Abstract
Cell shape changes within epithelia require the regulation of adhesive molecules that maintain tissue integrity. How remodelling of cell contacts is achieved while tissue integrity is maintained remains a fundamental question in morphogenesis. Dorsal Closure is a good system to study the dynamics of DE-Cadherin during morphogenesis. It relies on concerted cell shape changes of two epithelial sheets: amnioserosa cell contraction and epidermal cell elongation. To investigate the modulation of DE-Cadherin we performed antibody uptake experiments in live embryos during Dorsal Closure. We found that some antibodies access certain epitopes of the extracellular domain of native DE-Cadherin only in the amnioserosa and epidermal cells attached to the amnioserosa, which has never been observed in fixed DE-Cadherin in Drosophila embryos. These differences correlate with the different cell behaviour of these regions and therefore we suggest that DE-Cadherin exists in different forms that confer different adhesive strengths. We propose this to be a widespread mechanism for the differential modulation of adhesion during morphogenesis.
Introduction
The Cadherin protein family is a group of calcium dependent homophilic cell adhesion molecules that mediate adhesion between cells [1]. The signature of this protein family is an extracellular domain composed of “cadherin domains” that promote intercellular interactions, and an intracellular domain that serves as a link between the intercellular adhesion and the actin cytoskeleton through interactions with the catenins [1]. In epithelia, Cadherins localise at the Adherens Junctions (AJs) near the apical side of the cell and generate a continuum between the actin cytoskeleton of different cells allowing coordinated tissue deformation [2], [3], [4]. Although the dynamics of cytoskeletal activity during morphogenesis is being extensively studied [5], less is known about how adhesion is modulated during these processes. Biophysical models of morphogenetic processes predict that changes in adhesion are important in the modulation of the mechanical properties of epithelia [6]. This could be achieved by modulating the total amount of Cadherin, through the regulation of its expression [7], [8], [9], [10], or its steady-state levels at the membrane, through endocytosis and recycling [11], [12]. A third mechanism could target the adhesive properties of Cadherin, regulating its conformation, clustering state and other higher-order organizations [1].
Assessment of Cadherin adhesive properties in vivo during morphogenesis is difficult since genetic removal of Cadherin has a dramatic effect on tissue integrity. Dorsal Closure (DC) in Drosophila represents a good model to address DE-Cadherin modulation in vivo. DC is a process whereby two epithelia, the epidermis and the amnioserosa (AS), interact to cover a discontinuity on the dorsal epidermis of the Drosophila embryo [13], [14]. It is associated with cell shape changes and local cell interactions as generators of dynamical force fields that drive a patterned contraction of the AS and a correlated epidermis elongation [15], [16], [17]. Drosophila E-Cadherin, DE-Cadherin, encoded by the shotgun (shg) gene, provides an essential cell adhesion force, balancing the stresses generated during DC [18]. Removal of DE-Cadherin, both maternally and zygotically, results in the loss of epithelial integrity early in embryogenesis [10], [19]. However zygotic null mutants for shg receive maternal DE-Cadherin that allows the embryos to initiate DC with reduced levels of DE-Cadherin levels [18]. Interestingly, embryos mutant for null alleles of shg are rescued by ubi-DE-CadherinGFP expression and develop into normal adult flies [20] suggesting that any modulation of Cadherin activity during development might occur at the post-transcriptional level.
Here we investigate post-transcriptional modulations in DE-Cadherin during a morphogenetic process. Our study reveals surprising spatial differences in the configuration of the extracellular domain of DE-Cadherin which correlate with patterned cell shape changes during DC. We propose that these differences represent Cadherins with different adhesive properties.
Materials and Methods
Drosophila strains
Wild-type embryos were from the Oregon R strain, w ; shgR64/CyO strains (Tepass et al., 1996), ubi-DE–CadherinGFP [20], ShgR64 homozygous mutant embryos were selected from a cross between w; shgR64, UASactinGFP/CyO and w; shgR64, enGal4/CyO (N. Gorfinkiel).
In vivo hand-devitellinization
Our hand-devitillinization protocol follows published reports [21]. Embryos at DC stage were selected and aligned with the ventral region upward and anterior part towards the observer on top of a narrow stripe of double-sided tape. Sörensen phosphate buffer (SPB) was added to cover the aligned embryos. The vitelline membrane was pierced at the head with a glass needle that was moved to the posterior of the embryo; the movement is done without indenting deep in the embryo. The embryo was teased out of the vitelline membrane, away from the tape.
Antibody uptake assays
Hand-devitellinized embryos were transferred with a coated glass pipette into a coated glass dish with SPB at 4°C, then to another glass dish with 500 µl of cold SPB containing primary antibodies and incubated for 1 hour at 4°C, rinsed 3 times and finally washed 6 times for 2 minutes with SPB at 4°C. The embryos were either immediately fixed (time 0) or chased for 10, 30 minutes or 1 hour in Schneider's insect medium supplemented with 10% Fetal Calf Serum (FCS) and 1% L-Glutamine at 25°C. Fixation was performed in paraformaldehyde (PFA) 4% for 40 minutes at 25°C, wash-blocked (3 rinses plus four 10 minutes incubations) in BBT-BSA (BBS + CaCl2 1 mM + 0,1% Triton + 0,5% BSA). For further antibody labelling, embryos were incubated with other primary antibodies diluted in BBT-BSA for 2 hours at Room Temperature (RT), and thoroughly washed with BBT-BSA. Finally, embryos were incubated with 500 µl of BBT-BSA containing secondary antibodies at RT for 2 hours in the dark, rinsed 3 times and washed 4 times in BBT-BSA and then individually mounted in Vectashield.
