Abstract
We describe the isolation of an Arabidopsis gene that is closely related to the animal ZnT genes (Zn transporter). The protein encoded by the ZAT (Zn transporter of Arabidopsis thaliana) gene has 398 amino acid residues and is predicted to have six membrane-spanning domains. To obtain evidence for the postulated function of the Arabidopsis gene, transgenic plants with the ZAT coding sequence under control of the cauliflower mosaic virus 35S promoter were analyzed. Plants obtained with ZAT in the sense orientation exhibited enhanced Zn resistance and strongly increased Zn content in the roots under high Zn exposure. Antisense mRNA-producing plants were viable, with a wild-type level of Zn resistance and content, like plants expressing a truncated coding sequence lacking the C-terminal cytoplasmic domain of the protein. The availability of ZAT can lead to a better understanding of the mechanism of Zn homeostasis and resistance in plants.
Several heavy metals are essential during plant growth and development, but their excess can easily lead to toxic effects. Contamination of soils with heavy metals, either by natural causes or due to pollution, often has pronounced effects on the vegetation, resulting in the appearance of metallophytes, heavy-metal-tolerant plants. The precise mechanisms of uptake, transport, and accumulation of heavy metals in plants are poorly understood, but several genes likely to be involved in these processes have been described. Recently, a family of ZIP genes that are expressed in roots upon Zn deficiency was isolated from Arabidopsis (Grotz et al., 1998). The proteins encoded by the ZIP genes have eight predicted TM regions and a high degree of similarity to the ZRT genes from yeast that are involved in Zn uptake. Expression of the ZIP genes in yeast conferred Zn-uptake activities to these cells, demonstrating that they are probably functional homologs of the yeast ZRT genes (Grotz et al., 1998). The only other metal-transporting protein recently identified in plants belongs to the large family of cation-transporting P-type ATPases (Tabata et al., 1997), but these proteins are structurally very different from the metal-transporting proteins mentioned above.
Recent data have provided more insight into the mechanisms of heavy-metal tolerance. Metallophytes often exhibit tolerance to several different heavy metals, but all of these metals need not be present at toxic levels in their habitat (Schat and ten Bookum, 1992a; Schat and Vooijs, 1997, and refs. therein; Schat and Verkleij, 1998). Although such a feature is suggestive of a general mechanism of heavy-metal tolerance, recent genetic evidence has shown that a number of different mechanisms must exist, each with its own metal specificity (Schat and Vooijs, 1997). In Arabidopsis, a plant species with a typical level of tolerance to heavy metals, it has been demonstrated that the Cd-sensitive mutants cad1 and cad2 are defective in the synthesis of the metal-binding compound phytochelatin (Howden et al., 1995). cad1 plants were only slightly more sensitive to Cu and Zn, indicating that phytochelatin-mediated detoxification is not sufficient for Cu and Zn detoxification (Howden et al., 1995b). Metallothioneins appear to be of major importance for constitutive Cu tolerance in Arabidopsis (Zhou and Goldsbrough, 1994).
Aside from complexation of heavy metals by heavy-metal-binding proteins, there is evidence that transport-mediated sequestration can contribute to heavy-metal tolerance. In the Zn-tolerant plant Silene vulgaris it was shown that Zn transport across the tonoplast was about 2.5 times higher than in Zn-sensitive plants of the same species (Verkleij et al., 1998). The ZRC1 gene from the yeast Saccharomyces cerevisiae encodes a protein with six putative TM regions; when overexpressed, this gene confers elevated resistance to Zn and Cd (Kamizono et al., 1989). A structurally very similar gene, COT1, was later found to be involved in Co accumulation in yeast (Conklin et al., 1992).
Recently, several genes homologous to ZRC1 and COT1 have been described in mammalian cells. The first gene discovered, ZnT-1 (Zn transporter 1), was cloned by virtue of its ability to complement a mutated, Zn-sensitive cell line (Palmiter and Finley, 1995). Subsequently, ZnT-2 (Palmiter et al., 1996), ZnT-3 (Wenzel et al., 1997), and ZnT-4 (Huang and Gitschier, 1997) have been described. The ZnT-1 protein most likely transports Zn out of the cells (Palmiter and Finley, 1995), whereas ZnT-2 confers Zn resistance by facilitating vesicular sequestration (Palmiter et al., 1996a). Other proteins related to yeast ZRC1/COT1 and mammalian ZnT have been found in several bacteria; for example, the CzcD protein from Alcaligenes eutrophus might be involved in Zn efflux (Nies, 1992).
A family of proteins with six TM regions thus seems to be involved in the transport of heavy metals, mostly Zn, thereby conferring enhanced resistance toward these metals. To our knowledge, no plant homologs of this rather widespread gene family have yet been described. In this paper we describe an Arabidopsis cDNA clone encoding a protein closely related to the ZnT family of mammalian Zn transporters, demonstrating that plants do contain these types of genes. Experiments were performed to analyze the functional properties of the gene. We demonstrate that overexpression of the complete protein-coding domain results in enhanced Zn resistance and increased accumulation of Zn in the root. The relevance of these findings is discussed.
