Abstract
Asthma is etiologically and clinically heterogeneous, making the genomic basis of asthma difficult to identify. We exploited the strain-dependence of a murine model of allergic airway disease to identify different genomic responses in the lung. BALB/cJ and C57BL/6J mice were sensitized with the immunodominant allergen from the Dermatophagoides pteronyssinus species of house dust mite (Der p 1), without exogenous adjuvant, and the mice then underwent a single challenge with Der p 1. Allergic inflammation, serum antibody titers, mucous metaplasia, and airway hyperresponsiveness were evaluated 72 hours after airway challenge. Whole-lung gene expression analyses were conducted to identify genomic responses to allergen challenge. Der p 1–challenged BALB/cJ mice produced all the key features of allergic airway disease. In comparison, C57BL/6J mice produced exaggerated Th2-biased responses and inflammation, but exhibited an unexpected decrease in airway hyperresponsiveness compared with control mice. Lung gene expression analysis revealed genes that were shared by both strains and a set of down-regulated genes unique to C57BL/6J mice, including several G-protein–coupled receptors involved in airway smooth muscle contraction, most notably the M2 muscarinic receptor, which we show is expressed in airway smooth muscle and was decreased at the protein level after challenge with Der p 1. Murine strain–dependent genomic responses in the lung offer insights into the different biological pathways that develop after allergen challenge. This study of two different murine strains demonstrates that inflammation and airway hyperresponsiveness can be decoupled, and suggests that the down-modulation of expression of G-protein–coupled receptors involved in regulating airway smooth muscle contraction may contribute to this dissociation.
Keywords: asthma, airway hyperresponsiveness, inflammation, house dust mite, Der p 1
Clinical Relevance
The genomic basis of asthma in humans is obscure. Using mice with divergent responses to the house dust mite allergen Dermatophagoides pteronyssinus (Der p 1), we identified genomic factors that affect the response to Der p 1.
Identifying the genetic basis of asthma is difficult because asthma is etiologically and clinically heterogeneous (1, 2). The severity and persistence of asthma differ between patients, and some patients present with airway hyperresponsiveness (AHR) and elevated markers of inflammation and/or atopy, whereas others present only with AHR (3, 4). Gene–environment interactions are also likely to be important in the development of asthma (5). Thus, although recent genome-wide association studies (GWAS) were successful at beginning to identify common genetic variants associated with asthma, many GWAS studies are underpowered to detect genetic loci that are important in certain phenotypes of asthma, and those studies are also underpowered to detect gene–environment interactions because of the large sample sizes and detailed exposure assessments required. Hence other complementary approaches are needed.
Hallmark features of asthma can be modeled in the mouse (6), and that mouse strain (i.e., genome) strongly affects the phenotype (7–9). For example, the two most commonly used inbred strains, C57BL/6J and BALB/cJ, differ in ovalbumin models of allergic disease (10, 11). Furthermore, the link between allergic inflammation and AHR is also strain-dependent and model-dependent, with certain strains manifesting either or both phenotypes as a function of the induction of different allergen-response pathways (e.g., IL-4/CD4+ T-cell–dependent pathways (12) versus IL-5/eosinophil–dependent pathways (13)), and of the route or timing of exposure (9).
We therefore exploited the strain-dependence of murine models of allergic airway disease to help understand the genomic basis of the different etiologic pathways that lead to the primary phenotypes of airway inflammation and AHR. We used the immunodominant allergen from the Dermatophagoides pteronyssinus species of house dust mite (HDM), Der p 1, a relevant human allergen. As expected based on previous studies (8, 11), C57BL/6J mice exhibited a stronger inflammatory response than did BALB/cJ mice, yet showed a striking decrease in airway responsiveness to methacholine. Using gene expression analysis, we identified a set of down-regulated G-protein–coupled receptors (GPCRs) involved in airway smooth muscle contraction that may mediate this response. Our results imply that airway smooth muscle gene expression is an important determinant of the physiologic response to allergen, and serves as one explanation for strain-dependent differences in murine models of allergic airway disease.
Materials and Methods
Mice
C57BL/6J and BALB/cJ male mice were purchased from Jackson Laboratory (Bar Harbor, ME) and used beginning at age 7–8 weeks. Mice were housed 3–5 to a cage within an Association for Assessment and Accreditation of Laboratory Animal Care–approved facility.
Allergen Protocol
Mice were sensitized intraperitoneally with 10 μg low endotoxin Der p 1 (Indoor Biotechnology, Charlottesville, VA), without exogenous adjuvant, on Days 1 and 7 of the study. On Day 14, Der p 1–sensitized mice were challenged by orotracheal aspiration with 50 μg of Der p 1 in 40 μl of saline, or by saline alone (control mice). Seventy-two hours after the airway challenge, the mice were phenotyped.
Whole-Lung Lavage and Histology
Whole-lung lavage was performed immediately after the mice were killed. After lavage, the left lung was inflated with formalin at 25 cm of pressure, and then sectioned and stained for mucin, using the method of Evans and colleagues (14).
