Abstract
Rationale: Defining the biogeography of bacterial populations in human body habitats is a high priority for understanding microbial–host relationships in health and disease. The healthy lung was traditionally considered sterile, but this notion has been challenged by emerging molecular approaches that enable comprehensive examination of microbial communities. However, studies of the lung are challenging due to difficulties in working with low biomass samples.
Objectives: Our goal was to use molecular methods to define the bacterial microbiota present in the lungs of healthy individuals and assess its relationship to upper airway populations.
Methods: We sampled respiratory flora intensively at multiple sites in six healthy individuals. The upper tract was sampled by oral wash and oro-/nasopharyngeal swabs. Two bronchoscopes were used to collect samples up to the glottis, followed by serial bronchoalveolar lavage and lower airway protected brush. Bacterial abundance and composition were analyzed by 16S rDNA Q-PCR and deep sequencing.
Measurements and Main Results: Bacterial communities from the lung displayed composition indistinguishable from the upper airways, but were 2 to 4 logs lower in biomass. Lung-specific sequences were rare and not shared among individuals. There was no unique lung microbiome.
Conclusions: In contrast to other organ systems, the respiratory tract harbors a homogenous microbiota that decreases in biomass from upper to lower tract. The healthy lung does not contain a consistent distinct microbiome, but instead contains low levels of bacterial sequences largely indistinguishable from upper respiratory flora. These findings establish baseline data for healthy subjects and sampling approaches for sequence-based analysis of diseases.
Keywords: healthy lung colonization, microbiome, 16S rDNA, pyrosequencing
At a Glance Commentary
Scientific Knowledge on the Subject
Microbes are normally resident in many body sites with connections to the external environment, and it is increasingly clear that they play important roles in health, as well as disease when disrupted. The lung is classically thought to be sterile. New molecular techniques can comprehensively describe an entire population of bacteria without relying on the ability to culture specific organisms, and have suggested that the lung may harbor its own unique microbiome.
What This Study Adds to the Field
Bacteria are present in the lungs of healthy people at low levels compared to the upper respiratory tract, and are indistinguishable in community composition from upper airway microbiota. This finding suggests that they originate from the upper respiratory tract, most likely by microaspiration. There is no distinct lung-specific microbiome in healthy people, although very low levels of rare lung-specific bacteria may be present as well.
Intensive efforts are being directed at understanding microbial populations living in association with the human body, the human microbiome (1, 2). Microbial populations play important roles in health. In the gastrointestinal tract they instruct immune development, promote intestinal angiogenesis, and assist digestion (3–6). Disturbance of the healthy gut or genital tract microbiome, as with antibiotic therapy, can allow colonization with Clostridium difficile or Candida spp. Whether there is a normal microbiome in the lower respiratory tract (LRT) that contributes to health, or to disease when disrupted, has been unclear.
Traditional culture-based studies and classic teaching indicate that the healthy LRT is sterile (7–9), but recent culture-independent techniques have suggested otherwise (10, 11). Using molecular methods based on DNA sequencing or microarrays, it is possible to investigate microflora composition without requiring culture of individual microbes (12, 13). In such studies, DNA is prepared from the sampled microbiota, and composition analyzed by deep sequencing or DNA hybridization, allowing comprehensive profiling of full communities. Prior knowledge of organisms expected or the ability to culture and identify individual bacteria is not necessary. These approaches are being increasingly applied to the lung, and several reports have proposed that distinctive microbial populations reside in the healthy LRT.
However, studies of the lung microbiome face challenges not present in body sites that are easily accessible or contain high bacterial biomass such as skin, gut, oral cavity, or genital tract. Sampling by bronchoscopy requires passage through the upper respiratory tract (URT), which harbors large microbial populations and where contaminating organisms may be acquired. In addition, samples from lung may be of low microbial biomass, meaning that low-level admixture of sequences from dust, reagents, instruments, or other sources might also confound the data. To be definitive, molecular sampling needs to document the contribution of sequences from each of these sources.
In this study we performed intensive sampling of multiple sites along the respiratory tract of healthy individuals for comprehensive quantitative and qualitative molecular analysis using 16S Q-PCR and pyrosequencing. Low levels of bacterial 16S sequences were detected in the LRT, and community profiles were indistinguishable from the URT. Some organisms found in low-biomass lung samples matched environmental or control sequences. Unique LRT sequences were rare and inconsistent among subjects and samples. We conclude that there is no distinctive bacterial community in the healthy LRT, and the low amounts of bacterial sequences detectable in the LRT arise mainly from URT microbiota, likely through microaspiration along with bronchoscopic carryover during sampling.