The pulse-chases were done simultaneously, with ±6 embryos for each time point. The experiment was repeated 3 times. Thereafter all the experiments with different antibodies or mutant embryos were done using the same protocol without chase, always using DCAD2 as a control.
Antibodies
The following primary antibodies were used: rat anti-DE-Cadherin DCAD1 (T. Uemura) 1∶100, rat anti-DE-Cadherin DCAD2 (DSHB) 1∶200, rabbit anti-DE-Cadherin d-300 (Santa Cruz) 1∶100, goat anti-DE-Cadherin-intra dP-20 (Santa Cruz) 1∶200, rat anti-DE-Cadherin (V. Hartenstein) 1∶50, mouse anti-Notch-extra C458.24 (DSHB) 1∶50, rabbit anti-Scribble (C. Doe) 1∶1000, 1∶10. Secondary antibodies were from Molecular Probes.
Immunostainings
Embryos were fixed and stained as previously described (Kaltschmidt et al., 2002). Fluorescently labelled embryos were mounted in Vectashield (Vector) and examined under a Nikon D-Eclipse C1 confocal scanning unit, mounted on a Nikon Eclipse 90i microscope, using the EZ-C1 3.60 software and a 60x/1.40 NA Apo VC oil-immersion objective. Five-seven z-sections, 0.5 µm apart, were projected using ImageJ (http://rsb.info.nih.gov/ij/) and processed using Photoshop.
Quantification of fluorescence intensities
For quantification of fluorescence intensity, the polygon selection tool was used to draw around an object and the mean gray value was obtained using ImageJ. Notch and DCAD2 fluorescence intensity from different time points were compared using One-Way ANOVA and Tukey HSD Test for Post-ANOVA Pair-Wise Comparisons.
Time-lapse movies
Stage 13 Drosophila embryos carrying an ubi-DE-CadherinGFP construct (Oda and Tsukita, 2001), were dechorionated, mounted on coverslips with the dorsal side glued to the glass and covered with Voltalef oil 10S (Attachem). Imaging of the embryos was done using an inverted LSM 510 Meta laser-scanning microscope with a Plan-Apochromat 63x/1.4 oil-immersion objective. Embryos were maintained at 24°C during imaging and around 50 z-sections 1 µm apart were collected every 2 minutes. For Figure 1F, Ubi-DE-CadherinGFP Drosophila embryos at stage 13 were dechorionated, devitellinized and transferred into small dips in agar glass dishes with insect medium and imaged every 2 minutes, collecting around 50 z-sections 1 µm apart under a Nikon D-Eclipse C1 confocal scanning unit, mounted on a Nikon Eclipse 90i microscope, using the EZ-C1 3.60 software and a Nikon Fluor 40x10.80 water-immersion objective. Movies were assembled and processed ImageJ (http://rsb.info.nih.gov/ij/).
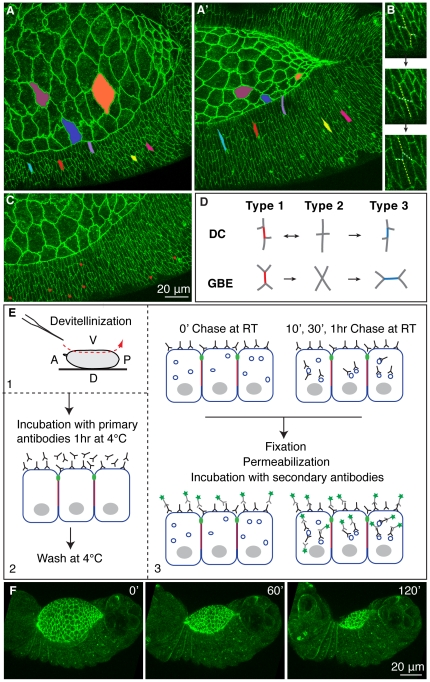
Figure 1. Cell shape changes and exchange of cell neighbours in DC.
(A–C) Stills from a time-lapse of an Ubi-DE-CadherinGFP embryo during DC. Colours identify cells that are followed during the process, highlighting cell shape changes (A,A′). Exchanges between neighbours (B) in the lateral epidermis (C). The number of cell neighbour exchanges is 6.4 ± 1.2 per segment (first 6 epidermal rows in 5 segments in 3 embryos over 90 minutes). (D) Pattern of neighbour exchange during DC and GBE. (E) Overview of the pulse-chase assay (antibodies appear outside the vesicles for simplicity). (F) Stills from a time-lapse of a hand-devitellinized Ubi-DE-CadherinGFP embryo (Movie S1).