MATERIALS AND METHODS
cDNA Isolation and Sequence Analysis
The cDNA sequence encoding ZAT (Zn transporter of Arabidopsis thaliana) was isolated fortuitously. A cDNA library prepared with a cDNA-synthesis kit (ZAP, Stratagene) from RNA isolated from auxin-treated root cultures of Arabidopsis ecotype C24 was differentially screened for cDNA clones corresponding to auxin-induced mRNAs (Neuteboom et al., 1999). While characterizing the clones obtained, we noticed that in one of these clones two cDNA sequences were present. Apparently due to a defective XhoI site, the cDNA sequence encoding ZAT preceded the cDNA sequence of an auxin-inducible mRNA. When the total cDNA insert of the original clone was used as a probe for northern-blot analysis, two signals were found. The signal corresponding to the mRNA of the lowest molecular mass was auxin inducible, whereas the other relatively weak signal corresponding to ZAT mRNA was not changed after auxin treatment. Probing northern blots with subcloned fragments confirmed that two cDNA inserts were present in the original clone, each giving rise to only one of the signals observed with the original clone (data not shown).
Sequence reactions on both strands were performed according to the dideoxy chain-termination method (Sanger et al., 1977) using the T7 sequencing mix (Pharmacia). DNA homology searches and sequence analysis were performed using the Basic Local Alignment Search Tool (BLAST, National Center for Biotechnology Information, Bethesda, MD), the sequence-analysis software package from the Genetics Computer Group (Devereux et al., 1984), the CBS SignalP version 1.1 World Wide Web Prediction Server (Nielsen et al., 1997), and miscellaneous links that can be found at the CMS Molecular Biology Resource home page (http://www.unl.edu/stc-95/ResTools/cmshp.html). Multiple-sequence alignments and the dendrogram were made with the SeqLab program using the PileUp function with the end-weight option (Genetics Computer Group).
Plasmid Constructs
A 1460-bp fragment was isolated from the original cDNA clone by digestion with BamHI (site in the SK+ polylinker) and BglII (site just downstream of the TAA termination codon). This ZAT coding sequence was cloned in the BamHI-digested yeast expression plasmid YEP112A1 (Riesmeyer et al., 1992). YEP112A1, YEP112A1-ZAT-1 sense, and YEP112A1-ZAT-1 antisense were used to transform yeast strain YPH250 (ura3, lys2, ade2, trp1, his3, and leu2; Sikorski and Hieter, 1989). Transformants were selected on minimal yeast medium (Zonneveld, 1986) lacking Trp.
For plant expression studies the same 1460-bp ZAT BamHI/BglII fragment was cloned in two orientations between the cauliflower mosaic virus 35S promoter and the octopine synthase terminator in BamHI-digested pART7 (Gleave, 1992), resulting in pART7-ZAT-S (sense) and pART7-ZAT-AS (antisense). Plasmid pART7-ZAT-S was digested with XbaI and religated. This resulted in pART7-ZAT-dC, a cDNA insert truncated just after the sixth TM domain. Expression cassettes present in pART7 were isolated as NotI fragments and cloned in similarly digested pART27 (Gleave, 1992) in Escherichia coli strain DH5α (Clontech, Palo Alto, CA). Plasmids with the octopine synthase terminator of the 35S expression cassette adjacent to the right border were selected and mobilized to Agrobacterium tumefaciens strain GV2260 (Deblaere et al., 1985) using a triparental mating procedure (Ditta et al., 1980). Transconjugants were selected on medium containing carbenicillin, rifampicin, and spectinomycin (100, 20, and 125 mg/L, respectively).
Transformation of Arabidopsis
The A. tumefaciens strains harboring the binary pART27 plasmid (control) or the pART27 derivatives with ZAT inserts were used for cocultivation experiments with root explants of Arabidopsis ecotype C24 essentially as described previously (Valvekens et al., 1988). Transgenic shoots were selected on kanamycin (50 mg/L). Seeds of T1 plants were germinated on one-half-strength Murashige-Skoog medium (Murashige and Skoog, 1962; but one-half strength for macrosalts) supplemented with 10 g/L Suc, 7 g/L agar, and 50 mg/L kanamycin at 21°C under a 16-h light/8-h dark regime. Kanamycin-resistant seedlings of randomly chosen independent primary transformants whose progeny segregated 3:1 for kanamycin resistance were allowed to set seed, and homozygous T3 lines were selected. For the ZAT-AS and ZAT-dC constructs four randomly chosen lines were used for further analysis, together with two vector controls. In a similar manner, two homozygous lines with one T-DNA locus were obtained for the ZAT-S construct derived from the only two fertile primary transformants obtained with that construct.
Heavy-Metal-Resistance Assays
Homozygous T3 seeds were sown on plates containing one-half-strength Murashige-Skoog medium supplemented with various concentrations of ZnSO4 or CdCl2 (Analar, British Drug House, Poole, UK). The standard one-half-strength Murashige-Skoog medium contains 30 μm Zn2+ and 0.75 mm SO42−. After 4 d at 4°C, seeds were germinated at 21°C under a 16-h light/8-h dark regime.
More detailed analyses of Zn resistance and content were done using the hydroponic testing system described by Schat and ten Bookum (1992b). Seeds were sown on moist peat and after cold stratification were transferred to a growth chamber (20°C/15°C day/night temperatures; 75% RH; 14-h light [250 μmol m−2 s−1]/10-h dark regime). About 12 d after germination the plants were transferred to polyethylene pots filled with 600 mL of continuously aerated nutrient solution (Schat and Kalff, 1992). After 1 week of preculture, during which time the solution was refreshed once, the plants were exposed to a series of Zn concentrations. The background solution was identical to the preculture solution, except for FeNaEDTA, which was omitted to prevent ZnEDTA complex formation, and the pH was set at 5.25 with 2 mm Mes/KOH. Before exposure to Zn the roots were stained black using the charcoal-suspension method (Schat and ten Bookum, 1992b). After 72 h of exposure, the length of the unstained apical segment of the longest lateral root was measured and the roots were subsequently desorbed in ice-cold 5 mm Pb(NO3)2 for 1 h. Then roots and shoots were harvested separately, frozen in liquid nitrogen, lyophilized, and stored under vacuum until analysis. Powdered lyophilized materials were digested in a 1:4 mixture (v/v) of 37% HCl and 65% HNO3 in closed Teflon bombs at 140°C for 7 h. The Zn concentration in the digests was measured using flame atomic absorption spectroscopy (model 1100, Perkin-Elmer). Root lengths and Zn contents were statistically analyzed using two-way analysis of variance, after ln transformation of the data.