Intracellular Cytokine Staining of Lung Lymphocytes and Flow Cytometry
Lung lymphocytes were isolated and stimulated with phorbol myristate acetate (10 ng/ml) and ionomycin (1 μg/ml) in the presence of brefeldin A (10 μg/ml). Cells were stained for the detection of intracellular IL-5, IL-13, IL-17, and IFN-γ (BD Biosciences, San Jose, CA), and the expression data for surface markers and intracellular cytokines were acquired on an LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Immunofluorescence Microscopy
Optimum cutting temperature compound-inflated, frozen lungs were sectioned at 8-μm thickness, fixed, and incubated with primary antibody to the muscarinic M2 receptor (10 μg/ml, catalogue number mAB367; Millipore, Billerica, MA), followed by goat anti-rabbit Alexa 555 (1:500) or primary antibody to the formyl peptide receptor (Fpr1; 4 μg/ml, catalogue number sc-13198; Santa Cruz Biotechnology, Santa Cruz, CA), followed by Alexa 555 secondary antibodies (1:500) and subsequently FITC-conjugated α–smooth muscle actin (1:250, catalogue number F3777; Sigma Chemical Co., St. Louis, MO). Sections were mounted in 4′,6-diamidino-2-phenylindole (DAPI)–containing medium, and imaged with a Zeiss LSM 510 confocal microscope (Thornton, NY).
Western Blot Analysis
Fifty micrograms of total lung protein were separated on 10–20% Tris–glycine gels, transferred to nitrocellulose membranes, and probed with primary antibodies for the M2 muscarinic receptor (Chrm2; 1:300 dilution, catalogue number AB-5166; Millipore) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:1,000 dilution, catalogue number sc-32233; Santa Cruz Biotechnology). Imaging and quantitation were performed using a Li-cor scanner (Li-Cor, Lincoln, NE).
Airway Hyperresponsiveness
We measured lung resistance using a Flexivent instrument (Scireq, Montreal, Quebec, Canada), with 10 mice per strain per group. Mice were anesthetized, tracheostomized, and paralyzed with pancuronium bromide to prevent voluntary respiration. Increasing doses of methacholine (0, 3, 6, 12, and 24 mg/ml) were administered by nebulization over a 10-second time period, and total lung resistance (RL) was monitored for 3 minutes per dose. Between each dose of methacholine, two inflations to total lung capacity were performed to normalize the lung.
RNA Isolation and Gene Expression Analyses
Total RNA from the lower right lobe was isolated (Qiagen RNEasy Kit, Valencia, CA), and 1 μg was used in protocols recommended by the manufacturer for GeneChip Mouse Exon 1.0 ST arrays (Affymetrix, Santa Clara, CA). The detailed array methods can be found in the online supplement and the accompanying Gene Expression Omnibus (GEO) accession (GSE19223).
Statistical Analysis
Student t tests were used to compare group means, and P < 0.05 was considered significant. Further details are provided in the online supplement.
Results
Increased Airway Inflammation in C57BL/6J Mice Compared with BALB/cJ Mice
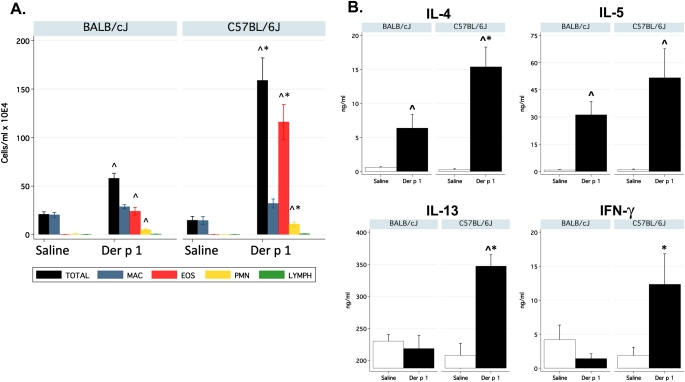
Seventy-two hours after airway challenge with Der p 1, the total number of white blood cells in whole-lung lavage fluid (WLLF) increased significantly in both BALB/cJ and C57BL/6J mice, compared with mice challenged with saline (Figure 1). However, C57BL/6J mice exhibited almost threefold more leukocytes than BALB/cJ mice after challenge with Der p 1 (159 ± 28 × 104 versus 58 ± 5 × 104, respectively; P = 8 × 10−4). In particular, Der p 1–induced airway eosinophilia was much greater in C57BL/6J mice compared with BALB/cJ mice. Within WLLF, Th2 cytokines were likewise greater in C57BL/6J mice compared with BALB/cJ mice (Figure 1B). C57BL/6J mice also manifested significantly greater concentrations of IFN-γ in WLLF.
Figure 1.
Increased airway inflammation in C57BL/6J mice. (A) Differential cell counts in whole-lung lavage fluid. (B) Cytokines in whole-lung lavage fluid (n = 6–8/group/strain). Der p 1, house dust mite allergen Dermatophagoides pteronyssinus. MAC = macrophage, EOS = eosinophils, PMN = neutrophils, LYMPH = lymphocytes. ^Significantly different from control mice. *Significantly different between strains.