Methods
Subjects and Sample Collection
Healthy subjects had no history of pulmonary disease or ongoing serious medical illnesses, and normal spirometry (see Table E1 in the online supplement). None had URT symptoms within 4 weeks or used antibiotics within 3 months. Subjects gave written informed consent. The protocol was approved by the University of Pennsylvania Institutional Review Board.
Flocked swabs (Copan Diagnostics, Murrieta, CA) were used to sample the nasopharynx (NP) and oropharynx (OP) as described (14), and a 10-ml saline oral wash (OW) was obtained by 90-second swish and gargle. Before bronchoscope insertion, 10 ml of saline was aspirated through the channel (Scope #1 and Scope #2 Pre-wash). After nebulized oropharyngeal anesthesia, bronchoscope #1 was introduced transorally to the glottis, where local anesthesia was administered and visible secretions aspirated. After removal, the scope tip was swabbed (Scope #1 Tip) and saline channel rinse obtained (Scope #1 Post-wash). Bronchoscope #2 was then introduced transorally, passed through the vocal cords without aspiration, and wedged in a right middle lobe bronchus. Fifty milliliters of saline were instilled and then aspirated (bronchoalveolar lavage [BAL]-A 1st Return), after which 100 ml were lavaged in the same location and collected separately (BAL-A 2nd Return). The bronchoscope was then wedged in an immediately adjacent bronchus, where 150 ml of saline was lavaged (BAL-B). After repositioning in the left mainstem bronchus, a double-sheathed wax-plug protected specimen brush (PSB) (Conmed, Utica, NY) was extended and left lower lobe mucosa sampled. Swabs and liquid samples were immediately placed on ice and frozen within 1 hour (−80°C).
DNA Extraction and 16S rDNA Quantification
DNA was isolated from swabs, unfractionated BAL and other samples using the PowerSoil DNA isolation kit (MoBio, Carlsbad, CA) in a BSL 2+ hood after treatment with DNAOff (MP Biomedicals, Solon, OH) and 30 minutes of ultraviolet irradiation. Swab tips were cut directly into bead tubes containing 60 μl of Solution C1 (PowerSoil manufacturer's 1st lysis solution). For liquid samples, 1.8 ml was centrifuged at 10,000 × g for 10 minutes at 4°C, pellets resuspended in 60 μl Solution C1, and transferred to bead tubes. Tubes were incubated at 65°C for 10 minutes, beadbeat for 2 minutes using Minibeadbeater-16 (BioSpec Products, Bartlesville, OK), and extracted per manufacturer protocol. Extracted DNA was stored at −20°C. Bacterial abundance was quantified by real-time PCR of 16S rDNA genes in triplicate 25 μl reactions using 1:10 or 1:20 dilutions of DNA samples, and primer/probe pairs and amplification conditions described in Reference 15.
Bacterial 16S rDNA Gene PCR Amplification, Pyrosequencing, and Sequence Analysis
We amplified bacterial 16S rDNA genes using barcoded broad-range V1V2 primers described in Reference 14. Duplicate 25 μl reactions were performed with AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA), pooled and purified, and pyrosequenced using primer A on a 454 Life Sciences FLX instrument (Branford, CT) as previously described in Reference 14. Reads were denoised at the flowgram level with DeNoiser (16), and integrated into the QIIME analysis pipeline (17), where they were clustered into OTUs at 97% sequence identity with UCLUST (18). OTUs were aligned to full-length 16S rDNA sequences with PyNAST (19), and removed if unalignable (minimum 75% identity, 150 nucleotides) or tagged as chimeric by ChimeraSlayer (20). Sequences were assigned taxonomy with RDP Classifier (50% confidence threshold) (21), and a phylogenetic tree generated de novo with FastTree2 (22). All programs are available online as indicated in References 16–22.
Statistical Methods
Using the QIIME pipeline (17), weighted UniFrac (23) was used to measure β diversity between bacterial communities as in Reference 24 and plotted in three-dimensional space using Principal Coordinate Analysis. The bias-corrected Chao1 metric (25, 26) was used to estimate the total number of OTUs per sample based on observed numbers of OTUs containing 1 or 2 reads. Comparisons of bacterial biomass between serial BAL specimens were analyzed with Page's trend test for ordered alternatives (27). We used Student's t test for significant differences of average between- versus within-group UniFrac with permutation (24).