Results and Discussion
During DC, contraction of the AS cells provides a tensile force that drives and maintains elongation of the epidermal epithelium [13], [15], [16], [22] (Figure 1A,A′). During epidermal elongation the average ratio between the Dorsal/Ventral (D/V) and Anterior/Posterior (A/P) cell axis changes from 1.5 to 5.2 (Young et al., 1993). There are two well defined domains in the epidermis: the Dorsal Most Epidermal (DME) cells, which form an interface between the AS and the epidermal sheet and bear the brunt to the forces of the process, and the lateral epidermis, where the elongation is associated with local cell rearrangements. Analysis of these rearrangements in the first 6 epidermal rows of 5 segments in 3 embryos over the period of 90 minutes reveals stereotyped exchange of neighbours (Figure 1B,C and Movie S1) and a number of 6.4 ± 1.2 cell neighbour exchanges per segment. During Germ Band Elongation (GBE) [23], [24] cells also undergo a sequence of cell contact changes termed T1>T2>T3 transition (Figure 1D,[23]). However, while in GBE the process is continuous and irreversible, T1>T2 type transitions are frequently maintained or reversed during DC, not leading to cell intercalation (Figure 1D). This difference might result from the fact that in GBE cell contact changes underlie tissue elongation that is driven by local forces [25] whereas in DC there is an external pulling force generated by AS contraction that drives epidermal cell elongation [15], [16], [17] and these transitions accommodate stresses associated with cell elongation in relation to neighbours. A similar conversion of junctions has been observed in the ventral epidermis of earlier Drosophila embryos during epidermal cell alignment along the D/V boundary [26] suggesting that they represent a theme in morphogenesis [24].
Cell shape changes and exchange of neighbours require modulation of the cell surface molecules, in particular of DE-Cadherin [27], [28]. As there are no reports of differential expression of DE-Cadherin in the epidermis during DC, we looked for dynamic changes in the cell surface pool of DE-Cadherin by labelling and chasing this pool of DE-Cadherin. We adapted an existing culture technique that retrieves the embryo from the vitelline membrane [21], allows progression of DC (Figure 1F) and makes the cells competent to take up dyes and antibodies (Figure 1E 2-E3). To test the assay, we pulse labelled embryos with antibodies against the extracellular domain of DE-Cadherin, and observed a translocation of the antibody into intracellular vesicles. In contrast, antibodies against the intracellular domain of the transmembrane receptor Notch or the intracellular protein Scribble did not show any signal (Figure S1A′,A″,B′,B″) which was only detected when cells were permeabilized before incubation (Figure S1C′,C″). These experiments validate this protocol for dynamic analysis of cell surface proteins.
Dynamic assays reveal that monoclonal antibody binding to native DE-Cadherin is patterned
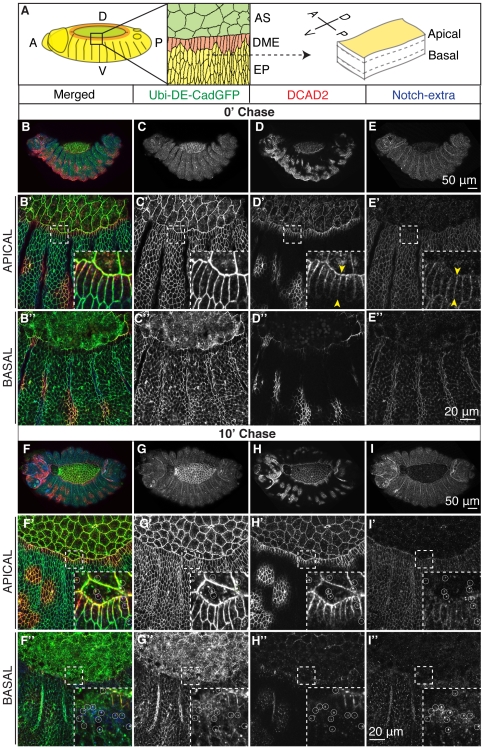
To study the dynamics of DE-Cadherin during DC, stage 13–14 Ubi-DE-CadherinGFP embryos were incubated at 4°C with the monoclonal antibody DCAD2, directed against the extracellular domain of DE-Cadherin, and with an antibody against the extracellular domain of Notch as a control, and chased for different time periods (0, 10, 30 minutes and 1 hour) at RT.
Embryos that have been fixed immediately after antibody loading, express DE-CadherinGFP in all cells (Figure 2C,C′,C″), however the binding of the DCAD2 antibody to the epidermal cells reveals a spatial pattern: the AS and the DME exhibit clear antibody binding which is absent in the lateral epidermal cells, except in small patches (Figure 2D,D′,D″). We have not investigated the nature of these patches but they could result e.g. from local rearrangements occurring underneath the epidermis that impinge into the epidermis tension and adhesion systems. The binding of DCAD2 to the AS and DME cells mainly, could be due to an inability of the antibodies to access epitopes on the surface of lateral epidermal cells. However, under the same experimental conditions, antibodies against the extracellular domain of Notch (Figure 2E, E′,E″), bind homogeneously to all the epidermal cells. Interestingly, in the DME cells, the borders that face lateral cells have less DCAD2 labelling than the others, and we observe a gradient of label along the D/V border (Figure 2D′ inset yellow arrows). In contrast, Notch antibodies bound homogeneously along the borders of all epidermal cells (Fig 2E′ inset yellow arrows). After 10 minutes of chase the pattern of DCAD2 binding was preserved and vesicles positive for DCAD2 and DE-CadherinGFP could be detected inside DCAD2 labelled cells (Figure 2G,G′,H,H′), which indicates that the pool of DE-Cadherin recognized by DCAD2 is dynamic. Vesicles containing Notch were observed in all epidermal cells at the same chase time, showing that both proteins are endocytosed in less then 10 minutes (Figure 2I,I″). Nevertheless, after 30 minutes of chase at room temperature, differences in the localization at the membrane of both proteins were accentuated. Even though both proteins seem to be equally abundant inside the cell, the majority of Notch protein detected by the antibody is cleared from the membrane over time, which contrasts with DE-Cadherin detected by the antibody, which stays at the membrane even with longer chases, indicating that both proteins have different turnovers at the cell surface (Figure S2C′,C″,D′,D″,I). These differences probably reflect their different functions. DE-Cadherin mediates cell-cell adhesion and Notch is a signalling molecule which undergoes endocytosis as part of its signalling activity [30], [31]. In agreement with this, after 1 hour chase, labelled Notch was undetectable (Figure S2H′,H″) but DCAD2 antibody was still detected at the membrane or in large vesicles, especially at the cell basal region (Figure S2G′,G″).