Enhanced Th2 Cytokine Production after Lymphocyte Stimulation in C57BL/6J Mice
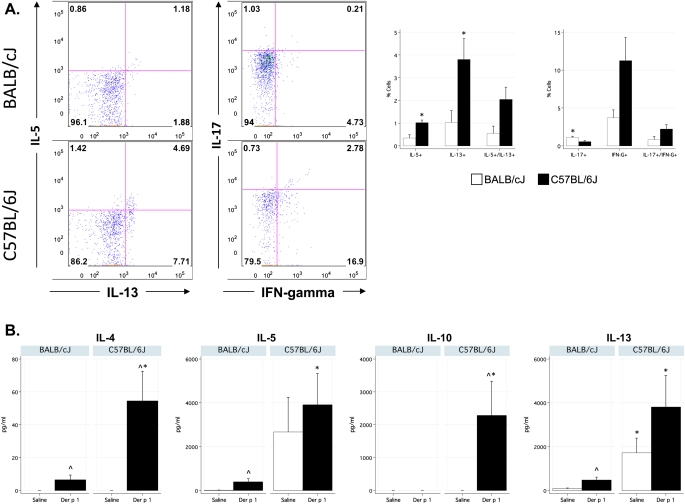
To determine the CD4+ lymphocyte phenotype after challenge with Der p 1, CD4+ lymphocytes were isolated from Der p 1–challenged BALB/cJ and C57BL/6J lung tissue, briefly stimulated in vitro, and analyzed for intracellular cytokine responses. C57BL/6J mice demonstrated a significantly higher frequency of Th2 cells (IL-5+, IL-13+, and IL-5+/IL-13+, Figure 2A) than did BALB/cJ mice. Notably, the percentage of CD4+ lymphocytes that expressed IL-13 and both IL-5+ and IL-13+ were significantly higher in the C57BL/6J strain among sensitized control mice (see Table E1 in the online supplement). C57BL/6J mice also showed an increase in IFN-γ, similar to the measurements in WLLF (Figure 1), although the difference did not reach significance. In general, BALB/cJ mice had fewer Th1 and Th2 cells. Numbers of Th17 cells, although low in frequency, were significantly higher in BALB/cJ mice, however.
Figure 2.
Increased Th2 responses in C57BL/6J mice after challenge with Der p 1. (A) Increased frequency of Th2 CD4+ lymphocytes in the lungs of Der p 1–challenged C57BL/6J mice. Left, Representative flow cytometry plots of CD4+ T cells. Numbers in plots represent the percentage of CD4+ T cells that were positive for each cytokine. Right, Summary data by strain (n = 4/group/strain). (B) Der p 1–specific cytokine release upon restimulation. Cells from mediastinal lymph nodes were isolated and cultured in the presence of PBS or 10 μg/ml Der p 1. Three days later, supernatants were collected and assayed. n = 6/group/strain. ^Significantly different from control mice. *Significantly different between strains.
To test antigen-specific cytokine responses, mediastinal lymph nodes from sensitized and challenged mice were restimulated in vitro with Der p 1. In general, C57BL/6J mice produced significantly more Th2 cytokines in response to Der p 1 (Figure 2B), correlating with higher frequencies of Th2 cells in the lung and higher frequencies of Th2 cytokines in WLLF. C57BL/6J mice also produced more Der p 1–specific IL-10, compared with BALB/cJ mice.
Increased Serum IgE and Der p 1–IgG1 in C57BL/6J Mice
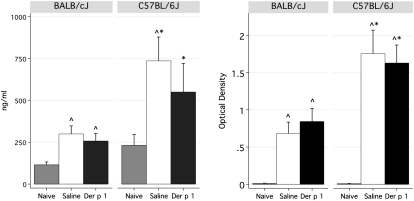
Total serum IgE increased in response to sensitization for both strains, and again was significantly higher in C57BL/6J mice (Figure 3), correlating with increased amounts of Th2 cells (Figure 2A) and IL-4 (Figures 1 and 2). Der p 1–specific IgE was not detectable in this acute, adjuvant-free model. However, titers of Der p 1–specific IgG1 were evident, and were significantly higher in C57BL/6J mice compared with BALB/cJ mice (Figure 3).
Figure 3.
Increased serum IgE and Der p 1–specific IgG1 in C57BL/6J mice. Left, Total IgE. Right, Der p 1–specific IgG1. Values for naive mice are shown for reference purposes (n = 6–8/group/strain). ^Significantly different from naive control mice. *Significantly different between strains.
Mucous Metaplasia Is Similar in BALB/cJ and C57BL/6J Mice
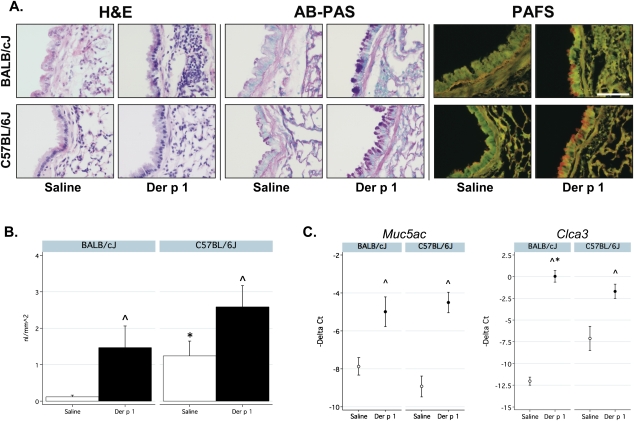
Alcian blue–periodic acid–Schiff staining of lungs showed increases in mucin production in allergen-challenged mice of both strains (Figure 4A). To quantify the degree of intracellular mucin production, we fluorescently stained fixed lung sections. Both strains showed evidence of increased mucin production in response to allergen challenge (Figure 4A), but the results did not differ by strain among allergen-challenged mice (Figure 4B). Interestingly, control C57BL/6J mice demonstrated higher concentrations of mucin compared with control BALB/cJ mice.
Figure 4.
