Abstract
We recently reported deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Functional analyses confirmed that PTPN2 acts as classical tumor suppressor repressing the proliferation of T cells, in part through inhibition of JAK/STAT signaling. We investigated the expression of PTPN2 in leukemia as well as lymphoma cell lines. We identified bi-allelic inactivation of PTPN2 in the Hodgkin’s lymphoma cell line SUP-HD1 which was associated with activation of the JAK/STAT pathway. Subsequent sequence analysis of Hodgkin’s lymphoma and T-cell non-Hodgkin’s lymphoma identified bi-allelic inactivation of PTPN2 in 2 out of 39 cases of peripheral T-cell lymphoma not otherwise specified, but not in Hodgkin’s lymphoma. These results, together with our own data on T-cell acute lymphoblastic leukemia, demonstrate that PTPN2 is a tumor suppressor gene in T-cell malignancies.
Keywords: mutation, phosphatase, T-cell NHL, tumor suppressor
Introduction
Protein tyrosine phosphatases are important enzymes that control the activity of multiple signaling pathways downstream of cytokine receptors and tyrosine kinases. In agreement with this pivotal function, mutations in tyrosine phosphatases have been associated with the development of cancer. In hematologic malignancies, activating mutations in PTPN11 have been associated with a set of myeloid malignancies next to precursor B-cell acute lymphoblastic leukemia, while so far inactivation of PTPN2 has only been identified in T-cell acute lymphoblastic leukemia (T-ALL).1–3
PTPN2 (also known as TC-PTP) is a classical protein tyrosine phosphatase that is highly expressed in the hematopoietic system.4,5 In line with its ubiquitous expression pattern, homozygous deletion of Ptpn2 from the mouse system is fatal and mice succumb rapidly after birth (3–5 weeks). Knockout mice display multiple phenotypic changes including severe defects in the blood system.6 Its pivotal role in hematopoiesis and the inflammatory phenotype of PTPN2 knockout mice together with the identification of cancer associated kinases and transcription factors as direct substrates of PTPN2, have pointed to a potential role of PTPN2 in tumorigenesis and the development of immune diseases.7, 8
We have previously identified focal deletions of the PTPN2 gene (chromosome 18p11) in 6% of T-ALL cases. Functional analysis confirmed PTPN2 to be a classical tumor suppressor gene repressing proliferation of T cells. Downregulation of PTPN2 expression provided a proliferation advantage to the leukemic cells due to increased sensitivity to cytokine stimulation and increased activation of the JAK/STAT pathway. In addition, loss of PTPN2 also had a direct effect on the activation of oncogenic kinases such as NUP214-ABL1, LCK, and mutant JAK1.3
In the current study, we screened a set of lymphoma cell lines for mutations or deletion of PTPN2 and we sequenced PTPN2 in Hodgkin’s lymphoma (HL) and T-cell non-Hodgkin’s lymphoma (T-NHL) samples.
Design and Methods
Patients’ samples and cell lines
After obtaining informed consent according to the guidelines of the local ethical committees, a series of 50 Hodgkin’s lymphomas (HL) and 69 T-cell non-Hodgkin’s lymphomas (T-NHL) were collected from the archives of the Department of Pathology of the University Hospital Leuven and the University of Würzburg. A review of the diagnosis of HL and T-NHL was based on morphology, immunophenotyping and molecular genetic and T-cell gene rearrangement studies, according to the World Health Organization criteria.9 Human cell lines were purchased from DSMZ (Braunschweig, Germany) and detailed information can be found at http://www.dsmz.de. All cells were cultured under standard conditions as previously described.10
Sequence analysis of PTPN2
Sequence reactions were performed on genomic DNA. All exons of PTPN2 (with exception of the GC-rich exon 1) were amplified in separate reactions with either specific Sanger sequencing primers (conventional sequence analysis) or amplicon fusion primers (454 deep sequence analysis). Next to the template specific sequence, fusion primers contained a directional titanium primer at the 5′-prime end followed by a multiplex identifier for barcode sample identification. Samples from 8 different patients were mixed and processed for 454 deep sequencing. The 454 sequence readings were analyzed using CLC Genomics Workbench software and were mapped to the human reference sequence (NCBI Build 36.1) using BWA. Variants were called with SAMTools (with a SNP-quality threshold of 20).11,12 For conventional sequence analysis of PTPN2 in T-NHL patients and cell lines, PCR amplicons were sent directly for sequencing to the VIB Genetics Service Facility (Antwerp, Belgium). The Mutation Nomenclature guidelines from the Human Genome Variation Society (HGVS) were followed to describe mutations at the cDNA and protein levels.
