplitude with 95% confidence intervals as error bars is include next to each circadian gene expression plot.
(TIF)
Rev-erbα is a core-clock gene and Dbp , and Pgc1α are clock-controlled genes. The expression of (A,D) Rev-erb, (B,E) Dbp, and (C,F) Pgc1α in the heart of young and adult WKY and SHR rats was measured by quantitative PCR. Samples were collected every 4 hours for 40 hours. Collections were performed under red light. The dark and light bars on the graph represent extrapolated subjective day and night as defined by ZT according to the prior L∶D cycle before release into DD. A plot of amplitude with 95% confidence intervals as error bars is include next to each circadian gene expression plot.
(TIF)
The circadian parameters of the core-clock genes Bmal1 , Clock , and Per2 in the heart of young and adult WKY and SHR calculated using JTK_CYCLE analysis.
(TIF)
The circadian parameters of the clock-controlled genes Rev-erb , Dbp , and Pgc1α in the heart of young and adult WKY and SHR calculated using JTK_CYCLE analysis.
(TIF)
Acknowledgments
We would like to thank John Hogenesch for kindly sharing and offering technical assistance in the use of the JTK_CYCLE software.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by R01AR055246 from NIAMS, RC1ES018636 from NIEHS, and a postdoctoral fellowship provided by the American Heart Association to Mitsunori Miyazaki (0825668D), http://www.heart.org/HEARTORG/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, et al. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci U S A. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H, Brewer JM, Champhekar A, Harris RBS, Bittman EL. Differential control of peripheral circadian rhythms by suprachiasmatic-dependent neural signals. Proc Natl Acad Sci U S A. 2005;102:3111–3116. doi: 10.1073/pnas.0409734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The Adaptive Value of Circadian Clocks: An Experimental Assessment in Cyanobacteria. Curr Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 4.Martino TA, Oudit GY, Herzenberg AM, Tata N, Koletar MM, et al. Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1675–R1683. doi: 10.1152/ajpregu.00829.2007. [DOI] [PubMed] [Google Scholar]
- 5.Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci U S A. 107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panda S, Antoch M, Miller B, Su A, Schook A, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 7.Weger BD, Sahinbas M, Otto GW, Mracek P, Armant O, et al. The Light Responsive Transcriptome of the Zebrafish: Function and Regulation. PLoS ONE. 2011;6(2):e17080. doi: 10.1371/journal.pone.0017080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, et al. Genome-Wide and Phase-Specific DNA-Binding Rhythms of BMAL1 Control Circadian Output Functions in Mouse Liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bur IM, Zouaoui S, Fontanaud P, Coutry N, Molino F, et al. The Comparison between Circadian Oscillators in Mouse Liver and Pituitary Gland Reveals Different Integration of Feeding and Light Schedules. PLoS ONE. 2010;5:e15316. doi: 10.1371/journal.pone.0015316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudic RD, Curtis AM, Cheng Y, FitzGerald G, Michael WY. Peripheral Clocks and the Regulation of Cardiovascular and Metabolic Function. Methods Enzymol. Academic Press; 2005. pp. 524–539. [DOI] [PubMed] [Google Scholar]