Results
Intensive Respiratory Tract Sampling
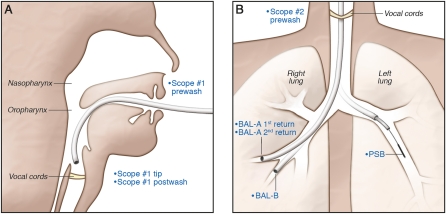
Respiratory tract microbiota were analyzed in six healthy individuals as outlined in Figure 1. First, NP and OP swabs and an OW were collected. One bronchoscope was inserted only to the level of the glottis, which provided samples approximating what a bronchoscope entering the LRT might carry down (28) (Scope #1 Tip and Post-wash). A second scope was then passed without suctioning, and sequential BAL done in two immediately adjacent regions of the right middle lobe (BAL-A and BAL-B), keeping the first aliquot of the initial BAL separate (BAL-A 1st and 2nd Returns) (29, 30). The lower left lobe bronchial mucosa was then sampled by protected specimen brush (PSB). To assess bacterial DNA that may exist in the bronchoscope channel, we collected saline washes of each bronchoscope before insertion into the subject (Scope #1 and #2 Pre-wash). Additional control samples including sterile swabs, brushes, lavage saline, and laboratory water were processed alongside biological samples.
Figure 1.
The two-bronchoscope procedure used to sample the respiratory tract. The samples collected are shown in blue lettering. (A) Sampling with the first bronchoscope. After nebulized oropharyngeal anesthesia, bronchoscope #1 was introduced transorally and advanced to the glottis, where local anesthesia was administered and visible secretions aspirated. The bronchoscope was removed and a sample obtained from the scope tip with a flocked swab (Scope #1 Tip) and from the channel by rinsing with lavage saline (Scope #1 Post-wash). (B) Sampling with the second bronchoscope. Bronchoscope #2 was introduced transorally, passed through the vocal cords without aspiration, and wedged in a right middle lobe segmental bronchus. Fifty milliliters of saline were instilled and then aspirated (BAL-A 1st Return), after which 100 ml of additional saline were instilled in the same location, without loss of wedge position, and aspirated (BAL-A 2nd Return). The bronchoscope was then repositioned and wedged in an immediately adjacent segmental bronchus, where 150 ml of saline were instilled and then aspirated (BAL-B). The bronchoscope was then repositioned in the left mainstem bronchus, and a protected specimen brush extended and used to sample the left lower lobe bronchial mucosa (PSB).
Quantifying Bacterial Abundance
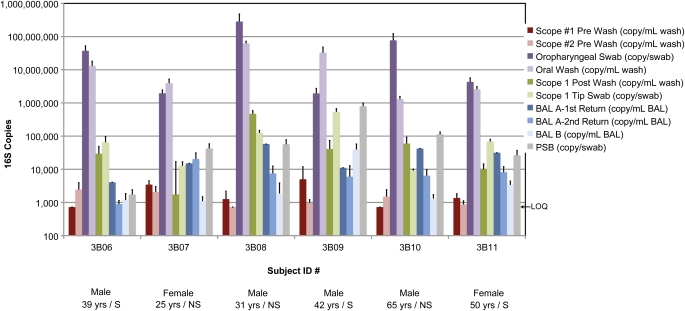
We quantified the bacterial biomass in each sample by 16S Q-PCR (Figure 2). When control water and lavage saline samples were processed through DNA extraction kits, three showed low but quantifiable levels of bacterial DNA by Q-PCR, three were indeterminate (near the threshold of detection), and two were below threshold and set to the lower limit of quantification (LOQ: 725 copies/ml). Using this estimate, we found an average of 914 16S DNA copies in these controls (range, 725–1,293). DNA extracted from two sterile swabs (one negative) and two brushes yielded an average of 1,356 16S copies (range, 1,191–1,622). Of 12 pre-procedure bronchoscopy channel washes, 9 had quantifiable 16S DNA while 3 were set to the LOQ. This yielded an average of 1,771 16S copies in Scope #1 and #2 Pre-washes (range, 734–5,004). Thus, although microbiologically sterile, instruments and reagents may contain low levels of bacterial DNA that may confound analysis of samples with low bacterial biomass.
Figure 2.
Quantification of bacterial 16S rDNA gene copies in the airway samples and controls. The subject studied is indicated along the x-axis by Subject ID. The 10 types of samples studied are summarized in the key to the right. Along the bottom is shown selected demographic information (S indicates smoker, NS nonsmoker). The y-axis shows the relative number of 16S gene copies detected using the Q-PCR assay. Error bars are shown as the standard error of the mean. All 16S copy numbers are normalized to the fractions collected (key at right) and are represented as either per ml of liquid sample (wash/BAL) or per whole swab/brush eluate. LOQ = lower limit of quantification, 725 16S rDNA copies/ml.