Mucous metaplasia in both strains. (A) Hematoxylin and eosin (H&E), alcian blue–periodic acid–Schiff (AB-PAS), and periodic acid–fluorescent Schiff (PAFS)–stained sections of representative murine lungs. Scale bar, 150 μm. (B) Fluorescence measurements of mucus production, using PAFS staining (n = 6–8/group/strain). (C) Expression of mucin 5, subtypes A and C (Muc5ac) and chloride channel calcium activated 3 (Clca3, also known as Gob5) by quantitative RT-PCR. Data are presented as negative Δ Ct (−1 Delta Ct) to facilitate comparisons with data in A. Fold-change results are shown in Table E5 in the online supplement. ^Significantly different from control mice. *Significantly different between strains.
We measured the gene expression of chloride channel calcium activated 3 (Clca3, also known as Gob5) and mucin 5, subtypes A and C (Muc5ac), using quantitative RT-PCR. Consistent with the data for mucin, the expression of Muc5ac increased significantly in both strains, but did not differ as a function of strain (Figure 4C). However, the allergen-induced expression of the chloride channel Clca3 did differ significantly by strain.
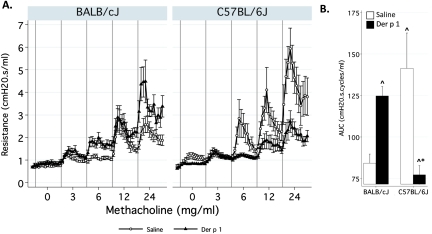
Resistance Is Increased in BALB/cJ Mice but Decreased in C57BL/6J Mice after Challenge with Der p 1
Total RL increased in response to challenge with Der p 1 in BALB/cJ mice (Figure 5). Area under the curve (AUC) analysis showed a significant increase of 48% (P = 0.001) in BALB/cJ mice. In contrast, C57BL/6J mice manifested a marked decrease in RL after challenge with Der p 1, and the AUC decreased by 45% (P = 0.006). Altogether, the results from this Der p 1 model show that AHR is coupled to inflammation in the BALB/cJ strain but not in the C57BL/6J strain, therefore providing an opportunity to investigate the different genomic response pathways that lead to, or protect from, the development of AHR.
Figure 5.
Resistance is increased in BALB/cJ mice, but decreased in C57BL/6J mice. (A) Total lung resistance (RL, cm H2O • second/ml) was measured using the forced oscillation technique (Flexivent). Gray vertical bars demarcate time periods (3 minutes each) spent measuring resistance at each concentration of methacholine. (B) Area under the curve (AUC) analysis of resistance data (n = 10–12/group/strain). ^Significantly different from control mice. *Significantly different between strains.
Gene Expression Analysis Reveals Shared and Distinct Patterns of Gene Expression
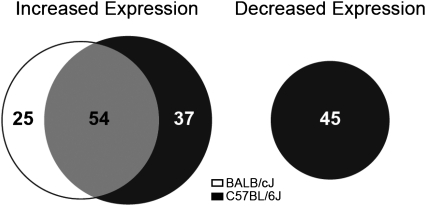
Challenge with Der p 1 caused significant changes in expression of 161 genes in lung tissue in either or both strains (Figure 6 and Table E2). Overall, 16% (25/161) of the genes identified by microarray were unique to BALB/cJ, whereas 51% (82/161) were unique to C57BL/6J. Hence 34% (54/161) of the genes were shared by both strains. The list of shared genes includes Arg1, Agr2, Clca3, Muc5ac, Ccl8, Ccl11, Chi3L3, Chi3L4, Chia, Scin, Retnla, Retnlb, Timp, Ear11, Mgl1, Saa3, Serpina3n, and Slc6a24, which were previously described in murine studies (15–22), adding further support to a role for these genes in allergic airway disease. A subset of these genes was evaluated and confirmed by quantitative RT-PCR (Table E4). Notably, among these shared genes, only Clca3 was differentially modulated by strain (P value for interaction = 4.9 × 10−4), with BALB/cJ mice showing a substantially higher increase in expression, correlating with the greater increase in mucin production (Figure 4) in that strain.
Figure 6.
Shared and distinct patterns of lung gene expression in BALB/cJ and C57BL/6J mice. The numbers in the venn diagram indicate the number of genes that were differentially expressed by Der p 1 treatment, and the size of the circles is proportional to the number of genes in each group. For the full list of differentially expressed genes, see Table E2 (n = 10–12/group/strain).
To illuminate the shared and distinct patterns of gene expression further, we performed pathway and gene ontology (GO) analysis (Table E5). Numerous pathways associated with cell-cycle regulation and mitosis were up-regulated in both strains, but were more significant in BALB/cJ mice, including pathways associated with cell-cycle progression and extracellular matrix remodeling. In contrast, chemokine receptor 3 (CCR3) signaling in eosinophils, IL-17 signaling, the production of arachidonic acid, and Toll-like receptor signaling were much more prominent in C57BL/6J mice, corroborating earlier findings of airway inflammation and cytokine production in this strain.