RNA Isolation and Northern-Blot Analysis
Aliquots of homozygous seeds were surface-sterilized for 1 min in 70% (v/v) ethanol followed by soaking for 10 min in a NaOCl solution (1% active ingredient supplemented with 0.05% [v/v] Tween 20 [Merck, Darmstadt, Germany]). Seeds were cultured in 250-mL flasks containing 50 mL of Gamborg B5 medium (Gamborg et al., 1968) on a gyratory shaker (100 rpm) at 21°C with a 16-h light/8-h dark cycle. After 14 d approximately 10 g fresh weight of total seedlings was used for RNA isolation (van Slogteren et al., 1983). Amounts of 30 μg of glyoxylated total RNA were electrophoresed on a 1.5% agarose gel and capillary blotted onto GeneScreen Plus membranes (NEN-DuPont) according to standard procedures (Sambrook et al., 1989).
For induction experiments seedlings were grown similarly in one-half-strength Murashige-Skoog medium (see above). Extra ZnSO4 was added to the flasks after 11 d of growth. Incubation in the flasks then continued for 6 or 24 h before mRNA isolation.
The cDNA insert was labeled by random priming. For generation of strand-specific RNA probes a 1460-bp BamHI/BglII fragment containing the complete ZAT coding sequence was cloned in BamHI-digested pBluescript SK+ plasmid (Stratagene). The plasmid with insert orientation giving rise to an antisense RNA probe with T7 RNA polymerase was linearized with BamHI, and the sense probe was generated from the plasmid with the reverse insert orientation after digestion with XbaI. Labeling reactions were performed using the TransProbe T kit (Pharmacia). Prehybridization was performed for 2 h at 42°C in 50% deionized formamide, 5× SSPE (1× SSPE = 180 mm NaCl, 1 mm EDTA, and 10 mm sodium phosphate, pH 6.5), 2.5% (w/v) SDS, followed by 24 h of hybridization in the same solution after addition of the probe. Blots were washed at 65°C in 0.1× SSPE, 0.1% SDS for a total of 1 h, and were then exposed to film (Fuji-RX, Fuji Photo Film Co. Ltd., Tokyo, Japan).
RESULTS
ZAT Encodes a Putative Zn Transporter
Sequence analysis of a fortuitously isolated Arabidopsis cDNA (see Methods for details) revealed that it encodes a protein of 398 amino acid residues. Several in-frame stop codons are present upstream of the ATG initiation codon, and the TAA stop codon is present well before the end of the cDNA sequence (Fig. 1). The cDNA sequence is therefore full length as far as the coding region is concerned. A BLAST search (February 24, 1998) with the protein sequence showed that the protein was related to a wide family of proteins that are (putatively) involved in the transport of Zn across biological membranes in various organisms. Except for a partial sequence from potato (accession no. U60071), which so far has been described only as a putative plant disease-resistance gene (Leister et al., 1996), no homologous plant protein sequences were detected, indicating that this type of protein has thus far not been found or recognized as such in plants. For reasons described below we have designated this protein ZAT.
Figure 1.
The Arabidopsis ZAT nucleotide sequence is shown with the amino acid sequence below (one letter code). ATG initiation and TAA termination codons are shown in boldface type. The cDNA sequence did not end in a poly(A+) tail, but occurred just after the putative polyadenylation signal fused with a second cDNA sequence (not shown). The XbaI site used for the C-terminal deletion just after the sixth TM domain (see Fig. 3) is indicated in boldface.
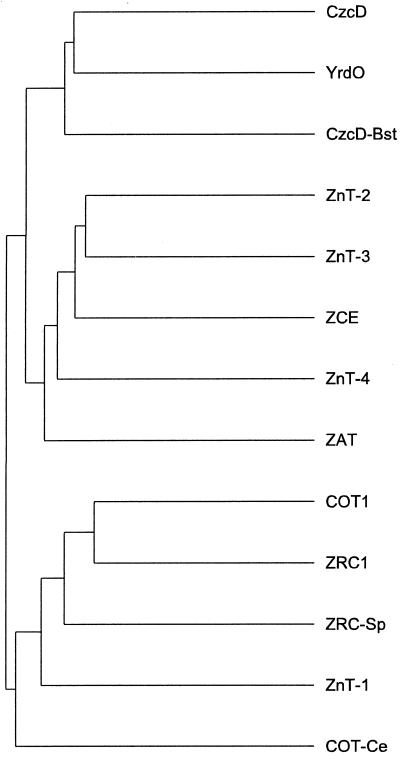
All related proteins are characterized by a structure with six potential TM regions and a long cytoplasmic C-terminal tail. When degrees of relatedness are visualized by a dendrogram it becomes clear that ZAT is most closely related to a subgroup of Zn transporters recently described for mammalian cells, ZnT-2, ZnT-3, and ZnT-4 (Palmiter et al., 1996a, 1996b; Huang and Gitschier, 1997, respectively) (Fig. 2). The levels of amino acid identity/similarity between ZAT and ZnT-2, ZnT-3, and ZnT-4 are 40.8%/52.5%, 37.4%/47.3%, and 35.3%/49.1%, respectively. These values correspond to 40% to 50% identity at the nucleotide level. Rat ZnT-2 facilitates Zn transport into an endosomal/lysosomal compartment and thus protects cells from Zn toxicity (Palmiter et al., 1996a). By analogy, it was hypothesized that the strongly related ZnT-3 and ZnT-4 proteins are involved in the same process (Huang and Gitschier, 1997). Three sequences from Caenorhabditis elegans encoding possible homologs that belong to the same group of proteins can be found with a BLAST search (only one of them is included in Fig. 2).