Oral cavity samples showed high bacterial biomass, with a mean 1.93 × 107 16S copies/ml in OW (range, 1.3 × 106 to 6.2 × 107) and 6.79 × 107 16S copies/ml in OP swab eluate (range, 1.94 × 106 to 2.58 × 108). Samples from Scope #1 had 135,637 16S copies/ml in the tip swab eluate (range, 9,127–533,021) and 102,064 16S copies/ml in the channel post-wash (range, 1,742–470,581). Thus, there is substantial bacterial DNA on a bronchoscope that reaches the glottis, as expected.
Lung communities sampled by serial BAL revealed a low mean copy number per ml of 26,923 for BAL-A 1st Return (range, 4,048–57,673), 8,209 for BAL-A 2nd Return (range, 892–20,313), and 8,024 for BAL-B (range, 1,091–39,334). We therefore asked if 16S DNA in BAL might be influenced by URT carryover. If carryover contributes to 16S copies in BAL, it would be highest in BAL-A 1st Return and diminish subsequently, as suggested by culture-based studies (29). Page's L trend values calculated from 16S copy numbers of BAL samples revealed a significant trend (L = 79, P < 0.05) to fewer copies across the three samples, demonstrating a serial decrement in biomass. Thus, levels of microbial DNA in BAL are low and affected by URT carryover. Since mean levels of bacterial DNA were quantitatively similar in BAL-A 2nd Return and BAL-B, carryover may be largely confined to the first return.
In contrast to BAL, the PSB brushings (estimated to sample ∼ 1 cm2 of respiratory mucosa) had an average of 172,271 16S copies (range, 1,740–792,258). In four of the six subjects, PSB samples had 104 to 105 copies, while one was considerably lower (1.7 × 103) and one was much higher (8.0 × 105). Although different types of samples are not entirely comparable, the data indicate a clear overall decrement in 16S copies from the upper to the lower respiratory tract across more similar samples, such as from OP swab to PSB, or OW to BAL and across serial BAL samples.
Bacterial Community Composition Analyzed by 454 Pyrosequencing
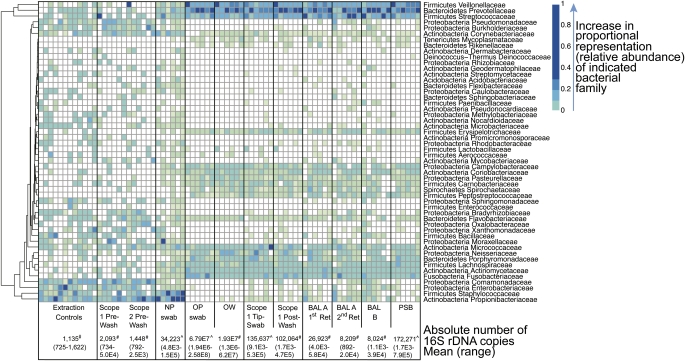
We surveyed the composition of respiratory tract bacteria by deep sequencing 16S rDNA, which generated over 417,300 raw partial (∼ 360 bp) 16S rDNA gene sequences. Quality filtering yielded more than 355,300 high-quality reads (mean, 4,555 reads/sample). The types and relative abundances of bacterial families populating each community are summarized in the heatmap in Figure 3. Each column shows a sample type, each row represents a bacterial family, and the color code shows relative abundance. The mean number of 16S copies recovered from each sample type is included at the bottom of the plot for comparison.
Figure 3.
Proportions of bacterial taxa in each sample type inferred from 16S rDNA pyrosequence data. Each column corresponds to an individual respiratory tract or control sample. The type of sample in each group is indicated in the boxes at the bottom of each group of columns. Each row corresponds to a specific bacterial family. OTUs were collected into families, so that some rows harbor multiple OTUs. OTUs that were not assigned at the family level are omitted. Rows were subjected to hierarchical clustering to emphasize families that show similar abundance patterns. The proportional representation (relative abundance) of each family is represented by the color code (key to the right). The absolute number of 16S copies determined from quantitative PCR of genomic DNA extracts are shown along the bottom. # indicates copies per ml; ^ indicates copies in the total volume eluted from the swab or brush.
Water control and lavage saline samples, while low in absolute numbers by Q-PCR, showed a consistent pattern with high relative abundance of organisms commonly residing on skin, including Staphylococcaceae and Propionbibacteriaceae (13, 31), as well as soil- and water-associated organisms such as Burkholderiaceae and Comamonadaceae. A similar distribution was seen in bronchoscope control samples (Scope #1 and #2 Pre-washes). Thus, despite stringent precautions, a low background of environmental organisms is detectable in lavage saline and pre-bronchoscopy channel specimens.