Strikingly, 45 genes showed significant decreases in expression in C57BL/6J mice that were not matched in BALB/cJ mice (Figure 6). The GO process analysis (Table E6) of these genes revealed a significant overrepresentation (P < 2 × 10−10) of genes encoding GPCRs implicated in the regulation of smooth muscle contraction, namely, Chrm2 (M2 muscarinic receptor), Pln (phospholamban), Fpr1 (Formyl peptide receptor), and Ptgfr (Prostaglandin F receptor). Quantitative RT-PCR confirmed the decrease in expression of these genes (Table E4). Note that BALB/cJ mice down-regulated Ptgfr, Pln, and Fpr1, but to a lesser extent. The formal test of strain–Der p 1 interactions using quantitative RT-PCR expression data was significant for Chrm2 and Fpr1, but not for Pln and Ptgfr (Table E4).
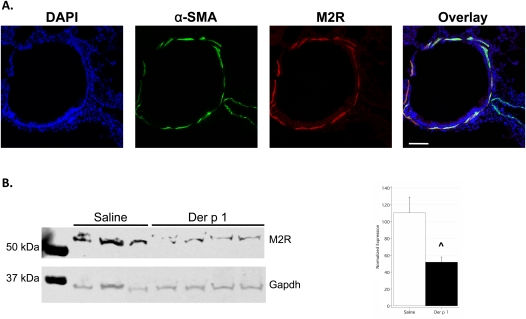
These results suggest that Chrm2 and Fpr1 are the most important GCPRs in terms of explaining the divergent physiologic responses to Der p 1 observed in C57BL/6J and BALB/cJ mice. Therefore, we sought to evaluate whether these two proteins are expressed at the putative site action, that is, airway smooth muscle (ASM). The expression of the M2 muscarinic receptor (M2R, encoded by Chrm2) in ASM was readily apparent (Figure 7A). The expression of Fpr1 was detected in white blood cells in the parenchyma (Figure E3), but not in ASM. Lastly, we measured M2R protein expression, using Western blot analysis, and detected a 57% decrease attributable to challenge with Der p 1 (Figure 7B), confirming the gene expression results.
Figure 7.
The M2 muscarinic receptor (M2R) is expressed in airway smooth muscle and is decreased after challenge with Der p 1. (A) Immunofluorescent detection of M2R in airway smooth muscle. Lung sections from C57BL/6J mice were stained using antibodies for α–smooth muscle actin (α-SMA) and M2R. Note the colocalization of α-SMA and the M2R in airway, but not in vascular smooth muscle. Scale bar, 100 μm. Similar results were obtained with BALB/cJ mice. (B) Western blot analysis of M2R expression in saline-challenged and Der p 1–challenged C57BL/6J mice. Densitometric analysis (right) shows a 57% reduction in expression, attributable to challenge with Der p 1. ^Significantly different from control mice. DAPI, 4′,6-diamidino-2-phenylindole; Gapdh, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
Our findings indicate that inflammation and airway hyperresponsiveness can be decoupled, and suggest that the transcriptional down-regulation of G-protein–coupled receptors involved in regulating the contraction of ASM may contribute to this dissociation. BALB/cJ mice produced classic Th2-mediated inflammatory responses coupled with AHR, and the degree of allergic inflammation correlated with the development of AHR, as reported previously (23). Gene expression changes in this strain included well-known allergen response genes, most notably a massive increase in Clca3 (gob5), a key mediator of goblet-cell hyperplasia. Conversely, C57BL/6J mice developed a stronger inflammatory response than did BALB/cJ mice, but exhibited a remarkable decrease in airway responsiveness to methacholine when challenged with Der p 1. These interstrain differences allowed us to identify GPCRs whose expression was significantly decreased after challenge with Der p 1 in C57BL/6J but not BALB/cJ mice. Hence our findings confirm that AHR and inflammation represent two distinct allergic airway disease phenotypes in mice (12, 24–26), and suggest that these differences are mediated by changes in expression of GPCRs.
The M2R is perhaps the strongest candidate, by virtue of its known role in mediating smooth muscle tone. Although the M3 muscarinic receptor (M3R) is better known for its direct role in the contraction of ASM, the M2R is expressed in ASM (Figure 7) (27), and plays a direct role in contraction, secondary to the M3 receptor (28). The M2R is also expressed at the parasympathetic prejunctional nerve terminal, but decreased expression at this site would ostensibly promote smooth muscle contraction because of its role in dampening signaling at the nerve terminal (29). Hence our data suggest that the effect we observed occurs in smooth muscle, and not in the nerve terminal.
Fpr1 is expressed in leukocytes (30), and is thought to modulate bronchoconstriction indirectly through the action of metabolites of arachidonic acid in guinea pigs and humans (31, 32). Previous reports documented the expression of Fpr1 in human bronchial ASM and in numerous other tissues (33), but we did not detect it in murine ASM using immunohistochemistry, arguing for an indirect role in the contraction of ASM. Although Pln is not differentially expressed as a function of strain–Der p 1 interaction, it is also noteworthy because it is known to modulate intracellular Ca2+ and the activity of the β–adrenergic receptor (34), and the ablation of Pln in mice results in decreased lung resistance (35).
Clearly, the detailed mechanisms leading to the down-regulation of these receptors merit further examination, and especially the question of whether this down-regulation represents some type of compensatory pathway.