Figure 2. In vivo antibody binding to native DE-Cadherin reveals patterned access to DE-Cadherin in the cell surface.
(A) Cartoon depicting the two tissues analyzed in the assays, the AS (green) and the epidermis (EP), which comprises the DME (orange), and the lateral epidermis (yellow). (B–I) Pulse-chase assay in Ubi-DE-Cadherin-GFP embryos using DCAD2 and Notch-extra antibodies. Yellow arrows highlight the binding of DCAD2 and Notch-extra antibody along the D/V contact of DME cells.
It has been reported that crosslinking of cell surface proteins by antibodies might trigger endocytosis [29]. While this remains a possibility, pulse-chase assays performed with Dextran revealed that DE-Cadherin is endocytosed in the epidermis and AS with Dextran (not shown), suggesting that an important fraction of what we observe is related to DE-Cadherin endocytosis.
It is noteworthy that the differential binding of DCAD2 to epidermal cells was also observed in wild-type embryos (Figure S3E-G′) and thus it is not a consequence of the expression of DE-CadherinGFP with the wild-type protein. The pattern has been seen in wild-type embryos in 76.4% ± 16.3 of the embryos at 0′ chase (n = 67 embryos from 5 independent experiments) and in ubi-DE-CadherinGFP embryos 73.2% ± 19.9 (n = 56 embryos from 8 independent experiments). The dynamics of this pattern could not be investigated in later embryos because cuticle secretion, which begins at stage 15, interferes with antibody binding.
Antibodies against different epitopes of DE-Cadherin bind differently along the epidermis
In contrast with the standard immunostaining protocols, which result in a homogeneous binding of DCAD2 to the epidermis, we incubate the embryos with the antibodies before fixation, which results in a patterned DCAD2 labelling of the epidermis. In fact, fixation after hand-devitellinization but before antibody incubation also disrupts the pattern (Figure 3A,A′). Formaldehyde, which was used to fix the embryos, crosslinks proteins during the process of fixation, that can lead to artefacts such as chemical modification of proteins, which then can affect the interaction of the antibody with the antigen [32]. Therefore, the pattern observed with DCAD2 could be associated with a particular epitope or form of DE-Cadherin that is disrupted upon fixation.
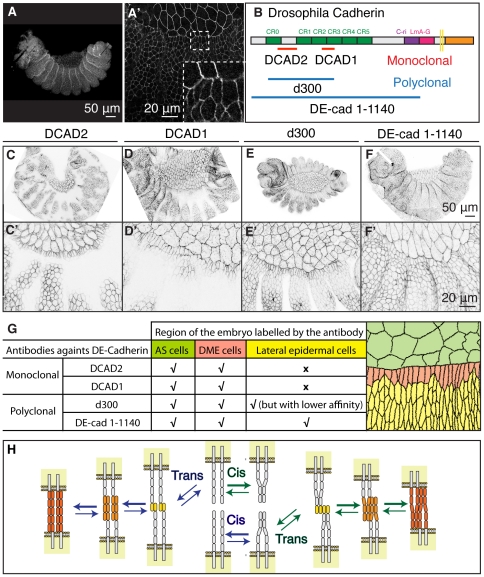
Figure 3. Antibodies against different epitopes of DE-Cadherin bind differently along the epidermis.
(A,A′) Hand-devitellinized embryos fixed and then incubated with DCAD2 at 0′ chase. (B) Structure of Drosophila E-Cadherin depicting the epitopes recognized by the antibodies used in this study [41], [42], [43]. (C–F) Confocal z-projections of Ubi-DE-CadherinGFP embryos pulsed with DCAD2, DCAD1, d300 and DE-cad 1-1140 antibodies, fixed without chase. (G) Summary of anti-DE-Cadherin antibodies binding patterns in AS and epidermal cells. (H) Current models for interactions between Cadherins (C, adapted from Leckband and Prakasam, 2006).
To investigate whether there are differences in the extracellular domain of native DE-Cadherin along the epidermis we used the same assay with antibodies against different regions of the DE-Cadherin extracellular domain (Figure 3B). Another monoclonal antibody, DCAD1, bound to the AS, DME cells and more ventral epidermal cells (Figure 3C,C′). The DCAD1 antibodies also recognized patches of the lateral epidermis. The polyclonal antibody d300 bound to all the epidermal cells, but with different affinities across the epidermis (Figure 3E,E′); the lateral epidermis, which was not labelled with DCAD2, was weakly labelled by d300. On the other extreme, a polyclonal antibody generated against the entire extracellular domain of DE-Cadherin, DE-cad 1–1140, labelled all epidermal cells homogeneously, though with a lower affinity when compared with the other antibodies against DE-Cadherin (Figure 3F,F′).
