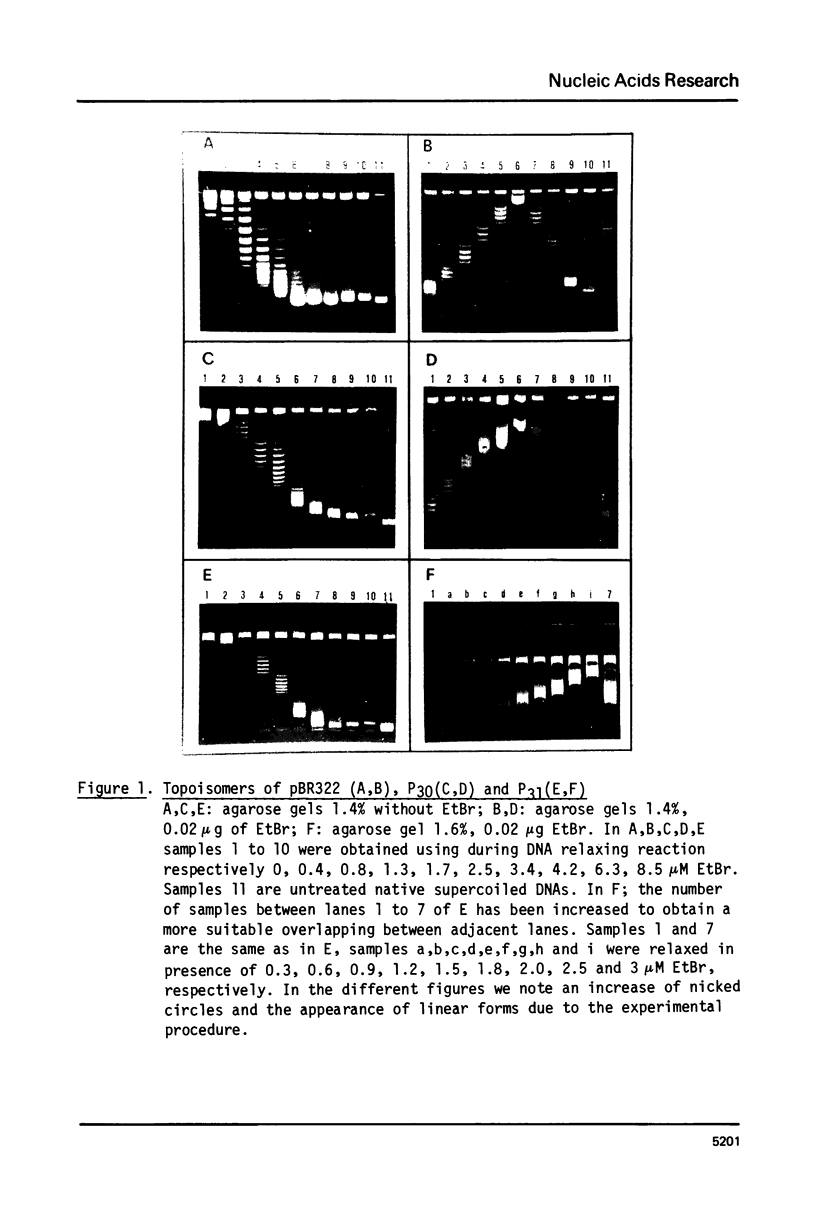
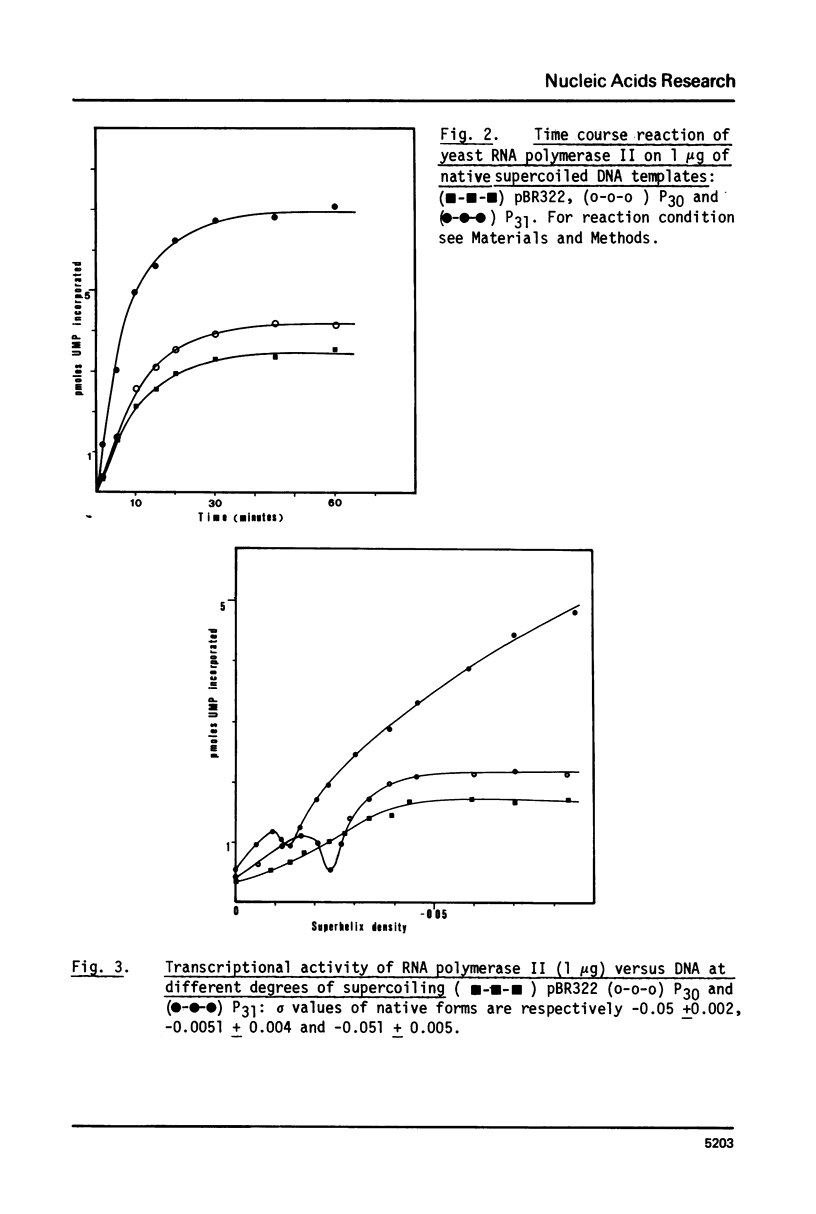
Abstract
Purified yeast RNA polymerase II was tested for transcriptional activity as a function of the degree of circular DNA supercoiling. Chimaeric plasmids P30 and P31 both containing inserts from the yeast transposable element TY1 cloned in pBR322 and the vector pBR322 were used as templates. For pBR322 the transcriptional activity increases about 4 fold from the fully relaxed covalently closed circles to the native supercoiled forms, further supercoiling having no effect on transcription. P30 shows a 5 fold increase of transcriptional activity reaching a plateau at the native supercoiled conformation. However, at an intermediate degree of supercoiling (sigma = 0.024), transcription decreases to a value close to zero. P31 too exhibits a conformation (sigma = 0.014) in which there is a drop of transcriptional activity. Furthermore, a 10 fold increase of transcription is obtained at the higher values of superhelix density. Both kinetic and autoradiographic experiments confirm the existence of DNA conformations that can inhibit "in vitro" transcription.