We examined 14 publically available gene expression datasets from the National Center for Biotechnology Information (NCBI) GEO of murine models of allergic asthma, to see if these genes were down-regulated in other models. We analyzed those data and did not find similar changes in gene expression in Chrm2 (Table E7). Pln or Ptgfr were also not differentially expressed in these datasets. However, we found some evidence for the decreased expression of Fpr1, especially in the study by Kunikata and colleagues (36), although none of the results for Fpr1 were statistically significant after correction for multiple testing. Moreover, Kunikata and colleagues (36) did not observe decreased AHR after allergen challenge in their study. One additional report in PubMed by Di Valentin and colleagues (17) reported a number of down-regulated genes that overlap with our results, namely, Igfbp6, Angpt1, Sult1d1, Fpr1, Pon1, Ms4a4 d, Galntl2, Aldh3a1, and Fmo3, of which Fpr1 is perhaps the gene of most interest in this overlap. Di Valentin and colleagues (17) repeatedly challenged BALB/cJ mice with ovalbumin (over the short, intermediate, and long terms), and those mice exhibited signs of inflammation and AHR. Therefore, the decreased expression of Fpr1 alone will apparently not yield a decrease in AHR, and the decreased expression of multiple genes involved in ASM contraction is necessary to yield decreased methacholine responsiveness.
Previous reports have shown that it is possible to increase AHR in HDM-sensitized and challenged C57LB/6J mice. Shibamori and colleagues (37) used Der f extract in repeated intranasal sensitizations, and observed increased AHR after a single challenge with 100 μg of Der f. Tournoy and Schou (26) achieved increased AHR in this strain by sensitizing mice with Der p 1 plus alum with a chronic challenge protocol (Days 14–20). Importantly, others showed that the addition of adjuvant, namely alum, initiates a distinct inflammatory response involving the inflammosome (38). Hammad and colleagues (39) used three intratracheal challenges with HDM extract, and observed increased AHR (contingent on Tlr4). In comparison to these studies, one or more of our model parameters varied, namely, the allergen (extract versus purified single protein), the route of sensitization and use of adjuvant, and the dose and number of challenges. Other investigators showed that these factors profoundly affect the type and intensity of phenotype (9, 40, 41)). Hence these differences seem likely to account for the discrepancy between our results and those of the cited works, and further experiments are required to delineate the dependence of the AHR phenotype on these model variables.
We focused our attention on the divergent physiologic consequences of allergen-induced inflammation in these two strains, rather than on baseline differences (which can be related to intrinsic AHR) (42, 43), because we are interested in identifying the genome-by-environment interactions that underlie allergic asthma. However, some of the phenotypes we evaluated were significantly different among sensitized control mice. This was the case for the proportion of CD4+ lymphocytes from the lung that expressed IL-13 and both IL-5 and IL-13 (Table E1). As a result, the percent change in the expression of these cytokines was actually higher in BALB/cJ mice. It is unclear which metric of cytokine expression, percent change or total amount, better describes the effect of allergen challenge. BALB/cJ mice would be considered higher responders in terms of percent change, whereas C57BL/6J mice would be considered higher responders in terms of total amount. Furthermore, in these experiments, C57BL/6J mice demonstrated higher levels of airway responsiveness to methacholine at baseline, in contrast to some of our previous results (8). This most likely occurred because airway responsiveness was only measured at one dose of methacholine in our previous work, whereas here a full dose–response was performed, and C57BL/6J mice were clearly more responsive at higher doses.
In conclusion, our results demonstrate the importance of genomic factors in determining responses to allergen sensitization and challenge, and the strain–Der p 1 interaction we identified offers new insights about the role of GCPRs in regulating ASM contraction, and about the impact of GCPRs on AHR. Our model and research strategy are well-suited to further, more detailed explorations of both the genomic pathways that lead to allergic phenotypes and the genetic basis of differential response. In particular, the question of which genes dictate different genomic responses is critically important. We plan to address this question using a quantitative trait locus mapping approach and a new murine genetics reference panel, known as the Collaborative Cross (44).
Supplementary Material
Acknowledgments
The authors thank Shelley Hoogstraten-Miller for significant training and input on the murine model, Abdel Elkahloun for microarray support, and Michael Erdos for substantial advice during the course of data generation and analysis. We would like to acknowledge the professional skills and advice of Dr. Christian A. Combs and Daniela Malide (Light Microscopy Core Facility, National Heart, Lung and Blood Institute, National Institutes of Health) regarding microscopy-related experiments performed in this paper. This work was supported by the intramural research programs of the National Human Genome Research Institute and the National Institute of Environmental Health Sciences, and by R01 HL080396 (NIH) and 10GRNT4200070 (Am. Heart Assoc.) to C. Evans.