Figure 2.
Dendrogram of the amino acid sequences of several (putative) Zn transporters. CzcD, Alcaligenes eutrophus (accession no. D67044); YrdO (CzcD), Bacillus subtilis (accession no. U93876 [U62055]); CzcD-Bst, Bacillus stearothermophilus (accession no. D87026); ZnT-2, Rattus norvegicus (accession no. S70632; Palmiter et al., 1996a); ZnT-3, Homo sapiens (accession no. U76010; Palmiter et al., 1996b); ZCE, C. elegans (accession no. T18D3.3 in Z68119); ZnT-4, H. sapiens (accession no. AF025409; Huang and Gitschier, 1997); ZAT, Arabidopsis (this study); COT1, Saccharomyces cerevisiae (accession no. P32798; Conklin et al., 1992); ZRC1, S. cerevisiae (accession no. P20107; Kamizono et al., 1989); ZRC-Sp, ZRC1 homolog, Schizosaccharomyces pombe (accession no. D89236); ZnT-1, R. norvegicus (accession no. S54303; Palmiter et al., 1995); COT-Ce, COT1 homolog in C. elegans (accession no. U23529). The distance between the branches on the horizontal axis are proportional to the degree of relatedness according to the program used, whereas the distances along the vertical axis are without any meaning.
Two other groups of structurally related proteins can be discerned (Fig. 2). ZnT-1, the first mammalian Zn transporter reported and found to be involved in Zn efflux (Palmiter and Findley, 1995), forms a subgroup together with Zn/Cd and Co transporters from yeast. The level of amino acid identity between these proteins and ZAT ranges between 20% and 30%. The partial potato protein sequence mentioned above seems to belong to this group (data not shown), but this classification might change when the complete sequence becomes available. C. elegans also contains a gene encoding a protein of this group. Metal-transporting proteins from bacteria form a third subgroup, possessing 30% to 37% amino acid identity with ZAT. The putative Zn-uptake carriers with eight predicted TM regions, such as the yeast ZRT genes (Zhao and Eide, 1996a, 1996b) and Arabidopsis ZIP genes (Grotz et al., 1998), are only distantly related to the genes mentioned and were not included in the analysis.
Alignment of the amino acid sequence of ZAT with other sequences of its subgroup showed that ZAT is deviating mainly due to the relatively large stretch of 81 amino acids between putative TM regions IV and V, which is more than twice the length of the corresponding region of its nearest homologs (Fig. 3). The His-rich region between TM regions IV and V is predicted to form a cytoplasmic loop in which the His residues are most likely responsible for the binding of Zn (Palmiter et al., 1996a, 1996b; Huang and Gitschier, 1997). ZAT contains 21 His residues in this region, but apart from the amino acid sequence HSH, there is little sequence identity with the ZnT-2, ZnT-3, and ZnT-4 proteins in this area. Most of the conserved amino acids are present within or just around the TM regions and also in the long cytoplasmic tails of the aligned proteins (Fig. 3). As far as the length of the putative cytoplasmic loop between TM regions IV and V is concerned, ZAT is more similar to the group of proteins constituting mammalian ZnT-1 and the yeast Zn/Cd and Co transporters, which also have a long loop sequence. However, in terms of conserved amino acids, the similarity is just as restricted as with the cytoplasmic loop of ZnT-2, ZnT-3, and ZnT-4 (comparison not shown). Within the CzcD group of bacterial cation-transport or efflux proteins, the putative TM regions IV and V are separated by only 12 to 13 amino acid residues, none of which is a His residue (not shown).
Figure 3.
Multiple alignment of representatives of (putative) Zn transporters most closely related to ZAT. The six putative TM regions are overlined. Amino acid residues that are identical in four or five of the sequences are shown in boldface. See legend to Figure 2 for further details.
ZAT from Arabidopsis is thus most closely related to the Zn transporters ZnT-2, ZnT-3, and ZnT-4 from mammals and the putative homologs of these proteins from C. elegans. Since expression of ZnT-2 was shown to result in enhanced Zn tolerance of cells, possibly by enhancing vesicular sequestration of Zn (Palmiter et al., 1996a), we pursued further analysis of ZAT along this line.
Ectopic Expression of ZAT
Expression of sense or antisense ZAT mRNA in the yeast S. cerevisiae under control of the yeast ADH promoter did not lead to a detectable change in the resistance of transgenic yeast cells toward Zn, Cd, or Co (data not shown). The yeast strain used was wild type for Zn, Cd, and Co tolerance and was therefore not as sensitive to changes in resistance to these metals as certain mutant yeast strains (Conklin et al., 1994). However, a marked change in Zn tolerance could be detected in wild-type yeast cells expressing ZnT-4 (Huang and Gitschier, 1997). Apparently, such a change in sensitivity was not achieved with ZAT under the conditions used.