- 13.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, et al. Harmonics of Circadian Gene Transcription in Mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woon PY, Kaisaki PJ, Braganca J, Bihoreau MT, Levy JC, et al. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci U S A. 2007;104:14412–14417. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mettauer B, Zoll J, Garnier A, Ventura-Clapier R. Heart failure: a model of cardiac and skeletal muscle energetic failure. Pflügers Arch European. 2006;452:653–666. doi: 10.1007/s00424-006-0072-7. [DOI] [PubMed] [Google Scholar]
- 16.Lunde PK, Sjaastad I, Schiøtz Thorud HM, Sejersted OM. Skeletal muscle disorders in heart failure. Acta Physiol Scand. 2001;171:277–294. doi: 10.1046/j.1365-201x.2001.00830.x. [DOI] [PubMed] [Google Scholar]
- 17.Harrington D, Anker SD, Chua TP, Webb-Peploe KM, Ponikowski PP, et al. Skeletal Muscle Function and Its Relation to Exercise Tolerance in Chronic Heart Failure. J Am Coll Cardiol. 1997;30:1758–1764. doi: 10.1016/s0735-1097(97)00381-1. [DOI] [PubMed] [Google Scholar]
- 18.Lipkin DP, Jones DA, Round JM, Poole-Wilson PA. Abnormalities of Skeletal Muscle in Patients with Chronic Heart Failure. Int J Cardiol. 1988;20:161. doi: 10.1016/0167-5273(88)90164-7. [DOI] [PubMed] [Google Scholar]
- 19.Harridge SD, Magnusson G, Gordon A. Skeletal muscle contractile characteristics and fatigue resistance in patients with chronic heart failure. Eur Heart J. 1996;17:896–901. doi: 10.1093/oxfordjournals.eurheartj.a014971. [DOI] [PubMed] [Google Scholar]
- 20.Drexler H, Riede U, Munzel T, Konig H, Funke E, et al. Alterations of skeletal muscle in chronic heart failure. Circ. 1992;85:1751–1759. doi: 10.1161/01.cir.85.5.1751. [DOI] [PubMed] [Google Scholar]
- 21.Lampert E, Mettauer B, Hoppeler H, Charloux A, Charpentier A, et al. Structure of Skeletal Muscle in Heart Transplant Recipients. J Amer Coll Cardiol. 1996;28:980–984. doi: 10.1016/s0735-1097(96)00272-0. [DOI] [PubMed] [Google Scholar]
- 22.Hambrecht R, Fiehn E, Yu J, Niebauer J, Weigl C, et al. Effects of Endurance Training on Mitochondrial Ultrastructure and Fiber Type Distribution in Skeletal Muscle of Patients With Stable Chronic Heart Failure. J Amer Coll Cardiol. 1997;29:1067–1073. doi: 10.1016/s0735-1097(97)00015-6. [DOI] [PubMed] [Google Scholar]
- 23.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An Efficient Nonparametric Algorithm for Detecting Rhythmic Components in Genome-Scale Data Sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Devel. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 25.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, et al. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 26.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, et al. The Circadian Gene Per1 Plays an Important Role in Cell Growth and DNA Damage Control in Human Cancer Cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 27.Pogue-Geile KL, Lyons-Weiler J, Whitcomb DC. Molecular overlap of fly circadian rhythms and human pancreatic cancer. Cancer Letters. 2006;243:55–57. doi: 10.1016/j.canlet.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 28.Ando H, Kumazaki M, Motosugi Y, Ushijima K, Maekawa T, et al. Impairment of Peripheral Circadian Clocks Precedes Metabolic Abnormalities in ob/ob Mice. Endocrinology. 2011;152:1347–1354. doi: 10.1210/en.2010-1068. [DOI] [PubMed] [Google Scholar]
- 29.Capaldo B, Lembo G, Napoli R, Rendina V, Albano G, et al. Skeletal muscle is a primary site of insulin resistance in essential hypertension. Metabolism. 1991;40:1320–1322. doi: 10.1016/0026-0495(91)90036-v. [DOI] [PubMed] [Google Scholar]
- 30.Wolfe RR. The underappreciated role of muscle in health and disease. The American Journal of Clinical Nutrition. 2006;84:475–482. doi: 10.1093/ajcn/84.3.475. DOI Electronic Resource Number. [DOI] [PubMed] [Google Scholar]