Nasopharyngeal samples showed a different community composition. We found an abundance of skin lineages as reported in previous molecular studies of the anterior nare and nostril including Staphylococcaceae, Propionbibacteriaceae, and Corynebacteriaceae (13, 31–34). In addition, NP samples contained organisms commonly found in the oral cavity, such as Streptococcaceae, Veillonellaceae, and Prevotellaceae (11, 13, 14, 32). This pattern differed from both environmental and oral samples.
Both OW and OP samples contained high relative abundance of Streptococcaceae and Veillonellaceae, as well as Fusobacteriaceae and Neisseriaceae, among other lineages. This pattern was similar to that reported previously in studies of oral flora (11, 13, 14, 32). Taxa characteristic of the control and nasal samples were less represented in oral communities.
Scope #1 Tip swab and Scope #1 Post-wash, representing communities on a bronchoscope that reaches the glottis, had types and distributions of bacterial lineages indistinguishable from oral specimens.
Finally, we found that samples from the lower respiratory tract, including all three BALs and the PSB, shared similar bacterial profiles as oral and peri-glottic samples. The types and distribution of bacteria was similar across all six individuals in each habitat when summarized by binning at the family level (Figure 3). Thus, from this analysis, no definitive distinction between the URT and LRT communities could be discerned.
Quantifying URT and LRT Community Relationships
The relationships between bacterial communities within each of the samples were then quantified by calculating the UniFrac distance between all pairs of samples. UniFrac scores the similarity between communities in terms of shared evolutionary history, whereby 16S sequences from each pair of communities are aligned on a common phylogenetic tree, and the branch length unique to each community computed (23). Communities that are similar to one another would contain closely related organisms, and would yield a lower UniFrac value. Thus, a single value summarizes the distance between a pair of communities. By computing all pairwise distances, we quantified how similar each community was to all others.
Visualization of the resulting UniFrac distance matrix by Principal Coordinate Analysis (Figure E1) revealed that oral/oropharyngeal (OW/OP), Scope #1 (Tip/Post-wash), and lung samples (BAL/PSB) all clustered together, consistent with sharing the same types and abundances of bacterial lineages. In contrast, both nasopharyngeal and control samples clustered separately.
We then analyzed within-group versus between-group UniFrac distances to ask whether each community type could be distinguished from every other type of community (Figure E2) (35). This analysis determines whether communities of a particular sample type (i.e., BAL-B) are more similar to each other (other BAL-Bs) than they are to each of the other sample types (i.e., extraction controls, Scope 1 samples, OWs, other BALs, PSB). Figure E2B shows that each of the lung sample types are as similar to the URT, Scope 1, and the other lung sample communities as they are to themselves, whereas they are significantly more similar to each other than they are to controls, Scope Pre-washes, or NP samples. Likewise, each of the communities from the URT and Scope 1 were as similar to the lung samples as they were to themselves. Thus, communities from lung, Scope 1, and OW/OP are closely related.
Comparing Community Relationships within Individual Subjects
Given the close relationship between lung and URT communities among our six subjects as a group, we then determined the relationship between communities within individual subjects. We first asked whether each subject's PSB was more closely related to his/her own Scope 1 samples than to any other subjects’ Scope 1 samples. Using the UniFrac metric, we found that every subject's PSB was more closely related to their own Scope 1 Tip-swab and Scope 1 Post-wash than to those of any other subject (P < 0.05 for both weighted and unweighted UniFrac; data not shown). We also used a nonphylogenetic index of shared OTUs, the Jaccard Index (36, 37), which also revealed that each PSB was more closely related to Scope 1 samples from that subject than from any other (P < 0.0002 for both binary and abundance-weighted Jaccard; data not shown). Finally, we applied an OTU network analysis (38) to lung PSB and peri-glottic Scope 1 Tip samples, to determine whether specific lineages were more likely to be shared by these two samples from the same subject than by samples from different subjects (Figure E3). This analysis revealed that organisms were far more likely to be shared by an individual's pair of PSB and Scope 1 Tip samples than by samples from different individuals (P = 0.0014, 720 permutations). Thus, each individual's lung community is tightly linked to their own URT microbiota.
Rare Distinctive Lineages in Lung Samples
We then asked whether it was possible to identify lineages that may be specific to the LRT within the background of URT-related organisms. As can be seen in the heatmap representation (Figure 3), while BAL and PSB closely match URT samples, several families appeared at relatively higher abundances in lung than in OP, OW, or Scope 1 samples. However, most of these taxa were also present at high relative abundances in control or pre-procedure channel wash samples, and their presence in lung was particularly associated with low bacterial biomass BAL. Some of these are mainly environmental (e.g., Sphingomonadaceae), whereas others are environmental but also have significant pathogenic potential (e.g., Pseudomonaceae, Burkholderiaceae, Staphylococcaceae). Some were also highly abundant in NP swabs (e.g., Propionibacteriaceae, Staphylococcaceae). Thus, their appearance in BAL cannot be ascribed to lung origin, because their presence in controls and association with low-biomass lung samples suggest that they may result from environmental sources, but appear relatively more abundant in lung samples that are less confounded by URT carryover.