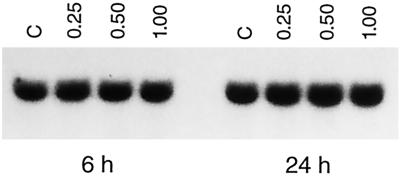
A genomic blot of Arabidopsis DNA probed under stringent conditions with the complete ZAT cDNA sequence exhibited a simple restriction enzyme digestion pattern, indicative of the presence of a single gene. Northern blots with RNA from different plant parts showed that mRNA was expressed throughout the plant at similar but low levels (data not shown). Treatment of seedlings with increasingly toxic levels of Zn for 6 and 24 h failed to induce ZAT mRNA (Fig. 4). To study the effect of altered levels of ZAT mRNA in Arabidopsis, we pursued expression of sense and antisense ZAT mRNA under control of the strong cauliflower mosaic virus 35S promoter in stably transformed Arabidopsis plant lines. In addition, a construct that should lead to overexpression of a ZAT protein lacking the C-terminal cytoplasmic domain was transformed in Arabidopsis. In animal cells it was shown that such a construct can lead to a toxic phenotype (Palmiter and Findley, 1995). We reproducibly found that the root-transformation protocol that worked very efficiently for the empty-vector control construct, as well as the ZAT-AS (antisense) and ZAT-dC (deletion of C-terminal cytoplasmic domain) constructs, was virtually ineffective for the ZAT-S (sense) construct. Only a few transgenic calli were obtained with ZAT-S, and only two independent calli formed shoots that could set seed.
Figure 4.
Nothern blots of RNA isolated from seedlings treated in liquid medium with increasing concentrations of ZnSO4 for 6 and 24 h. Lanes C, Control seedlings (30 μm Zn already present in the medium); lanes 0.25, 0.50, and 1.00, seedlings with different millimolar concentrations of extra ZnSO4 added. Exposure was for 2 d at −70°C with an intensifying screen.
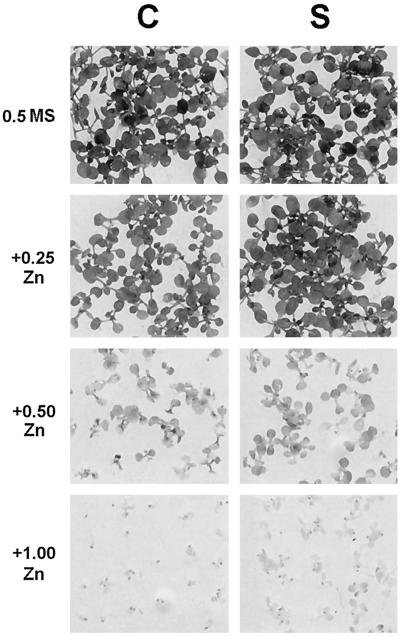
Homozygous T3 seed lines containing one T-DNA locus were used for Zn- and Cd-resistance tests. Two control lines, four ZAT-AS lines, and four ZAT-dC lines were randomly chosen, along with the only two ZAT-S lines and the nontransformed C24 ecotype. All lines were phenotypically normal in vitro and in the greenhouse. In the first test, seedlings were grown on plates supplemented or not with 0.25 mm ZnSO4 or 0.1 mm CdCl2. Only the two ZAT-S lines were nearly completely resistant to the extra 0.25 mm Zn. No differences were observed for the Cd treatment. The two ZAT-S lines, the two control lines, and C24 were tested again on medium containing a series of added Zn (0.25, 0.5, and 1.0 mm) or Cd (25, 50, and 100 μm) concentrations and compared with growth on the normal medium (containing 30 μm Zn as a trace element). Again, the two ZAT-S lines were nearly fully resistant to added 0.25 mm Zn, a concentration that resulted in chlorosis and slower development in the controls (Fig. 5; note the full expansion of the true leaves of the ZAT-S line at 0.25 mm extra Zn, whereas in the control most of the true leaves were smaller than the cotyledons).
Figure 5.
Zn-resistance test of representative Arabidopsis transgenic lines. C, Control; S, sense. Seedlings were germinated and grown on solid medium (one-half-strength Murashige-Skoog medium). 0.5 MS, Control seedlings (30 μm Zn already present in the medium); 0.25, 0.50, and 1.00, seedlings with different millimolar concentrations of extra ZnSO4 added.
When 0.5 or 1.0 mm extra Zn was added, development of both the control and the ZAT-S line was severely inhibited. Although the ZAT-S lines were still growing slightly better than the controls at these high Zn concentrations (Fig. 5), it was evident that the gain in Zn resistance of the ZAT-S lines was relatively modest. No differences in Cd sensitivity were observed, although the concentration range used now covered the complete spectrum of low (25 μm) to high (100 μm) toxicity (the latter concentration completely inhibited root development).