- 31.Gray SD, Carlsen RC, Deng J. Soleus muscle contractile properties in hypertensive rats. Amer J Physiol - Regulatory, Integrative and Comparative Physiology. 1994;267:R735–R739. doi: 10.1152/ajpregu.1994.267.3.R735. [DOI] [PubMed] [Google Scholar]
- 32.Hulman S, Falkner B, Freyvogel N. Insulin resistance in the conscious spontaneously hypertensive rat: Euglycemic hyperinsulinemic clamp study. Metabolism. 1993;42:14–18. doi: 10.1016/0026-0495(93)90165-k. [DOI] [PubMed] [Google Scholar]
- 33.Atrakchi A, Gray SD, Carlsen RC. Development of soleus muscles in SHR: relationship of muscle deficits to rise in blood pressure. Amer J Physiol - Cell Physiol. 1994;267:C827–C835. doi: 10.1152/ajpcell.1994.267.3.C827. [DOI] [PubMed] [Google Scholar]
- 34.Cabassi A, Vinci S, Quartieri F, Moschini L, Borghetti A. Norepinephrine Reuptake Is Impaired in Skeletal Muscle of Hypertensive Rats In Vivo. Hypertension. 2001;37:698–702. doi: 10.1161/01.hyp.37.2.698. [DOI] [PubMed] [Google Scholar]
- 35.Pickar JG, Carlsen RC, Atrakchi A, Gray SD. Increased Na(+)-K+ pump number and decreased pump activity in soleus muscles in SHR. Amer J Physiol - Cell Physiol. 1994;267:C836–C844. doi: 10.1152/ajpcell.1994.267.3.C836. [DOI] [PubMed] [Google Scholar]
- 36.Ameen M, Davies JE, Ng LL. A Comparison of Free Intracellular Calcium and Magnesium Levels in the Vascular Smooth Muscle and Striated Muscle Cells of the Spontaneously Hypertensive and Wistar Kyoto Normotensive Rat. Ann N Y Acad Sci. 1991;639:550–553. doi: 10.1111/j.1749-6632.1991.tb17348.x. [DOI] [PubMed] [Google Scholar]
- 37.Bortolotto SK, Stephenson DG, Stephenson GM. Fiber type populations and Ca2+-activation properties of single fibers in soleus muscles from SHR and WKY rats. Amer J Physiol - Cell Physiol. 1999;276:C628–C637. doi: 10.1152/ajpcell.1999.276.3.C628. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi N, DeLano FA, Schmid-Schonbein GW. Oxidative Stress Promotes Endothelial Cell Apoptosis and Loss of Microvessels in the Spontaneously Hypertensive Rats. Arterioscler Thromb Vasc Biol. 2005;25:2114–2121. doi: 10.1161/01.ATV.0000178993.13222.f2. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan MJ, Duscha BD, Klitgaard H, Kraus WE, Cobb FR, et al. Altered expression of myosin heavy chain in human skeletal muscle in chronic heart failure. Med Sci Sports Exerc. 1997;29:860–866. doi: 10.1097/00005768-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Lowrey PL, Takahashi JS. Mammalian Circadian Biology: Elucidating Genome-Wide Levels of Temporal Organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akashi M, Takumi T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 42.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, et al. A Functional Genomics Strategy Reveals Rora as a Component of the Mammalian Circadian Clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 44.Lau P, Nixon SJ, Parton RG, Muscat GE. RORα Regulates the Expression of Genes Involved in Lipid Homeostasis in Skeletal Muscle Cells. J Biol Chem. 2004;279:36828–36840. doi: 10.1074/jbc.M404927200. [DOI] [PubMed] [Google Scholar]
- 45.Ramakrishnan SN, Lau P, Burke LJ, Muscat GE. Rev-erbα Regulates the Expression of Genes Involved in Lipid Absorption in Skeletal Muscle Cells. J Biol Chem. 2005;280:8651–8659. doi: 10.1074/jbc.M413949200. [DOI] [PubMed] [Google Scholar]
- 46.Vissing K, Andersen JL, Harridge SD, Sandri C, Hartkopp A, et al. Gene expression of myogenic factors and phenotype-specific markers in electrically stimulated muscle of paraplegics. J Appl Physiol. 2005;99:164–172. doi: 10.1152/japplphysiol.01172.2004. [DOI] [PubMed] [Google Scholar]