Therefore, we asked whether there were taxa present in lung samples but absent in both URT and environmental control samples. Sequences reads were clustered at 97% identity and analyzed as “operational taxonomic units (OTUs)” (Table E2), representing our most fine-grained taxonomic classification. Out of 3,431 total OTUs, 410 were only detected in BAL or PSB specimens (Table E3). Over 87% of these OTUs were identified by only a single sequence read, and thus it is uncertain whether they are derived from rare organisms or from sequencing errors that eluded quality filters. Seven OTUs were identified as lung-specific and supported by more than 10 sequence reads. Each of these seven taxa was unique to a single individual's lung samples, and five of the seven were found in only one of the four LRT sample types from that individual, again raising questions about authenticity. However, the lung-specific lineage with the highest sequence count (32 sequences) was found in all three BAL and PSB samples from one individual (subject 3B10) but not in URT, Scope Pre-washes, or environmental controls. This OTU was identified as T. whipplei, a member of the Tropheryma genus.
A comparison of the observed to calculated estimate of total number of taxa in our controls demonstrated that we identified approximately 83 to 93% of the theoretical maximum number of bacterial lineages present (Table E4), suggesting that there may be 4 to 10 taxa in control samples that were not detected. Thus, it is possible that some of these rare LRT-distinctive lineages may also have been found in controls had those samples been sequenced to saturation. We conclude that at most a handful of OTUs out of 3,431 detected in lung are unique to LRT samples, and none are shared among different subjects.
Discussion
A principal challenge in defining the lung microbiome is collecting samples that faithfully reflect lung-derived bacteria. Molecular-based studies of the LRT using bronchoscopic collection of BAL or mucosal brushes have reported that airways of healthy individuals contain a characteristic distribution of organisms, often at very high levels (10, 11). However, these studies lacked separate analysis of upper airway microbiota, as well as environmental admixture controls, that we show here are needed to assign microbial populations to the lower airway. From intensive sampling of these six subjects, we conclude that despite the presence of detectable 16S DNA the healthy lung lacks a consistent and unique microbiome.
In designing our sampling strategy, we started with the assumption that the LRT microbiome would likely be very low biomass, especially compared with the URT. Scope #1 samples were intended to represent flora that could be carried from the upper to the lower respiratory tract by a bronchoscope. If the second bronchoscope does bring down upper airway organisms, then we would expect BAL communities to be similar to Scope #1 samples, and to see a decrease in bacterial quantity across serial BALs as the flora is serially washed from the scope. This was exactly what was seen. Such carryover would likely be more significant with a standard single-scope bronchoscopy.
Our study therefore raises the question of whether low levels of URT organisms are genuinely present in the LRT. The protected specimen brush sample of the relatively proximal endobronchial mucosa, performed after multiple BALs on the opposite side, had substantial levels of bacterial DNA, suggesting that that bacterial material originated from the LRT airway surface rather than entirely from bronchoscope carryover. However, the PSB-derived bacterial communities still retained the same relative composition as upper airway samples. Previous molecular studies show that microbial community structures are exquisitely sensitive to differences in environment niche, such that bacterial populations are clearly distinguishable when obtained from different regions of the skin or gastrointestinal tract or even gingival crevices of different teeth (13, 31, 39). Therefore, it is unlikely that LRT community profiles reflect an independent self-sustaining lung microbiome that is indistinguishable from URT communities, and most likely reflect transient URT-derived bacteria. Low levels of microaspiration normally occur in healthy people (40–42), consistent with this finding. It is unclear whether the bacterial DNA derived from live or dead bacteria.
While we have established here that there is not a distinct LRT microbiome common among our subjects, it is important to determine if any bacterial lineages detected in the LRT represent authentic unique lung inhabitants. One lung-specific OTU, found in a single subject but in multiple samples from that subject, corresponded to the causative agent of Whipple's disease, which can involve the lungs. This organism was recently reported in BAL from patients with pneumonia or interstitial lung disease (43, 44), and may represent genuine detection of a bacteria resident of the LRT in this subject. Six other OTUs were present in fewer reads in LRT samples from individual subjects but absent in URT and environmental samples. However, they comprise only 6 OTUs versus 3,431 in the full set of lung samples and thus, if authentic LRT residents, they are extremely low abundance; none were found in more than one individual, and most were in only one of four LRT samples from that subject. One limitation of our study is the relatively small number of individuals sampled, and thus it will be important to extend this type of analysis to additional subjects.