Full text
PDF






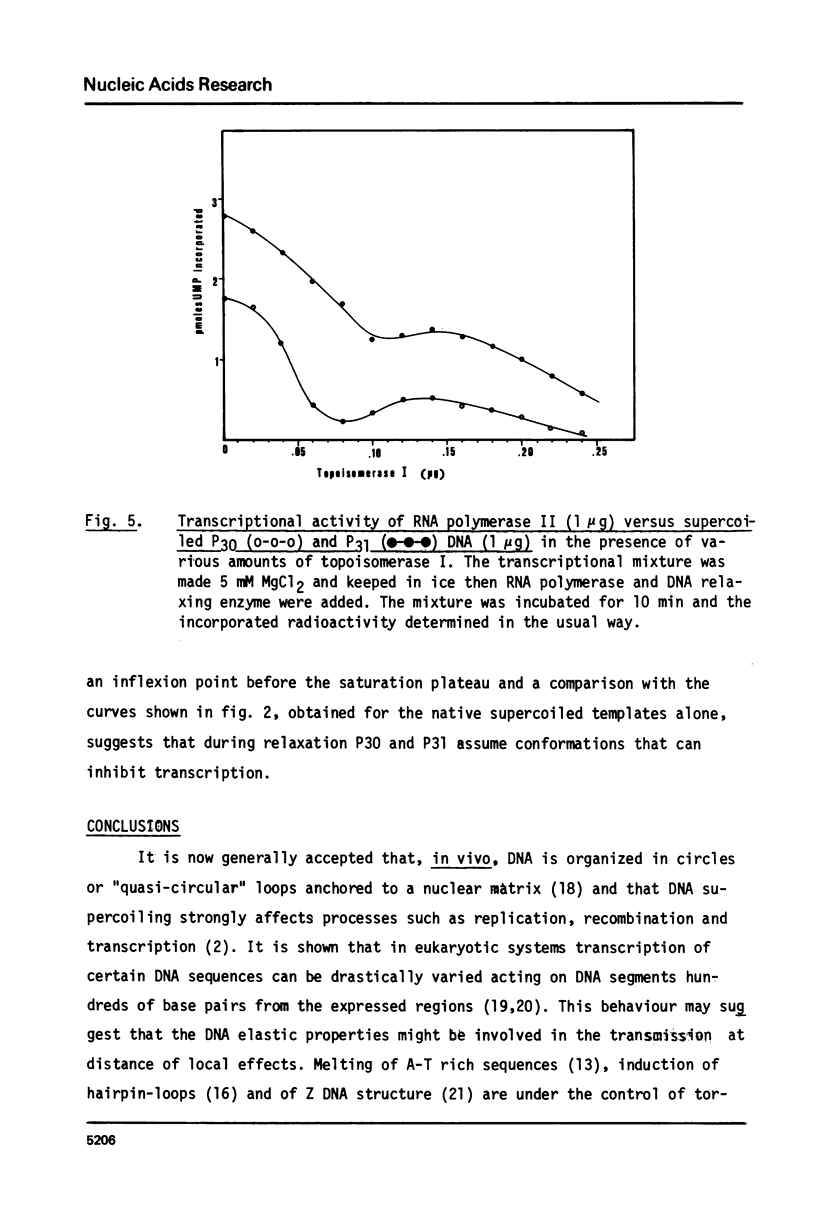
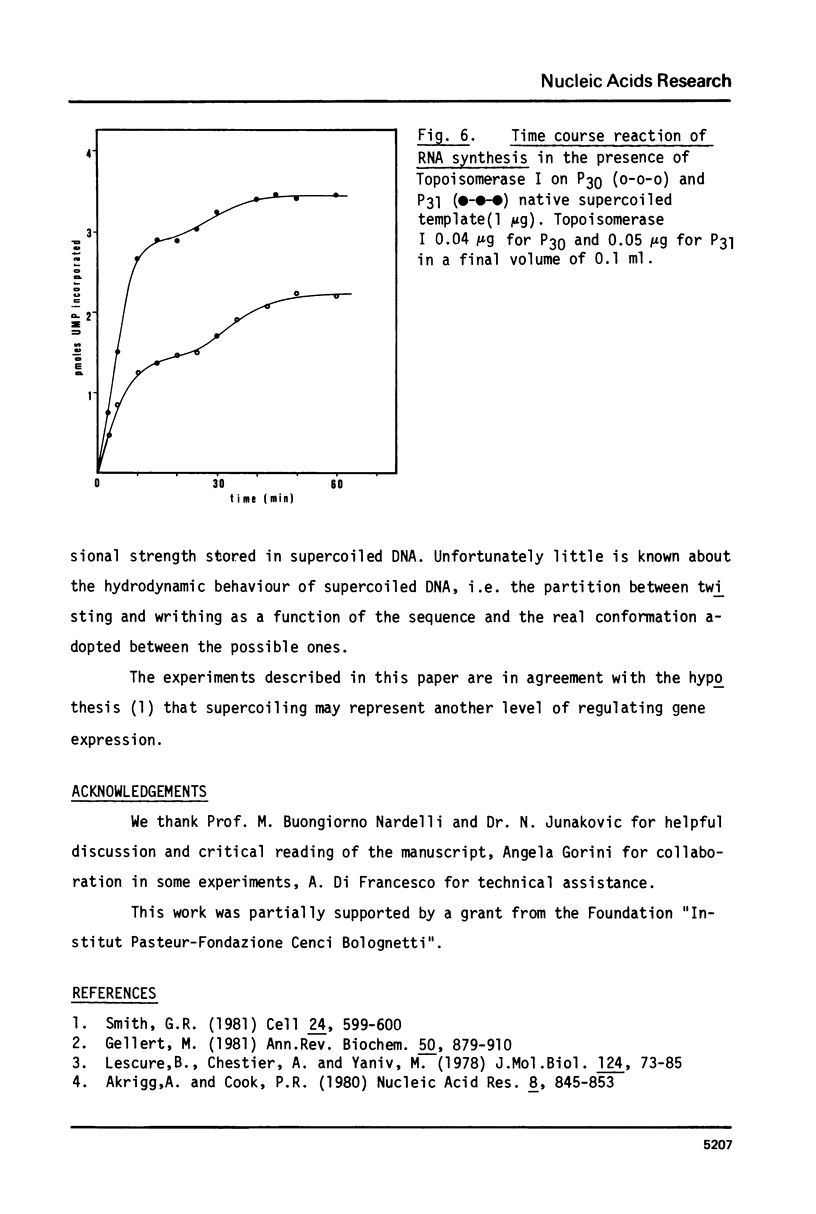

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akrigg A., Cook P. R. DNA gyrase stimulates transcription. Nucleic Acids Res. 1980 Feb 25;8(4):845–854. [PMC free article] [PubMed] [Google Scholar]
- Ballario P., Buongiorno-Nardelli M., Carnevali F., Di Mauro E., Pedone F. Selective in vitro transcription by purified yeast RNA polymerase II on cloned 2 micron DNA. Nucleic Acids Res. 1981 Aug 25;9(16):3959–3978. doi: 10.1093/nar/9.16.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina-Stein M., Vogel T., Singer D. S., Singer M. F. H5 Histone and DNA-relaxing enzyme of chicken erythrocytes. Interaction with superhelical DNA. J Biol Chem. 1976 Dec 10;251(23):7363–7366. [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Davies D. R., Zimmerman S. A new twist for DNA? Nature. 1980 Jan 3;283(5742):11–12. doi: 10.1038/283011a0. [DOI] [PubMed] [Google Scholar]
- Dezélée S., Sentenac A. Role of DNA-RNA hybrids in eukaryotes. Purification and properties of yeast RNA polymerase B. Eur J Biochem. 1973 Apr 2;34(1):41–52. doi: 10.1111/j.1432-1033.1973.tb02726.x. [DOI] [PubMed] [Google Scholar]
- Elder R. T., St John T. P., Stinchcomb D. T., Davis R. W., Scherer S., Davis R. W. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. II. Recombination and expression of Ty1 and adjacent sequences. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):581–591. doi: 10.1101/sqb.1981.045.01.075. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Broach J. R., Hicks J. B. Regulation of transcription in expressed and unexpressed mating type cassettes of yeast. Nature. 1981 Jan 22;289(5795):239–244. doi: 10.1038/289239a0. [DOI] [PubMed] [Google Scholar]