These results indicate the existence of a spatial pattern of accessibility to the extracellular domain of DE-Cadherin in the epidermis at stages 13–15 (Figure 3G). These differences could result from different DE-Cadherin homophilic binding states, from different organization of DE-Cadherin at the cell surface or binding to other proteins at the cell surface.
We do not present direct data showing that these antibodies recognize different DE-Cadherin forms, nevertheless there are examples of antibodies that bind to different conformations of molecules involved in adhesion [33], [34], [35]. Furthermore, there is evidence for different configurations of Cadherin at the cell surface (Figure 3H) [1], [36], [37]. During the establishment of Cadherin mediated adhesion, dimerization of Cadherin molecules precedes trans interactions, but the lateral dimers can dissociate to form adhesive trans-homophilic bonds or remain dimerized in the trans interactions. Initially, N-terminal domain interactions are thought to have an important role in adhesion and binding selectivity in the trans interactions. However, these adhesive complexes can progress to associations involving further ectodomains that strengthens the adhesive bonds [36]. Since our results suggest that different regions of the extracellular domain of DE-Cadherin are more exposed in certain regions of the epidermis, we propose that the different antibodies recognize DE-Cadherins engaged in binding that involves a different number of extracellular domains. This also agrees with the existence of different pools of Cadherin with different adhesive properties as shown in the Drosophila epidermis [38].
Genetic reduction of DE-Cadherin increases DCAD2 binding to epidermal cells
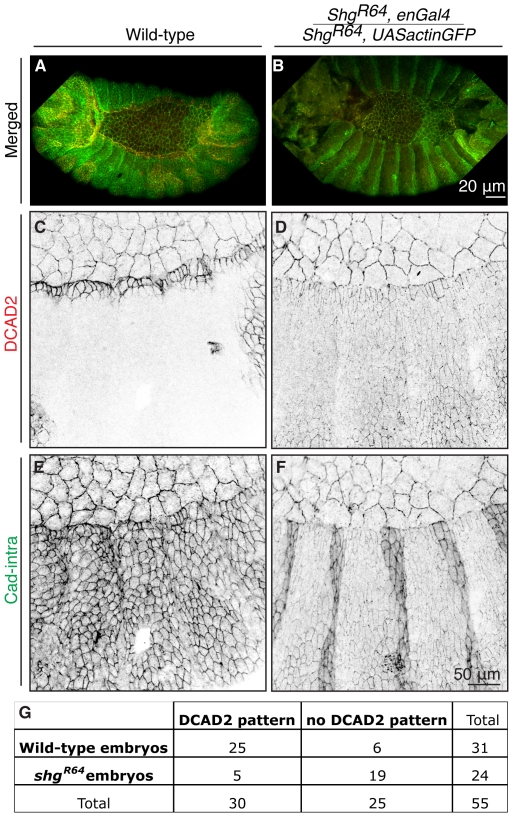
The observed surface pattern of DCAD2 binding correlates with different cell behaviours during DC [18]. To experimentally assess the functional role of the observed DCAD2 pattern along the epidermis, we performed the same assay in shgR64 zygotic mutant embryos, which only have the maternal contribution of DE-Cadherin [18]. We reasoned that if the differences observed were linked to differential engagement in adhesion, altering the levels of Cadherin would alter this balance and therefore the pattern.
Cad-intra antibody (Figure 4 A,E) shows that the levels of Cadherin in wild-type are homogeneous along the epidermis but DE-Cadherin is detected differently on the extracellular domain by DCAD2 (Figure 4C). In the shgR64 mutant, Cad-intra antibody clearly shows that the total levels of DE-Cadherin are lower than in wild-type (Figure 4B,F), which explains why DCAD2 antibody labelling is weak in the epidermis (Figure 4D). Interestingly, in shgR64 mutant embryos DCAD2 antibody binds to DE-Cadherin in all epidermal cells (Figure 4D). We quantified the number of embryos exhibiting the DCAD2 labelling pattern in wild-type embryos and shgR64, UASactinGFP/shgR64, enGal4 embryos, applied the χ2-test and found that the differences between wild-type and mutant embryos are statistically highly significant (p<0.0001; Figure 4G). In this attempt to manipulate the adhesive strength, lowering the amount of DE-Cadherin levels in the embryo could have led to epitope exposure and higher antibody accessibility, nevertheless when we tried an antibody against FasII, a component of the Septate Junctions that lies below the AJs, the binding was homogeneous (not shown), suggesting that the different binding of DCAD2 in wild-type or in the shgR64 mutants is not result of different accessibility of the antibody to the protein epitope.
Figure 4. Genetic Reduction of DE-Cadherin increases DCAD2 binding to epidermal cells.