Traditional culture-based bronchoscopic studies of lower respiratory tract microbiota were highly attuned to carryover issues. These studies typically excluded organisms normally present at high levels in the URT or restricted analysis to potential pulmonary pathogens, and utilized quantitative thresholds for “clinically significant” numbers of organisms (45–49). Unlike quantitative thresholds empirically determined for diagnosis of pneumonia, however, there are no validated criteria for defining colonization or identifying normal microbial populations of the lower airways. Some culture-based studies attempted to minimize URT carryover using the two-scope approach or separate analysis of BAL 1st Return (28–30, 50). As we have shown, culture-independent characterization of LRT microflora does not obviate the need for similar careful sampling and interpretation. Despite these caveats, molecular methods have a powerful advantage in analyzing lower respiratory tract inhabitants. Traditional culture is incomplete, highly selective, and semi-quantitative at best. In contrast, molecular analysis characterizes entire populations and, while not completely unbiased in DNA extraction or amplification efficiency, can accurately define relative abundances of all bacteria in distinct communities. Thus, molecular community characterization can compare different niches and identify organisms present in one site at a higher relative abundance than another. Similarly, our results do not imply that the presence of an organism in both the URT and LRT precludes biological significance, particularly for lung pathogens that may colonize the URT or that may be expectorated and secondarily found in the URT.
Thus, bacterial populations in the healthy LRT largely reflect URT organisms, likely resulting from transient entry rather than independent communities with indistinguishable structure. Authentic lung bacteria can be molecularly identified given appropriate attention to minimize URT carryover and define its impact. In addition, low-biomass niches such as the lung require scrupulous attention to environmental admixture including Q-PCR determination of low- and high-biomass samples, and sequencing of otherwise unquantifiable amplification products. Using these approaches, it will be possible to define the role of LRT microbiota in lung health and disease, and to identify genuine LRT inhabitants.
Supplementary Material
Acknowledgments
The authors are grateful to the research subjects who volunteered for this study, W. Russell and D. Frame for critical study assistance, and to members of the Collman and Bushman laboratories for helpful discussions.
Footnotes
Supported by the Lung HIV Microbiome Project Grant U01 HL98957 from the National Institutes of Health to R. G. C. and F. D. B., and assisted and supported by the Penn Center for AIDS Research. The authors also acknowledge the Penn Genome Frontiers Institute and a grant with the Pennsylvania Department of Health; the Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusion. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors' contributions: E.S.C, R.G.C., I.F. and F.D.B. designed research; A.R.H., A.Y. and A.S.F collected samples; E.S.C. performed research; E.S.C., K.B., R.G.C., and F.D.B. analyzed data; and E.S.C., R.G.C., and F.D.B. wrote the paper.
These investigators were co-senior authors.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org.
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, et al. A catalog of reference genomes from the human microbiome. Science 2010;328:994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, et al. The NIH human microbiome project. Genome Res 2009;19:2317–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abt MC, Artis D. The intestinal microbiota in health and disease: the influence of microbial products on immune cell homeostasis. Curr Opin Gastroenterol 2009;25:496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science 2005;308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell 2010;140:859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science 2001;292:1115–1118 [DOI] [PubMed] [Google Scholar]
- 7.Laurenzi GA, Potter RT, Kass EH. Bacteriologic flora of the lower respiratory tract. N Engl J Med 1961;265:1273–1278 [DOI] [PubMed] [Google Scholar]
- 8.Niederman MS, Sarois GA. Respiratory tract infections. Philadelphia: Lippincott Williams & Wilkins; 2005 [Google Scholar]
- 9.Pecora DV. A comparison of transtracheal aspiration with other methods of determining the bacterial flora of the lower respiratory tract. N Engl J Med 1963;269:664–666 [DOI] [PubMed] [Google Scholar]
- 10.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 2011;6:e16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010;5:e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Relman DA. New technologies, human-microbe interactions, and the search for previously unrecognized pathogens. J Infect Dis 2002;186:S254–S258 [DOI] [PubMed] [Google Scholar]