- Lescure B., Chestier A., Yaniv M. Transcription of polyoma virus DNA in vitro. II. Transcription of superhelical and linear polyoma DNA by RNA polymerase II. J Mol Biol. 1978 Sep 5;124(1):73–85. doi: 10.1016/0022-2836(78)90148-1. [DOI] [PubMed] [Google Scholar]
- Lescure B., Williamson V., Sentenac A. Efficient and selective initiation by yeast RNA polymerase B in a dinucleotide-primed reaction. Nucleic Acids Res. 1981 Jan 10;9(1):31–45. doi: 10.1093/nar/9.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. K., Burgess R. R. Transcription of simian virus 40 DNA by wheat germ RNA polymerase II. Priming of RNA synthesis by the 3'-hydroxyl of DNA at single strand nicks. J Biol Chem. 1980 May 25;255(10):4928–4936. [PubMed] [Google Scholar]
- Lilley D. M. In vivo consequences of plasmid topology. Nature. 1981 Jul 23;292(5821):380–382. doi: 10.1038/292380a0. [DOI] [PubMed] [Google Scholar]
- Shure M., Vinograd J. The number of superhelical turns in native virion SV40 DNA and minicol DNA determined by the band counting method. Cell. 1976 Jun;8(2):215–226. doi: 10.1016/0092-8674(76)90005-2. [DOI] [PubMed] [Google Scholar]
- Smith G. R. DNA supercoiling: another level for regulating gene expression. Cell. 1981 Jun;24(3):599–600. doi: 10.1016/0092-8674(81)90085-4. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]