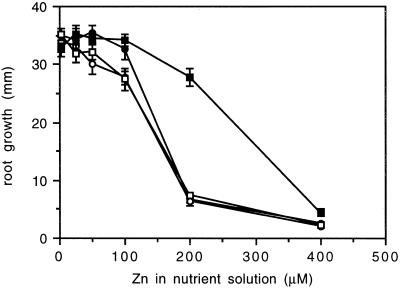
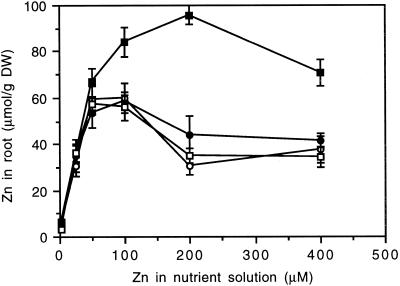
One each of the lines with control, ZAT-AS, ZAT-S, and ZAT-dC was subjected to more detailed testing for Zn resistance and accumulation using a hydroponic system. As inferred from the root-growth response (Fig. 6), only the ZAT-S line exhibited altered Zn sensitivity, being virtually resistant to a 200 μm concentration of external Zn. At 200 μm the difference from the other lines was highly significant (P < 0.001). However, a 400 μm concentration of external Zn inhibited ZAT-S root growth about as severely as in the other lines. Therefore, the increase in Zn resistance in the ZAT-S line was clear, but was limited to a rather narrow concentration range. The ZAT-S line also showed a different pattern of Zn accumulation in the root (Fig. 7). At up to 50 μm external Zn there were no significant differences between lines, but at higher concentrations the ZAT-S line showed a significantly higher root Zn content than the other lines. No significant differences were detected in shoot Zn content between the different lines (data not shown). Control, ZAT-AS, and ZAT-dC lines were not significantly different in the measured parameters. A less-detailed analysis with a second line of each transgenic type, including the second ZAT-S line, yielded very similar results, confirming the enhanced Zn resistance and root Zn content due to the ZAT-S construct.
Figure 6.
Arabidopsis root growth during 72 h of exposure to Zn in hydroponic culture. •, Control; ○, ZAT-AS; ▪, ZAT-S; □, ZAT-dC. Data points are the means of 15 plants. Vertical bars represent ±se.
Figure 7.
Arabidopsis root Zn content after 72 h of exposure to Zn in hydroponic culture. •, Control; ○, ZAT-AS; ▪, ZAT-S; □, ZAT-dC. Data points are the means of five pooled samples of three plants each. Vertical bars represent ±se. DW, Dry weight.
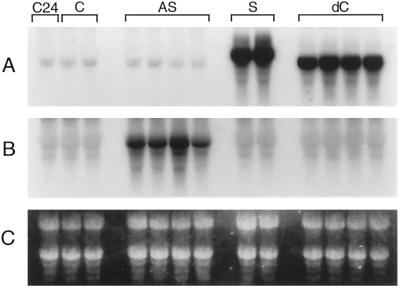
RNA was isolated from all the plant lines and analyzed on northern blots with strand-specific probes (Fig. 8). The antisense probe detected a high level of extra sense ZAT mRNA in the two ZAT-S lines and a similarly high level of the truncated ZAT-dC mRNA in the four ZAT-dC lines at a lower-molecular-mass position (Fig. 8A). A difference of 264 bases is expected between the mRNA produced by these constructs. The endogenous ZAT mRNA ran at a lower-molecular-mass position than the ectopically expressed ZAT-S mRNA, and apparently was of the same length as the ZAT-dC mRNA. The cauliflower mosaic virus 35S expression cassette used therefore leads to a mRNA about 260 bases longer than the endogenous ZAT mRNA. Although all four ZAT-AS lines produced high levels of antisense mRNA (Fig. 8B), this did not lead to significant changes in the level of endogenous ZAT mRNA detected with the antisense probe in the same plant lines (Fig. 8A).
Figure 8.
Northern blots probed with strand-specific RNA probes. A, Antisense probe (showing sense mRNA). B, Sense probe (showing antisense RNA). C, Ethidium-bromide-stained total RNA present on the original gels. Lane C24, Nontransformed C24 ecotype; lane C, vector-transformed control; lane AS, antisense construct line; lane S, sense construct line; lane dC, C-terminal deletion construct line. Exposure was for 1 d without an intensifying screen.
DISCUSSION
We have shown that Arabidopsis expresses a gene designated ZAT that is most closely related to the ZnT-2, ZnT-3, and ZnT-4 family of Zn-transporter genes that so far have been described only in mammals and in the nematode C. elegans. More generally, ZAT has significant similarity to a variety of genes that encode proteins with six predicted TM regions and a long C-terminal cytoplasmic domain (see legend to Fig. 2). This structure is characteristic for a wide family of proteins that are involved in the transport of certain heavy metals, mostly Zn, across cellular membranes. In plants such proteins have not been described previously. A partial sequence from potato that has been reported as a putative disease-resistance gene (Leister et al., 1996) does, however, indicate that these proteins are present in more plant species. It should be noted that the ZAT-related proteins are structurally different from the ZRT1 and ZRT2 proteins, which have eight predicted TM regions and mediate Zn uptake in yeast cells (Zhao and Eide, 1996a, 1996b). Recently, a family of Zn transporters related to the yeast ZRT genes was identified in Arabidopsis. These ZIP genes were indeed able to confer Zn-uptake activities to yeast cells, and for this reason can be regarded as functional homologs of the yeast ZRT genes (Grotz et al., 1998).
For the mammalian genes most related to ZAT, experimental evidence strongly suggests that their encoded proteins are involved in the facilitation of vesicular sequestration of Zn, which can result in increased resistance to otherwise toxic levels of extracellular Zn. Overexpression of ZnT-2 in a Zn-sensitive hamster kidney cell line resulted in elevated levels of intravesicular Zn and an increase in Zn tolerance when cells were grown at a high Zn concentration (Palmiter et al., 1996a). A similar experiment with ZnT-3 did not result in these effects (Palmiter et al., 1996b), but the ZnT-3 protein was found to be localized in synaptic vesicle membranes, suggesting that it is also involved in the vesicular sequestration of Zn in certain cells (Wenzel et al., 1997). ZnT-4 was able to confer enhanced Zn resistance to yeast (Huang and Gitschier, 1997).