- 13.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science 2009;326:1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD, Collman RG. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS ONE 2010;5:e15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol 2010;3:148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 2010;7:668–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–2461 [DOI] [PubMed] [Google Scholar]
- 19.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010;26:266–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. Chimeric 16S rRNA sequence formation and detection in sanger and 454-pyrosequenced PCR amplicons. Genome Res 2011;21:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005;71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 2007;73:1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chao A. Non-parametric estimation of the number of classes in a population. Scand J Stat 1984;11:265–270 [Google Scholar]
- 26.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 1987;43:783–791 [PubMed] [Google Scholar]
- 27.Page EB. Ordered hypotheses for multiple treatments: a significance test for linear ranks. J Am Stat Assoc 1963;58:216–230 [Google Scholar]
- 28.Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006;173:991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qvarfordt I, Riise GC, Andersson BA, Larsson S. Lower airway bacterial colonization in asymptomatic smokers and smokers with chronic bronchitis and recurrent exacerbations. Respir Med 2000;94:881–887 [DOI] [PubMed] [Google Scholar]
- 30.Soler N, Ewig S, Torres A, Filella X, Gonzalez J, Zaubet A. Airway inflammation and bronchial microbial patterns in patients with stable chronic obstructive pulmonary disease. Eur Respir J 1999;14:1015–1022 [DOI] [PubMed] [Google Scholar]
- 31.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, et al. Topographical and temporal diversity of the human skin microbiome. Science 2009;324:1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio 2010;1(3):e00129-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, Becker K, Pieper DH. A poke into the diversity and associations within human anterior nare microbial communities. ISME J 2010;7:839–851 [DOI] [PubMed] [Google Scholar]
- 34.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. The human nasal microbiota and Staphylococcus aureus carriage. PLoS ONE 2010;5:e10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, Lozupone CA, Hamady M, Knight R, Bushman FD. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog 2008;4:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao A, Chazdon RL, Colwell RK, Shen T-J. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett 2005;8:148–159 [Google Scholar]
- 37.Jaccard P. The distribution of the flora in the alpine zone. New Phytol 1912;11:37–50 [Google Scholar]
- 38.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preza D, Olsen I, Willumsen T, Grinde B, Paster BJ. Diversity and site-specificity of the oral microflora in the elderly. Eur J Clin Microbiol Infect Dis 2009;28:1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beal M, Chesson A, Garcia T, Caldito G, Stucker F, Nathan CO. A pilot study of quantitative aspiration in patients with symptoms of obstructive sleep apnea: comparison to a historic control group. Laryngoscope 2004;114:965–968 [DOI] [PubMed] [Google Scholar]
- 41.Gleeson K, Eggli DF, Maxwell SL. Quantitative aspiration during sleep in normal subjects. Chest 1997;111:1266–1272 [DOI] [PubMed] [Google Scholar]
- 42.Huxley EJ, Viroslav J, Gray WR, Pierce AK. Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am J Med 1978;64:564–568 [DOI] [PubMed] [Google Scholar]
- 43.Bousbia S, Papazian L, Auffray JP, Fenollar F, Martin C, Li W, Chiche L, La Scola B, Raoult D. Tropheryma whipplei in patients with pneumonia. Emerg Infect Dis 2010;16:258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, Pace NR. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA 2007;104:20529–20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorpe JE, Baughman RP, Frame PT, Wesseler TA, Staneck JL. Bronchoalveolar lavage for diagnosing acute bacterial pneumonia. J Infect Dis 1987;155:855–861 [DOI] [PubMed] [Google Scholar]
- 46.Cantral DE, Tape TG, Reed EC, Spurzem JR, Rennard SI, Thompson AB. Quantitative culture of bronchoalveolar lavage fluid for the diagnosis of bacterial pneumonia. Am J Med 1993;95:601–607 [DOI] [PubMed] [Google Scholar]
- 47.Kirkpatrick MB, Bass JB Jr. Quantitative bacterial cultures of bronchoalveolar lavage fluids and protected brush catheter specimens from normal subjects. Am Rev Respir Dis 1989;139:546–548 [DOI] [PubMed] [Google Scholar]
- 48.Ortqvist A, Kalin M, Lejdeborn L, Lundberg B. Diagnostic fiberoptic bronchoscopy and protected brush culture in patients with community-acquired pneumonia. Chest 1990;97:576–582 [DOI] [PubMed] [Google Scholar]
- 49.Souweine B, Veber B, Bedos JP, Gachot B, Dombret MC, Regnier B, Wolff M. Diagnostic accuracy of protected specimen brush and bronchoalveolar lavage in nosocomial pneumonia: impact of previous antimicrobial treatments. Crit Care Med 1998;26:236–244 [DOI] [PubMed] [Google Scholar]
- 50.Pang JA, Cheng AF, Chan HS, French GL. Quantitative bacterial cultures of bronchoalveolar lavage fluids. Am Rev Respir Dis 1989;140:854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.