Abstract
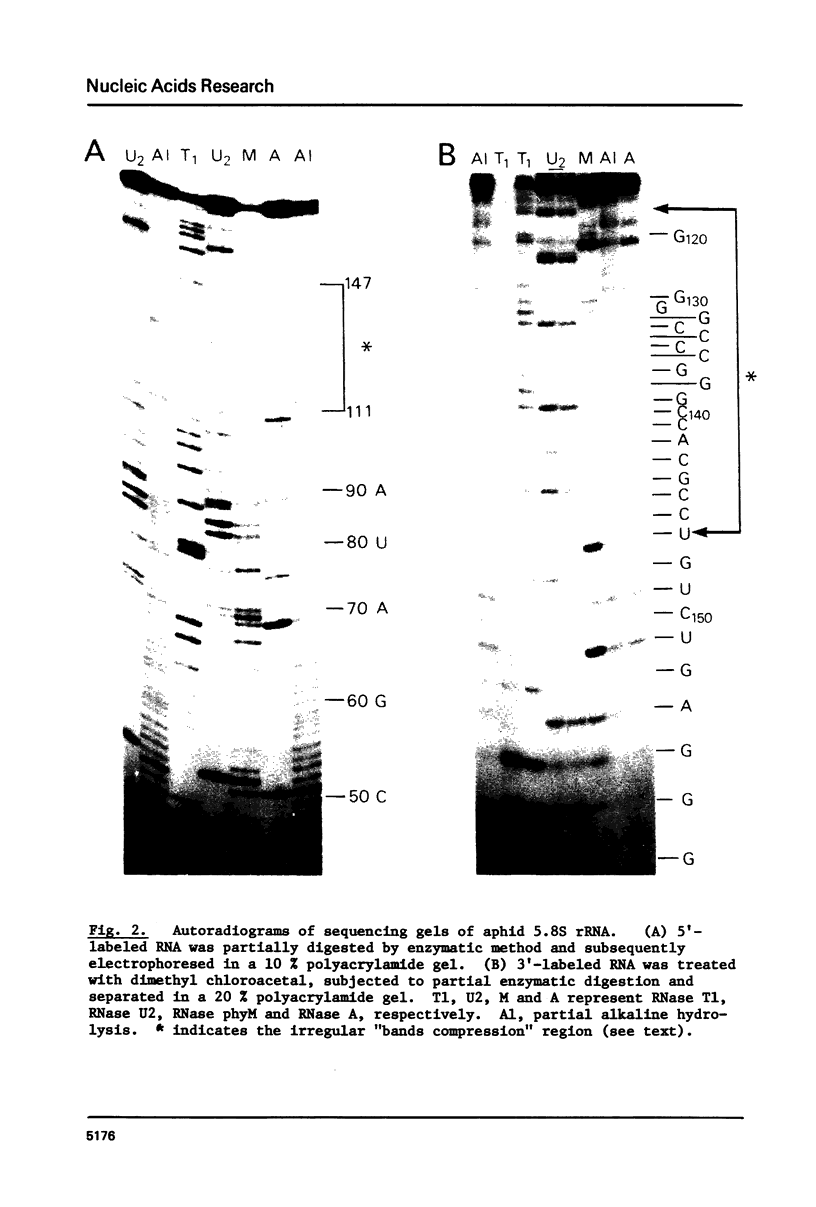
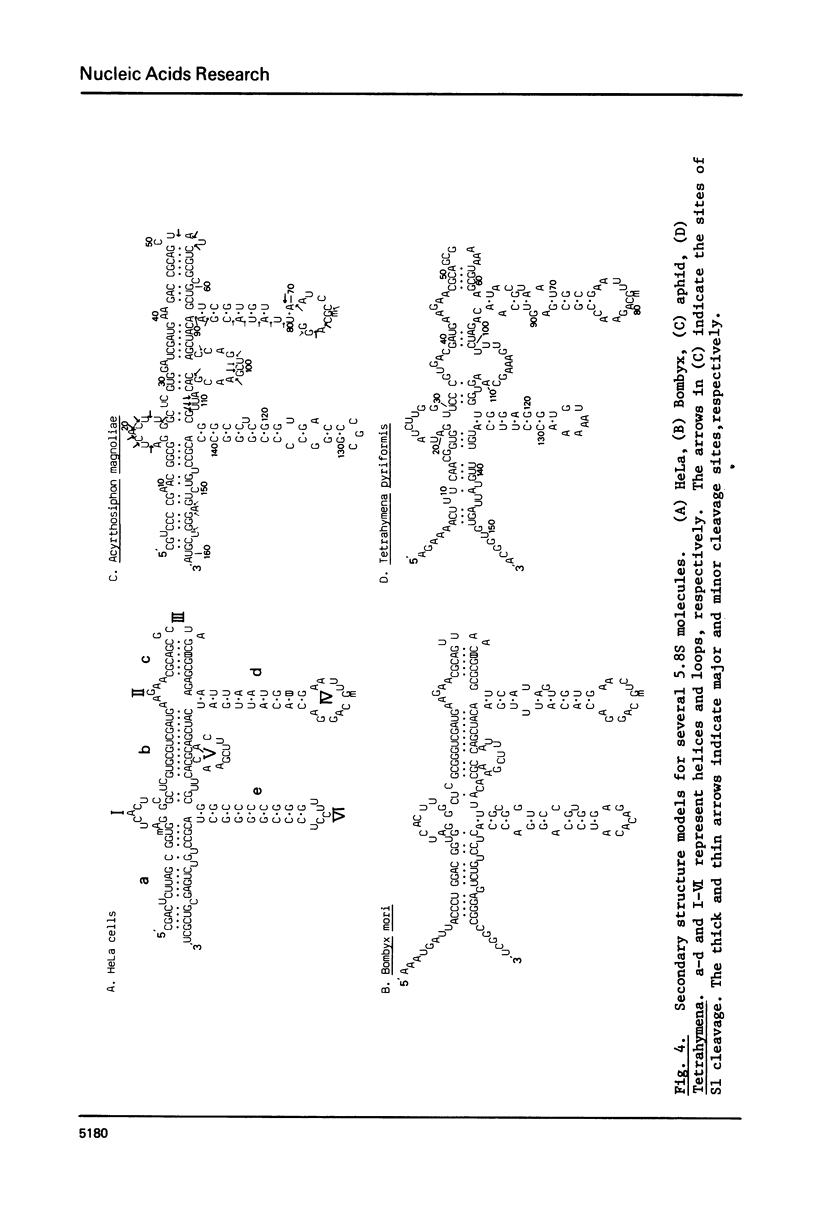
The Tetrahymena 5.8S rRNA is 154 nucleotides long, the shortest so far reported except for the split 5.8S rRNAs of Diptera (m5.8S plus 2S rRNA). In this molecule several nucleotides are deleted in the helix e (GC-rich stem) region. Upon constructing the secondary structure in accordance with "burp-gun" model, the Tetrahymena 5.8S rRNA forms a wide-open "muzzle" of the terminal regions due to both extra nucleotides and several unpaired bases. The aphid 5.8S rRNA consists of 161 nucleotides and can form stable helices in both terminal and helix e regions. As a whole, the secondary structure of Tetrahymena 5.8S rRNA resembles that of Bombyx 5.8S molecule while the aphid 5.8S rRNA shares several structural features with the HeLa 5.8S molecule. Likely, the 5.8S rRNA attached to the 28S rRNA with the hidden break differs in structure from those interacting with the 28S partners without the break. Nucleotide sequences of 5.8S rRNA in insects as well as in protozoans are not so conservative evolutionarily as in vertebrates.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Addison W. R., Gillam I. C., Tener G. M. The nucleotide sequence of tRNA4Val of Drosophila melanogaster. Chloroacetaldehyde modification as an aid to RNA sequencing. J Biol Chem. 1982 Jan 25;257(2):674–677. [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S RNA sequences and their precursors. Nucleic Acids Res. 1982 Jan 22;10(2):r93–115. doi: 10.1093/nar/10.2.762-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H., Kawata Y., Ishikawa H. Primary and secondary structure of 5.8S rRNA from the silkgland of Bombyx mori. Nucleic Acids Res. 1982 Apr 10;10(7):2415–2418. doi: 10.1093/nar/10.7.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Klotz L. C., Blanken R. L., Loeblich A. R., 3rd An evaluation of the phylogenetic position of the dinoflagellate Crypthecodinium cohnii based on 5S rRNA characterization. J Mol Evol. 1981;17(6):334–337. doi: 10.1007/BF01734355. [DOI] [PubMed] [Google Scholar]