Projection of confocal z-sections of wild-type (A, C, E) and shgR64 homozygous mutant (B, D, F) embryos pulsed with DCAD2 at 0′ chase. ShgR64 homozygous mutant embryos result from a cross between shgR64,enGal4 and shgR64,UASactinGFP, therefore a stronger staining on the engrailed domain is detected in F. (G) Contingency table with the number of embryos exhibiting DCAD2 labelling pattern in wild-type embryos and shgR64, UASactinGFP/shgR64, enGal4 embryos. The χ2-test revealed that the differences between wild-type and mutant embryos are extremely statistically significant (p<0.0001).
Our interpretation for the homogeneous binding of DCAD2 to all epidermal cells in the shgR64 mutants is that in the mutant all DE-Cadherin is engaged in strong homophilic adhesion, to compensate for DE-Cadherin reduced levels and to avoid epidermal cells from falling apart. Accordingly with this consideration DCAD2-labelled-DE-Cadherin would be engaged in stronger adhesion. Although, there is no direct data that shows whether the differences in antibody binding reflect strong Cadherin-Cadherin interactions or weak Cadherin-Cadherin interactions, the results obtained in shgR64 mutant suggest that DE-Cadherin exists in different forms that confer different adhesive strengths during DC in the Drosophila embryo.
Downregulation of Cadherin mediated adhesion and changes of adhesive activity with no detectable changes in the levels of Cadherin were observed during Xenopus laevis development. Importantly, this change in Cadherin activity also altered the binding of antibodies to native Cadherin [39], [40]. Our results suggest that these adhesive Cadherin properties are conserved and provide direct evidence for the first time for a spatial cellular organization of Cadherin during a morphogenetic process. The pattern observed along the epidermis correlates with differential cell behaviour during DC. The DME cells are attached to the AS, remain tightly bound to each other and bear most of the mechanical stress of the process (Figure 5A). Therefore, they have strong staining, the stronger the closer to the LE. The lateral epidermal cells, that undergo continuous cell rearrangement and might be in a more fluid phase, show less staining than DME cells and the AS (Figure 5B). Moreover, the binding of DCAD2 to the AS is similar to the DME cells and cell intercalation has not been observed in the AS [17]. Finally, DE-Cadherin level reduction results in a more homogeneous binding and we suggest that in this situation the little DE-Cadherin available is engaged in stronger adhesion. This would implicate that in wild-type DE-Cadherin molecules in the lateral epidermis, that are not recognized by DCAD2, are engaged in weaker adhesion. Altogether, our results suggest that structural differences in the extracellular domain of Cadherin can mediate differential cell adhesion during development, allowing distinct cell behaviour required for morphogenesis.
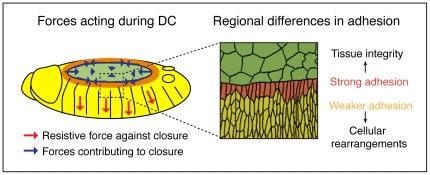
Figure 5. Forces driving DC.
AS contraction, actin purse string and zippering contribute positively to closure, in contrast to the resistive force exerted by the epidermis . Model of regional differences in adhesion along the epidermis.
Supporting Information
Validation of the live pulse-chase assay in embryos. Pulse-chase assays with 0′ chase (first raw) and 30′ chase (second raw) were performed with antibodies against the extracellular domain of DE-Cadherin (A,B), the intracellular domain of Notch (A′,B′) and Scribble, an intracellular protein (A″,B″). Arrowheads show intracellular puncta positive for DCAD2 that result from endocytosis occurred during the 30′ of chase (B). The intracellular antibodies (against Notch-intra and Scribble) were not able to access the interior of the cell. In the third raw, embryos were fixed and permeabilized before incubation with the referred antibodies; under these conditions the antibodies against intracellular epitopes in Notch (C′) and Scribble (C″), can bind and reveal patterns of expression. Confocal sections from the z-stack projected in B (5 µm along the z-axis).
(TIF)
Pulse-chase assay in ubi-DE-CadherinGFP embryos with DCAD2 and Notch-extra antibodies. Pulse-chase assay of DE-Cadherin and Notch in ubi-DE-CadherinGFP embryos with DCAD2 and Notch-extra antibodies. After 30′ of chase at RT the DCAD2 pattern is still maintained (C, C′), but Notch levels continue to decrease in the cell membrane of the AS and epidermis (D, D′). The vesicles of Notch tend to be bigger and more basal (D′,D″). With 1 hour of chase at RT, DCAD2 is still present at the membrane of AS and DME cells and also in large cytoplasmic vesicles (G,G′,G″). Notch is cleared from the membrane and the number and size of vesicles is greatly reduced (H,H′,H″). (I) Quantitative comparison of DCAD2 and Notch labelling at the cell membrane of LE cells over time. A significant difference occurs in Notch between 0′ chase and 10′ chase (p<0.01, n0′ = 30 and n10′ = 30) and 0′ chase and 30′ chase (p<0.01, n0′ = 30 and n30′ = 20) but not in DCAD2 (error bars show the SD).
(TIF)
DCAD2 pattern is also observed in wild-type Drosophila embryos. Using the standard staining protocol for Drosophila embryos, in which fixation and permeabilization precedes antibody incubation, DCAD2 binds homogeneously to the epidermis and AS, regardless of the DC stage (A–D′). The pattern of DCAD2 observed in ubi-DE-CadherinGFP expressing embryos is also observed in wild-type embryos at different time points of the pulse-chase (E′, F′,G′).
(TIF)
Time-lapse of a hand-devitellinized Ubi-DE-CadherinGFP embryo.