Footnotes
This work was supported by the Intramural Research Program of the National Human Genome Research Institute at the National Institutes of Health (F.S.C.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2010-0315OC on March 18, 2011
Author Disclosure: D.A.S. received expert witness fees from Wallace and Graham, Brayton and Purcell, Weitz and Luxemberg, and Waters and Kraus, and is a full-time employee of National Jewish Health. The rest of the authors have no financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Kauffmann F, Dizier M-H, Pin I, Paty E, Gormand F, Vervloet D, Bousquet J, Neukirch F, Annesi I, Oryszczyn M-P, et al. Epidemiological study of the genetics and environment of asthma, bronchial hyperresponsiveness, and atopy: phenotype issues. Am J Respir Crit Care Med 1997;156:S123–S129 [DOI] [PubMed] [Google Scholar]
- 2.Pillai SG, Tang Y, van den Oord E, Klotsman M, Barnes K, Carlsen K, Gerritsen J, Lenney W, Silverman M, Sly P, et al. Factor analysis in the genetics of asthma international network family study identifies five major quantitative asthma phenotypes. Clin Exp Allergy 2008;38:421–429 [DOI] [PubMed] [Google Scholar]
- 3.Nadif R, Siroux V, Oryszczyn M-P, Ravault C, Pison C, Pin I, Kauffmann F. Heterogeneity of asthma according to blood inflammatory patterns. Thorax 2009;64:374–380 [DOI] [PubMed] [Google Scholar]
- 4.Douwes J, Gibson P, Pekkanen J, Pearce N. Non-eosinophilic asthma: importance and possible mechanisms. Thorax 2002;57:643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez FD. Gene–environment interactions in asthma: with apologies to William of Ockham. Proc Am Thorac Soc 2007;4:26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkelman FD, Wills-Karp M. Usefulness and optimization of mouse models of allergic airway disease. J Allergy Clin Immunol 2008;121:603–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brewer JP, Kisselgof AB, Martin TR. Genetic variability in pulmonary physiological, cellular, and antibody responses to antigen in mice. Am J Respir Crit Care Med 1999;160:1150–1156 [DOI] [PubMed] [Google Scholar]
- 8.Whitehead GS, Walker JKL, Berman KG, Foster WM, Schwartz DA. Allergen-induced airway disease is mouse strain dependent. Am J Physiol Lung Cell Mol Physiol 2003;285:L32–L42 [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Lamm WJ, Albert RK, Chi EY, Henderson WR, Lewis DB. Influence of the route of allergen administration and genetic background on the murine allergic pulmonary response. Am J Respir Crit Care Med 1997;155:661–669 [DOI] [PubMed] [Google Scholar]
- 10.Gueders M, Paulissen G, Crahay C, Quesada-Calvo F, Hacha J, Van Hove C, Tournoy K, Louis R, Foidart J-M, Nol A, et al. Mouse models of asthma: a comparison between C57BL/6 and BALB/c strains regarding bronchial responsiveness, inflammation, and cytokine production. Inflamm Res 2009;58:845–854 [DOI] [PubMed] [Google Scholar]
- 11.Van Hove CL, Maes T, Cataldo DD, Gueders MM, Palmans E, Joos GF, Tournoy KG. Comparison of acute inflammatory and chronic structural asthma-like responses between C57BL/6 and BALB/c mice. Int Arch Allergy Immunol 2009;149:195–207 [DOI] [PubMed] [Google Scholar]
- 12.Locksley RM, Wiener-Kronish JP, Matthay MA, Erle DJ, Warnock ML, Folkesson HG, Corry DB. Interleukin 4, but not interleukin 5 or eosinophils, is required in a murine model of acute airway hyperreactivity. J Exp Med 1996;183:109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Dimina D, Macias M, Ochkur S, McGarry M, O'Neill K, Protheroe C, Pero R, Nguyen T, Cormier S, et al. Defining a link with asthma in mice congenitally deficient in eosinophils. Science 2004;305:1773–1776 [DOI] [PubMed] [Google Scholar]
- 14.Evans CM, Williams OW, Tuvim MJ, Nigam R, Mixides GP, Blackburn MR, DeMayo FJ, Burns AR, Smith C, Reynolds SD, et al. Mucin is produced by Clara cells in the proximal airways of antigen-challenged mice. Am J Respir Cell Mol Biol 2004;31:382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Follettie MT, Ellis DK, Donaldson DD, Hill AA, Diesl V, DeClercq C, Sypek JP, Dorner AJ, Wills-Karp M. Gene expression analysis in a murine model of allergic asthma reveals overlapping disease and therapy dependent pathways in the lung. Pharmacogenomics J 2006;6:141–152 [DOI] [PubMed] [Google Scholar]
- 16.Lewis CC, Yang JY, Huang X, Banerjee SK, Blackburn MR, Baluk P, McDonald DM, Blackwell TS, Nagabhushanam V, Peters W, et al. Disease-specific gene expression profiling in multiple models of lung disease. Am J Respir Crit Care Med 2008;177:376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Valentin E, Crahay C, Garbacki N, Hennuy B, Gueders M, Noel A, Foidart J-M, Grooten J, Colige A, Piette J, et al. New asthma biomarkers: lessons from murine models of acute and chronic asthma. Am J Physiol Lung Cell Mol Physiol 2009;296:L185–L197 [DOI] [PubMed] [Google Scholar]
- 18.Lewis CC, Aronow B, Hutton J, Santeliz J, Dienger K, Herman N, Finkelman FD, Wills-Karp M. Unique and overlapping gene expression patterns driven by IL-4 and IL-13 in the mouse lung. J Allergy Clin Immunol 2009;123:795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SG, Choi JW, Kim H, Roh GS, Bok J, Go MJ, Kwack K, Oh B, Kim Y. Genome-wide profiling of antigen-induced time course expression using murine models for acute and chronic asthma. Int Arch Allergy Immunol 2008;146:44–56 [DOI] [PubMed] [Google Scholar]