ZAT mRNA seemed to be expressed constitutively throughout the plant and was not induced by increasing Zn concentrations. Overexpression of ZAT mRNA in transgenic Arabidopsis plants resulted in a modest but significant increase in Zn tolerance and an enhanced accumulation of Zn in the root at high external Zn concentrations. These latter data indicate that the ZAT protein performs a function similar to the animal ZnT-2 protein. The ZAT protein might therefore be involved in the sequestration of Zn, thereby contributing to Zn resistance. In plants Zn can be transported into the vacuole, as was found in the Zn-tolerant species Silene vulgaris. Transport of Zn across the tonoplast was found to be 2.5 times higher than in Zn-sensitive plants of the same species (Verkleij et al., 1998). It is tempting to speculate that this elevated Zn transport is due to a ZAT-like protein, but at the moment we lack data that ZAT is indeed located in the tonoplast membrane.
Although it is likely that ZnT/ZAT-like proteins are involved in transport of Zn, the mechanism by which they do so is far from clear. The proteins lack an obvious nucleotide-binding domain indicative of an active transporter. Ion transport across a membrane is thus probably coupled to some other energetically favorable reaction and therefore more likely to be some kind of facilitated diffusion (Palmiter et al., 1996a). Bacterial CzcD proteins are related to ZnT/ZAT proteins (Fig. 2), but their role is still obscure. It has been postulated that the CzcD protein performs a regulatory role for the metal-transporting, membrane-bound CzcCBA protein complex (Nies, 1992), whereas the CzcA protein is the most likely candidate as the central cation-proton antiporter (Rensing et al., 1997). However, CzcA is without further similarity to ZnT/ZAT and related proteins. By analogy to the bacterial CzcCBA system, we therefore speculated that ZnT proteins may be part of a larger complex in which they determine ion specificity or regulate Zn transport, rather than being directly involved in transport (Palmiter et al., 1996a). However, it should be noted, as mentioned in “Results,” that bacterial CzcD proteins lack the His-rich loop between TM regions IV and V that is present in the other putative transporters. If such a loop is indeed essential for Zn transport, the CzcD proteins will necessarily have to operate in a mechanistically different manner than the eukaryotic Zn transporters or Zn-transporting systems.
Whatever the exact molecular mechanism, it is clear that Zn resistance mediated by ZAT overexpression is correlated with increased root Zn content and is certainly not based on a reduction of the net Zn influx. When compared at similar degrees of root-growth inhibition, root Zn contents were about 2-fold higher in the ZAT-S plants than in the other lines (Figs. 6 and 7). The increase in Zn resistance in ZAT-S plants can be estimated to be about 2-fold as well. Considering the huge amount of sense ZAT mRNA in the transgenic ZAT-S plants (Fig. 8A), these differences are small. The simplest explanation is that the extra mRNA is poorly translated, but an alternative reason for this discrepancy has to be considered. When the ZAT protein is part of a larger protein complex, as discussed above, overexpression of only one of the components of the complex will have only marginal effects. Only when the endogenous ZAT concentration is relatively low and the other components are more abundant will extra ZAT protein lead to more functional complexes. Considering the difficulties in acquiring transgenic calli and shoots after transformation with the ZAT-S construct, it could be that the same situation exists during the initial stages of callus formation and/or regeneration. Cells may get intoxicated by an excessive accumulation of Zn in the vacuole.
The C-terminal intracellular domain seems to be important for ZAT function, because the ZAT-dC construct lacking this domain was not able to confer Zn resistance or Zn accumulation. A toxic effect of the C-terminal deletion was not observed, in contrast to a similar construct made for rat ZnT-1 (Palmiter and Findley, 1995). This could mean that the inactive C-terminal deletion mutant of ZAT is not able to intoxicate multimeric protein complexes that contain ZAT or a related protein. Alternatively, it could mean that intoxication or disruption of such protein complexes are without effect under the conditions used. The lack of effect of antisense ZAT mRNA expression on Zn resistance and accumulation seems to suggest the latter possibility. However, for unknown reasons, the level of endogenous ZAT mRNA in the ZAT-AS lines appeared to be unaffected by the high level of antisense mRNA, so the lack of effect has to be interpreted with caution.
The precise role of the ZAT gene in normal, wild-type Zn homeostasis remains elusive. However, based on the data presented here, we propose that it is involved in the vesicular/vacuolar sequestration of Zn, thereby contributing to Zn tolerance in plants. Further study of ZAT, together with the recently cloned ZIP genes that are likely to be responsible for Zn uptake (Grotz et al., 1998), will lead to a better understanding of Zn homeostasis and tolerance in higher plants.
The accession number for the nucleotide sequence of the ZAT cDNA reported in this paper is AF072858.
ACKNOWLEDGMENTS
We thank Riet Vooijs for technical assistance, Dr. Frank van Iren for stimulating discussions, Drs. Werner Pansegrau and Dorien Postma-Haarsma for help with protein alignments, and Bram Wetselaar and Elly Schrijnemakers for plant care in the greenhouse. We also thank Peter Hock for preparation of the figures and Adri't Hooft and Peter van Mulken for photography.