(AVI)
Acknowledgments
We want to thank Tadashi Uemura, Volker Hartenstein, and Francois Schweisguth for providing flies and antibodies. We are also grateful to Nicole Gorfinkiel for constant support during the project and Fan Cheng, Joaquin de Navascués and Paula Coelho for comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by Fundação para a Ciência e Tecnologia from Portugal (AMM) and the Wellcome Trust (AMA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005. pp. 622–634. [DOI] [PubMed]
- 2.Colas JF, Schoenwolf GC. Towards a cellular and molecular understanding of neurulation. Dev Dyn. 2001. pp. 117–145. [DOI] [PubMed]
- 3.Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, et al. folded gastrulation, cell shape change and the control of myosin localization. Development. 2005. pp. 4165–4178. [DOI] [PubMed]
- 4.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009. pp. 445–457. [DOI] [PubMed]
- 5.Kasza KE, Zallen JA. Dynamics and regulation of contractile actin-myosin networks in morphogenesis. Curr Opin Cell Biol. 2010. [DOI] [PMC free article] [PubMed]
- 6.Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol. 2007;8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 7.Duband JL, Monier F, Delannet M, Newgreen D. Epithelium-mesenchyme transition during neural crest development. Acta Anat (Basel) 1995. pp. 63–78. [DOI] [PubMed]
- 8.Haag TA, Haag NP, Lekven AC, Hartenstein V. The role of cell adhesion molecules in Drosophila heart morphogenesis: faint sausage, shotgun/DE-cadherin, and laminin A are required for discrete stages in heart development. Developmental Biology. 1999. pp. 56–69. [DOI] [PubMed]
- 9.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007. pp. 415–428. [DOI] [PubMed]
- 10.Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Török T, et al. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes & Development. 1996. pp. 672–685. [DOI] [PubMed]
- 11.Bryant DM, Stow JL. The ins and outs of E-cadherin trafficking. Trends Cell Biol. 2004. pp. 427–434. [DOI] [PubMed]
- 12.D'Souza-Schorey C. Disassembling adherens junctions: breaking up is hard to do. Trends Cell Biol. 2005. pp. 19–26. [DOI] [PubMed]
- 13.Jacinto A, Woolner S, Martin P. Dynamic analysis of dorsal closure in Drosophila: from genetics to cell biology. Developmental Cell. 2002. pp. 9–19. [DOI] [PubMed]
- 14.Martinez Arias A. Development and patterning of the larval epidermis of Drosophila In The Development of Drosophila melanogaster; In: Bates aAMA M, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 15.Hutson MS, Tokutake Y, Chang M-S, Bloor JW, Venakides S, et al. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 2003. pp. 145–149. [DOI] [PubMed]
- 16.Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. The Journal of Cell Biology. 2000. pp. 471–490. [DOI] [PMC free article] [PubMed]
- 17.Gorfinkiel N, Blanchard GB, Adams RJ, Martinez Arias A. Mechanical control of global cell behaviour during dorsal closure in Drosophila. Development. 2009. pp. 1889–1898. [DOI] [PMC free article] [PubMed]
- 18.Gorfinkiel N, Arias AM. Requirements for adherens junction components in the interaction between epithelial tissues during dorsal closure in Drosophila. Journal of Cell Science. 2007. pp. 3289–3298. [DOI] [PubMed]
- 19.Uemura T, Oda H, Kraut R, Hayashi S, Kotaoka Y, et al. Zygotic Drosophila E-cadherin expression is required for processes of dynamic epithelial cell rearrangement in the Drosophila embryo. Genes & Development. 1996. pp. 659–671. [DOI] [PubMed]
- 20.Oda H, Tsukita S. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. Journal of Cell Science. 2001. pp. 493–501. [DOI] [PubMed]
- 21.Broadie KS, Skaer H, Bate M. Whole-embryo culture of Drosophila: development of embryonic tissues in vitro. Roux's Archives of Developmental Biology. 1992;201:364–375. doi: 10.1007/BF00365124. [DOI] [PubMed] [Google Scholar]
- 22.Homsy JG, Jasper H, Peralta XG, Wu H, Kiehart DP, et al. JNK signaling coordinates integrin and actin functions during Drosophila embryogenesis. Dev Dyn. 2006;235:427–434. doi: 10.1002/dvdy.20649. [DOI] [PubMed] [Google Scholar]
- 23.Bertet C, Sulak L, Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004. pp. 667–671. [DOI] [PubMed]
- 24.Pilot F, Lecuit T. Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev Dyn. 2005;232:685–694. doi: 10.1002/dvdy.20334. [DOI] [PubMed] [Google Scholar]
- 25.Rauzi M, Lenne PF, Lecuit T. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature. 2010;468:1110–1114. doi: 10.1038/nature09566. [DOI] [PubMed] [Google Scholar]
- 26.Simone RP, DiNardo S. Actomyosin contractility and Discs large contribute to junctional conversion in guiding cell alignment within the Drosophila embryonic epithelium. Development. 2010;137:1385–1394. doi: 10.1242/dev.048520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lecuit T. Adhesion remodeling underlying tissue morphogenesis. Trends Cell Biol. 2005. pp. 34–42. [DOI] [PubMed]