- 20.Walker JK, Ahumada A, Frank B, Gaspard R, Berman K, Quackenbush J, Schwartz DA. Multistrain genetic comparisons reveal CCR5 as a receptor involved in airway hyperresponsiveness. Am J Respir Cell Mol Biol 2006;34:711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest 2003;111:1863–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumes DJ, Connolly A, Dent LA. Expression of survivin in lung eosinophils is associated with pathology in a mouse model of allergic asthma. Int Immunol 2009;21:633–644 [DOI] [PubMed] [Google Scholar]
- 23.Takeda K, Haczku A, Lee JJ, Irvin CG, Gelfand EW. Strain dependence of airway hyperresponsiveness reflects differences in eosinophil localization in the lung. Am J Physiol Lung Cell Mol Physiol 2001;281:L394–L402 [DOI] [PubMed] [Google Scholar]
- 24.Humbles A, Lloyd C, McMillan S, Friend D, Xanthou G, McKenna E, Ghiran S, Gerard N, Yu C, Orkin S, et al. A critical role for eosinophils in allergic airways remodeling. Science 2004;305:1776–1779 [DOI] [PubMed] [Google Scholar]
- 25.Mathur M, Herrmann K, Li X, Qin Y, Weinstock J, Elliott D, Monahan J, Padrid P. Trfk-5 reverses established airway eosinophilia but not established hyperresponsiveness in a murine model of chronic asthma. Am J Respir Crit Care Med 1999;159:580–587 [DOI] [PubMed] [Google Scholar]
- 26.Tournoy K, Schou P. Airway eosinophilia is not a requirement for allergen-induced airway hyperresponsiveness. Clin Exp Allergy 2000;30:79–85 [DOI] [PubMed] [Google Scholar]
- 27.Jositsch G, Papadakis T, Haberberger R, Wolff M, Wess J, Kummer W. Suitability of muscarinic acetylcholine receptor antibodies for immunohistochemistry evaluated on tissue sections of receptor gene–deficient mice. Naunyn Schmiedebergs Arch Pharmacol 2009;379:389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Struckmann N, Schwering S, Wiegand S, Gschnell A, Yamada M, Wess KW, Jr, Haberberger RV. Role of muscarinic receptor subtypes in the constriction of peripheral airways: studies on receptor-deficient mice. Mol Pharmacol 2003;64:1444–1451 [DOI] [PubMed] [Google Scholar]
- 29.Coulson FR, Fryer AD. Muscarinic acetylcholine receptors and airway diseases. Pharmacol Ther 2003;98:59–69 [DOI] [PubMed] [Google Scholar]
- 30.Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: a promiscuous subfamily of G protein–coupled receptors controlling immune responses. Cytokine Growth Factor Rev 2006;17:501–519 [DOI] [PubMed] [Google Scholar]
- 31.Hamel R, Ford-Hutchinson AW, Lord A, Cirino M. Bronchoconstriction induced by n-formyl–methionyl–leucyl–phenylalanine in the guinea pig: involvement of arachidonic acid metabolites. Prostaglandins 1984;28:43–56 [DOI] [PubMed] [Google Scholar]
- 32.Armour CL, Black JL, Johnson PR, Vincenc KS, Berend N. Formyl peptide–induced contraction of human airways in vitro. J Appl Physiol 1986;60:141–146 [DOI] [PubMed] [Google Scholar]
- 33.Becker EL, Forouhar FA, Grunnet ML, Boulay F, Tardif M, Bormann BJ, Sodja D, Ye RD, Woska JR, Murphy PM. Broad immunocytochemical localization of the formylpeptide receptor in human organs, tissues, and cells. Cell Tissue Res 1998;292:129–135 [DOI] [PubMed] [Google Scholar]
- 34.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol 2003;4:566–577 [DOI] [PubMed] [Google Scholar]
- 35.McGraw DW, Fogel KM, Kong S, Litonjua AA, Kranias EG, Aronow BJ, Liggett SB. Transcriptional response to persistent {beta}2–adrenergic receptor signaling reveals regulation of phospholamban, which alters airway contractility. Physiol Genomics 2006;27:171–177 [DOI] [PubMed] [Google Scholar]
- 36.Kunikata T, Yamane H, Segi E, Matsuoka T, Sugimoto Y, Tanaka S, Tanaka H, Nagai H, Ichikawa A, Narumiya S. Suppression of allergic inflammation by the prostaglandin E receptor subtype EP3. Nat Immunol 2005;6:524–531 [DOI] [PubMed] [Google Scholar]
- 37.Shibamori M, Ogino K, Kambayashi Y, Ishiyama H. Intranasal mite allergen induces allergic asthma-like responses in NC/NGA mice. Life Sci 2006;78:987–994 [DOI] [PubMed] [Google Scholar]
- 38.Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 2008;453:1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammad H, Chieppa M, Perros F, Willart MA, Germain RN, Lambrecht BN. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat Med 2009;15:410–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, Toll-like receptor 4–dependent T helper cell Type 2 responses to inhaled antigen. J Exp Med 2002;196:1645–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson RH, Whitehead GS, Nakano H, Free ME, Kolls JK, Cook DN. Allergic sensitization through the airway primes Th17-dependent neutrophilia and airway hyperresponsiveness. Am J Respir Crit Care Med 2009;180:720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hadeiba H, Corry DB, Locksley RM. Baseline airway hyperreactivity in A/J mice is not mediated by cells of the adaptive immune system. J Immunol 2000;164:4933–4940 [DOI] [PubMed] [Google Scholar]
- 43.Wagers SS, Haverkamp HC, Bates JHT, Norton RJ, Thompson-Figueroa JA, Sullivan MJ, Irvin CG. Intrinsic and antigen-induced airway hyperresponsiveness are the result of diverse physiological mechanisms. J Appl Physiol 2007;102:221–230 [DOI] [PubMed] [Google Scholar]
- 44.Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 2004;36:1133–1137 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.