Abbreviation:
- TM
transmembrane
LITERATURE CITED
- Conklin D, Culbertson MR, Kung C. Interactions between gene products involved in divalent cation transport in Saccharomyces cerevisiae. Mol Gen Genet. 1994;244:303–311. doi: 10.1007/BF00285458. [DOI] [PubMed] [Google Scholar]
- Conklin DS, McMaster JA, Culbertson MR, Kung C. COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:3678–3688. doi: 10.1128/mcb.12.9.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, de Greve H, Deboeck F, Schell J, van Montagu M, Leemans J. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 1985;13:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Stanfield S, Corbin D, Helinski DR. Broad host range DNA cloning system from gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA. 1998;95:7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R, Andersen CR, Goldsbrough PB, Cobett CS. A cadmium-sensitive, glutathione-deficient mutant of Arabidopsis thaliana. Plant Physiol. 1995a;107:1067–1073. doi: 10.1104/pp.107.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R, Goldsbrough PB, Andersen CR, Cobett CS. Cadmium-sensitive, cad1 mutants of Arabidopsis thaliana are phytochelatin deficient. Plant Physiol. 1995b;107:1059–1066. doi: 10.1104/pp.107.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Gitschier J. A novel gene involved in zinc transport is deficient in the lethal milk mouse. Nat Genet. 1997;17:292–297. doi: 10.1038/ng1197-292. [DOI] [PubMed] [Google Scholar]
- Kamizono A, Nishizawa M, Teranishi Y, Murata K, Kimura A. Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1989;219:161–167. doi: 10.1007/BF00261172. [DOI] [PubMed] [Google Scholar]
- Leister D, Ballvora A, Salamini F, Gebhardt C. A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet. 1996;14:421–429. doi: 10.1038/ng1296-421. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Neuteboom LW, Ng JMY, Kuyper M, Clijdesdale OR, Hooykaas PJJ, van der Zaal BJ. Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formation. Plant Mol Biol. 1999;39:273–287. doi: 10.1023/a:1006104205959. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nies DH. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J Bacteriol. 1992;174:8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Cole TB, Findley SD. ZnT-2, a mammalian protein that confers resistance to zinc by facilitating vesicular sequestration. EMBO J. 1996a;15:1784–1791. [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Cole TB, Quaife CJ, Findley SD. ZnT-3, a putative transporter of zinc into synaptic vesicles. Proc Natl Acad Sci USA. 1996b;93:14934–14939. doi: 10.1073/pnas.93.25.14934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Findley SD. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 1995;14:639–649. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Pribyl T, Nies DH. New functions for the three subunits of the CzcCBA cation-proton antiporter. J Bacteriol. 1997;179:6871–6879. doi: 10.1128/jb.179.22.6871-6879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 1992;11:4705–4713. doi: 10.1002/j.1460-2075.1992.tb05575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook L, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat H, Kalff MMA. Are phytochelatins involved in differential metal tolerance or do they merely reflect metal-imposed strain? Plant Physiol. 1992;99:1475–1480. doi: 10.1104/pp.99.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat H, ten Bookum WM (1992a) Metal-specificity of heavy metal tolerance syndromes in higher plants. In AJM Baker, RD Reeves, J Proctor, eds, The Vegetation of Ultramaffic (Serpentine) Soils. Intercept, Andover, UK, pp 337–353
- Schat H, ten Bookum WM. Genetic control of copper tolerance in Silene vulgaris. Heredity. 1992b;68:219–229. [Google Scholar]
- Schat H, Verkleij JAC (1998) The role of non-woody plants in phytorestoration: possibilities to exploit adaptive heavy metal tolerance. In J Vangronsveld, C Cunningham, eds, In Situ Phytoremediation of Heavy Metals Contaminated Sites. Springer Verlag and R.G. Landes Co., Austin, TX, pp 51–65
- Schat H, Vooijs R. Multiple tolerance and co-tolerance to heavy metals in Silene vulgaris: a co-segragation analysis. New Phytol. 1997;136:489–496. doi: 10.1046/j.1469-8137.1997.00756.x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K, Kashiwagi S, Mori H, Ueguchi C, Mizuno T. Cloning of a cDNA encoding a putative metal-transporting P-type ATPase from Arabidopsis thaliana. Biochim Biophys Acta. 1997;1326:1–6. doi: 10.1016/s0005-2736(97)00064-3. [DOI] [PubMed] [Google Scholar]
- Valvekens D, van Montagu M, van Lijsebettens M. Agrobacterium tumefaciens mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slogteren GMS, Hoge JHC, Hooykaas PJJ, Schilperoort RA. Clonal analysis of heterogenous crown gall tumour tissues induced by wild-type and shooter mutant strains of Agrobacterium tumefaciens: expression of T-DNA genes. Plant Mol Biol. 1983;2:321–333. doi: 10.1007/BF01578594. [DOI] [PubMed] [Google Scholar]
- Verkleij JAC, Koevoets PLM, Blake-Kalff MMA, Chardonnens AN. Evidence for an important role of the tonoplast in the mechanism of naturally selected zinc tolerance in Silene vulgaris. J Plant Physiol. 1998;153:188–191. [Google Scholar]
- Wenzel HJ, Cole TB, Born DE, Schwartzkroin PA, Palmiter RD. Ultrastructural localization of Zn transporter-3 (ZnT-3) to synaptic vesicle membranes within mossy fiber boutons in the hippocampus of mouse and monkey. Proc Natl Acad Sci USA. 1997;94:12676–12681. doi: 10.1073/pnas.94.23.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci USA. 1996a;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem. 1996b;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- Zhou J, Goldsbrough PB. Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell. 1994;6:875–884. doi: 10.1105/tpc.6.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonneveld BJM. Cheap and simple yeast media. J Microbiol Methods. 1986;4:287–291. [Google Scholar]