- 47.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martino TA, Tata N, Belsham DD, Chalmers J, Straume M, et al. Disturbed Diurnal Rhythm Alters Gene Expression and Exacerbates Cardiovascular Disease With Rescue by Resynchronization. Hypertension. 2007;49:1104–1113. doi: 10.1161/HYPERTENSIONAHA.106.083568. [DOI] [PubMed] [Google Scholar]
- 49.Maywood ES, O'Neill J, Wong GK, Reddy AB, Hastings MH, et al. Circadian timing in health and disease Prog Brain Res. Elsevier; 2006. pp. 253–269. [DOI] [PubMed] [Google Scholar]
- 50.Filipski E, Delaunay F, King VM, Wu MW, Claustrat B, et al. Effects of Chronic Jet Lag on Tumor Progression in Mice. Cancer Res. 2004;64:7879–7885. doi: 10.1158/0008-5472.CAN-04-0674. [DOI] [PubMed] [Google Scholar]
- 51.Yasuniwa Y, Izumi H, Wang KY, Shimajiri S, Sasaguri Y, et al. Circadian Disruption Accelerates Tumor Growth and Angio/Stromagenesis through a Wnt Signaling Pathway. PLoS ONE. 2010;5:e15330. doi: 10.1371/journal.pone.0015330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durgan DJ, Young ME. The Cardiomyocyte Circadian Clock: Emerging Roles in Health and Disease. Circ Res. 2010;106:647–658. doi: 10.1161/CIRCRESAHA.109.209957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorrez L, Van Deun K, Tranchevent LC, Van Lommel L, Engelen K, et al. Using Ribosomal Protein Genes as Reference: A Tale of Caution. PLoS ONE. 2008;3:e1854. doi: 10.1371/journal.pone.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geyer CJ. Stat 5102 Notes: Nonparametric Tests and Confidence Intervals. 4/13/2003. from, http://www.stat.umn.edu/geyer/old03/5102/notes/rank.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bmal1 , Clock , Per2 and Rora are core-clock genes. Expression of (A) Bmal1, (B) Clock, (C) Per2, and (D) Rora from the gastrocnemius of WKY and SHR in heart failure (80 weeks) was determined by quantitative PCR. (*p<0.05).
(TIF)
The expression of the skeletal muscle genes (A) Myod1 , (B) Mck , (C) Mef2c , and (D) Ttn from the gastrocnemius of WKY and SHR in heart failure (80 weeks) was determined by quantitative PCR. (*p<0.05).
(TIF)
The expression of (A,D) Bmal1 , (B,E) Clock , and (C,F) Per2 from the heart of young and adult WKY and SHR was determined by quantitative PCR. Samples were collected every 4 hours for 40 hours. Collections were performed under total red light. The dark and light bars on the graph represent extrapolated subjective day and night as defined by ZT according to the prior L∶D cycle before release into DD. A plot of amplitude with 95% confidence intervals as error bars is include next to each circadian gene expression plot.
(TIF)
Rev-erbα is a core-clock gene and Dbp , and Pgc1α are clock-controlled genes. The expression of (A,D) Rev-erb, (B,E) Dbp, and (C,F) Pgc1α in the heart of young and adult WKY and SHR rats was measured by quantitative PCR. Samples were collected every 4 hours for 40 hours. Collections were performed under red light. The dark and light bars on the graph represent extrapolated subjective day and night as defined by ZT according to the prior L∶D cycle before release into DD. A plot of amplitude with 95% confidence intervals as error bars is include next to each circadian gene expression plot.
(TIF)
The circadian parameters of the core-clock genes Bmal1 , Clock , and Per2 in the heart of young and adult WKY and SHR calculated using JTK_CYCLE analysis.
(TIF)
The circadian parameters of the clock-controlled genes Rev-erb , Dbp , and Pgc1α in the heart of young and adult WKY and SHR calculated using JTK_CYCLE analysis.
(TIF)