- 28.Cavey M, Lecuit T. Molecular bases of cell-cell junctions stability and dynamics. Cold Spring Harb Perspect Biol. 2009;1:a002998. doi: 10.1101/cshperspect.a002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsh EW, Leopold PL, Jones NL, Maxfield FR. Oligomerized transferrin receptors are selectively retained by a lumenal sorting signal in a long-lived endocytic recycling compartment. J Cell Biol. 1995;129:1509–1522. doi: 10.1083/jcb.129.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders PGT, Muñoz-Descalzo S, Balayo T, Wirtz-Peitz F, Hayward P, et al. Ligand-independent traffic of Notch buffers activated Armadillo in Drosophila. PLoS Biol. 2009. e1000169. [DOI] [PMC free article] [PubMed]
- 31.Fortini ME, Bilder D. Endocytic regulation of Notch signaling. Curr Opin Genet Dev. 2009;19:323–328. doi: 10.1016/j.gde.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendersen DS. Totowa, New Jersey: Human Press; 2004. Drosophila Cytogenetics Protocols (Methods in Molecular Biology). [Google Scholar]
- 33.Chigaev A, Waller A, Amit O, Halip L, Bologa CG, et al. Real-time analysis of conformation-sensitive antibody binding provides new insights into integrin conformational regulation. J Biol Chem. 2009;284:14337–14346. doi: 10.1074/jbc.M901178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 35.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 36.Leckband D, Prakasam A. MECHANISM AND DYNAMICS OF CADHERIN ADHESION. Annu Rev Biomed Eng. 2006. pp. 259–287. [DOI] [PubMed]
- 37.Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci USA. 2009. pp. 109–114. [DOI] [PMC free article] [PubMed]
- 38.Cavey M, Rauzi M, Lenne P-F, Lecuit T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 2008. pp. 751–756. [DOI] [PubMed]
- 39.Brieher WM, Gumbiner BM. Regulation of C-cadherin function during activin induced morphogenesis of Xenopus animal caps. The Journal of Cell Biology. 1994. pp. 519–527. [DOI] [PMC free article] [PubMed]
- 40.Zhong Y, Brieher WM, Gumbiner BM. Analysis of C-cadherin regulation during tissue morphogenesis with an activating antibody. The Journal of Cell Biology. 1999. pp. 351–359. [DOI] [PMC free article] [PubMed]
- 41.Fung S, Wang F, Chase M, Godt D, Hartenstein V. Expression profile of the cadherin family in the developing Drosophila brain. J Comp Neurol. 2008;506:469–488. doi: 10.1002/cne.21539. [DOI] [PubMed] [Google Scholar]
- 42.Oda H, Tsukita S. Nonchordate classic cadherins have a structurally and functionally unique domain that is absent from chordate classic cadherins. Dev Biol. 1999;216:406–422. doi: 10.1006/dbio.1999.9494. [DOI] [PubMed] [Google Scholar]
- 43.Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of the live pulse-chase assay in embryos. Pulse-chase assays with 0′ chase (first raw) and 30′ chase (second raw) were performed with antibodies against the extracellular domain of DE-Cadherin (A,B), the intracellular domain of Notch (A′,B′) and Scribble, an intracellular protein (A″,B″). Arrowheads show intracellular puncta positive for DCAD2 that result from endocytosis occurred during the 30′ of chase (B). The intracellular antibodies (against Notch-intra and Scribble) were not able to access the interior of the cell. In the third raw, embryos were fixed and permeabilized before incubation with the referred antibodies; under these conditions the antibodies against intracellular epitopes in Notch (C′) and Scribble (C″), can bind and reveal patterns of expression. Confocal sections from the z-stack projected in B (5 µm along the z-axis).
(TIF)
Pulse-chase assay in ubi-DE-CadherinGFP embryos with DCAD2 and Notch-extra antibodies. Pulse-chase assay of DE-Cadherin and Notch in ubi-DE-CadherinGFP embryos with DCAD2 and Notch-extra antibodies. After 30′ of chase at RT the DCAD2 pattern is still maintained (C, C′), but Notch levels continue to decrease in the cell membrane of the AS and epidermis (D, D′). The vesicles of Notch tend to be bigger and more basal (D′,D″). With 1 hour of chase at RT, DCAD2 is still present at the membrane of AS and DME cells and also in large cytoplasmic vesicles (G,G′,G″). Notch is cleared from the membrane and the number and size of vesicles is greatly reduced (H,H′,H″). (I) Quantitative comparison of DCAD2 and Notch labelling at the cell membrane of LE cells over time. A significant difference occurs in Notch between 0′ chase and 10′ chase (p<0.01, n0′ = 30 and n10′ = 30) and 0′ chase and 30′ chase (p<0.01, n0′ = 30 and n30′ = 20) but not in DCAD2 (error bars show the SD).
(TIF)
DCAD2 pattern is also observed in wild-type Drosophila embryos. Using the standard staining protocol for Drosophila embryos, in which fixation and permeabilization precedes antibody incubation, DCAD2 binds homogeneously to the epidermis and AS, regardless of the DC stage (A–D′). The pattern of DCAD2 observed in ubi-DE-CadherinGFP expressing embryos is also observed in wild-type embryos at different time points of the pulse-chase (E′, F′,G′).
(TIF)
Time-lapse of a hand-devitellinized Ubi-DE-CadherinGFP embryo.
(AVI)