- Ishikawa H. Arthropod ribosomes. Integrity of ribosomal ribonucleic acids from aphids and water fleas. Biochim Biophys Acta. 1976 Jul 2;435(3):258–268. doi: 10.1016/0005-2787(76)90107-6. [DOI] [PubMed] [Google Scholar]
- Ishikawa H. Re-joining of the 18S fragments dissociated from the 28S ribosomal RNA of insect: a structural role of 5.8S RNA. Biochem Biophys Res Commun. 1979 Sep 27;90(2):417–424. doi: 10.1016/0006-291x(79)91251-8. [DOI] [PubMed] [Google Scholar]
- Kao T. H., Crothers D. M. A proton-coupled conformational switch of Escherichia coli 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3360–3364. doi: 10.1073/pnas.77.6.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata Y., Ishikawa H. Nucleotide sequence and thermal property of 5S rRNA from the elder aphid. Acyrthosiphon magnoliae. Nucleic Acids Res. 1982 Mar 25;10(6):1833–1840. doi: 10.1093/nar/10.6.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. M., Cox R. A. The nucleotide sequence at the 3'-end of Neurospora crassa 25S-rRNA and the location of a 5.8S-rRNA binding site. Nucleic Acids Res. 1981 Mar 11;9(5):1111–1121. doi: 10.1093/nar/9.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Kato M., Sugisaki H., Nishimura S. Nucleotide sequence of starfish initiator tRNA. Nucleic Acids Res. 1979 Aug 10;6(11):3459–3469. doi: 10.1093/nar/6.11.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay R. M., Spencer D. F., Doolittle W. F., Gray M. W. Nucleotide sequences of wheat-embryo cytosol 5-S and 5.8-S ribosomal ribonucleic acids. Eur J Biochem. 1980 Dec;112(3):561–576. doi: 10.1111/j.1432-1033.1980.tb06122.x. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Structural analyses of mammalian ribosomal ribonucleic acid and its precursors. Nucleotide sequence of ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1975 Nov 25;250(22):8591–8597. [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O. Role of the 5'-terminal sequence in the RNA binding site of yeast 5.8 S rRNA. FEBS Lett. 1980 Jun 16;115(1):71–76. doi: 10.1016/0014-5793(80)80729-0. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Sogin M. L. Nucleotide sequence of Dictyostelium discoideum 5.8S ribosomal ribonucleic acid: evolutionary and secondary structural implications. Biochemistry. 1982 May 11;21(10):2335–2343. doi: 10.1021/bi00539a010. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M. A., Walker T. A., Pace N. R. Independent binding sites in mouse 5.8S ribosomal ribonucleic acid for 28S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2329–2335. doi: 10.1021/bi00539a009. [DOI] [PubMed] [Google Scholar]
- Schnare M. N., Gray M. W. Nucleotide sequence of an exceptionally long 5.8S ribosomal RNA from Crithidia fasciculata. Nucleic Acids Res. 1982 Mar 25;10(6):2085–2092. doi: 10.1093/nar/10.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberklang M., Prochiantz A., Haenni A. L., Rajbhandary U. L. Studies on the sequence of the 3'-terminal region of turnip-yellow-mosaic-virus RNA. Eur J Biochem. 1977 Feb;72(3):465–478. doi: 10.1111/j.1432-1033.1977.tb11270.x. [DOI] [PubMed] [Google Scholar]
- Ursi D., Vandenberghe A., De Wachter R. The sequence of the 5.8 S ribosomal RNA of the crustacean Artemia salina. With a proposal for a general secondary structure model for 5.8 S ribosomal RNA. Nucleic Acids Res. 1982 Jun 11;10(11):3517–3530. doi: 10.1093/nar/10.11.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., van Heerikhuizen H., Planta R. J. The nucleotide sequence of the intergenic region between the 5.8S and 26S rRNA genes of the yeast ribosomal RNA operon. Possible implications for the interaction between 5.8S and 26S rRNA and the processing of the primary transcript. Nucleic Acids Res. 1981 Oct 10;9(19):4847–4862. doi: 10.1093/nar/9.19.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. A., Johnson K. D., Olsen G. J., Peters M. A., Pace N. R. Enzymatic and chemical structure mapping of mouse 28S ribosomal ribonucleic acid contacts in 5.8S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2320–2329. doi: 10.1021/bi00539a008. [DOI] [PubMed] [Google Scholar]