Transfection, virus production and retroviral transduction
In this study, 293T cells were used for virus production and transfection experiments to evaluate the activity of identified PTPN2 variants. Twenty-four hours after seed out, cells were transfected using TurbofectTM in vitro Transfection Reagent (Fermentas). For virus production, one μg of plasmid DNA was mixed with 1 μg of retroviral envelope vector. Transfection mixture was refreshed 24 h post transfection and harvested another 24 h later. Cells were transduced as previously described.10 All re-expression experiments were performed using the bicistronic, retroviral vector pRetroX-PTuner (Clontech). Protein expression was induced for 24 h (Shield1 concentration: 500nM) before samples were lysed for protein analysis.3
Results and Discussion
Genetic and functional characterization of PTPN2 in leukemia and lymphoma cell lines
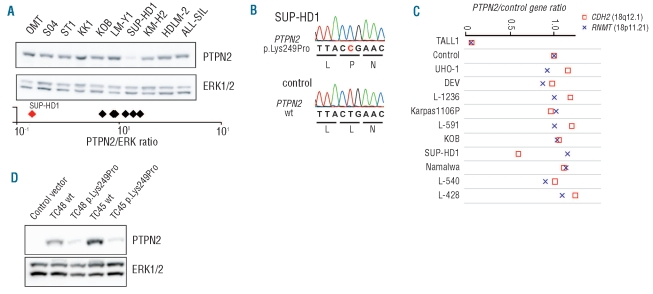
Based on our study of PTPN2 deletions in T-ALL, we sought evidence for the involvement of PTPN2 in the pathogenesis of other hematologic malignancies. We screened a panel of cell lines including T-ALL, adult T-cell leukemia/lymphoma (ATL) and Hodgkin’s lymphoma (HL) cell lines for variations in PTPN2 at the protein and DNA level. Among the studied cell lines, the SUP-HD1 Hodgkin’s lymphoma cell line displayed very low PTPN2 protein levels (Figure 1A). Subsequent genetic analysis confirmed that the SUP-HD1 cell line harbored biallelic inactivation of PTPN2. We identified deletion of one PTPN2 allele and a missense mutation in exon 7 of the other allele, resulting in a p.Lys249Pro substitution, in the SUP-HD1 cell line (Figure 1B and C). The mutation (Lysine to Proline) occurs in an α-helical region, a substitution known to break the helix. We tested whether the proline mutation indeed caused instability of the PTPN2 protein in both isoforms TC45 and TC48, known to have a different subcellular localization.4,13 Both isoforms, wild-type or those containing the p.Lys249Pro mutation, were cloned and stably expressed in the murine pro-B cell line Ba/F3. Our data confirmed that the mutant proteins were expressed at low levels, whereas expression of wild-type isoforms showed comparable levels to endogenous expression observed in other cell lines (Figure 1A and D).
Figure 1.
Genetic inactivation of PTPN2 in HL cell line SUP-HD1. (A) Western blot analysis of PTPN2 expression in human HL and T-ALL cell lines. Quantification of protein blots showed reduced abundance of PTPN2 protein in the SUP-HD1 cell line compared to other cell lines. Normalized quantification values are shown below the immunoblot (logarithmic scale). ERK1/2 is shown as loading control and was used for normalization. (B) Sequence of part of exon 7 of PTPN2. Upper panel: mutated sequence (SUP-HD1); lower panel wild-type (wt) sequence (control; T-ALL cell line without PTPN2 alteration). Hemizygous missense mutation resulted in the amino acid substitution p.Lys249Pro. (C) Quantitative PCR analysis of the PTPN2 copy number status in human cell lines (KOB (ATL), Namalwa (Burkitt’s lymphoma), Karpas1106P (B-cell lymphoma), all others (HL)) depicted monoallelic loss of PTPN2 in SUP-HD1 cells. Analysis of publically available genomics data of the SUP-HD1 cell line (GSE22208) showed heterozygous loss of a 3.5 Mb region on chromosome 18 including the PTPN2 gene (del(18p)(p11.21p11.21).14 Primer sets for CDH2 (□, 18q12.1) and RNMT (×, 18p11.21) were used as control genes and all values were normalized to a control. A genomic DNA sample from an individual (TALL1) harboring a homozygous deletion of PTPN2 was included as positive control and has been previously described.3 (D) Western blot analysis of whole cell lysates prepared from Ba/F3 cells stably expressing either human wild-type (wt) or mutated variants (p.Lys249Pro) of indicated PTPN2 isoforms (TC45 and TC48). PTPN2 protein levels were detected with human specific antibody allowing the discrimination between endogenous murine and reconstituted human PTPN2. Ba/F3 cells transduced with the empty vector were used as negative control. Blots were stripped and re-probed with ERK1/2 to ensure equal loading.
From the other cell lines, HDLM-2 and KM-H2 showed loss of one chromosome 18 (where PTPN2 is located),14 but did not have reduced PTPN2 protein expression (Figure 1A). The cell line L-540, a HL cell line derived from a patient with HL of the nodular sclerosis subtype and assigned to the T-cell lineage,15 showed a heterozygous variation in exon 6 of PTPN2 (p.Val234Ile). We could not, however, detect decreased phosphatase activity of the PTPN2 p.Val234Ile variant (Online Supplementary Figure S1), and this variation was recently identified as a polymorphism present in the normal population (www.ensem-bl.org). Taken together, these data illustrate that PTPN2 was inactivated in the SUP-HD1 cell line, but not in other lymphoma cell lines.
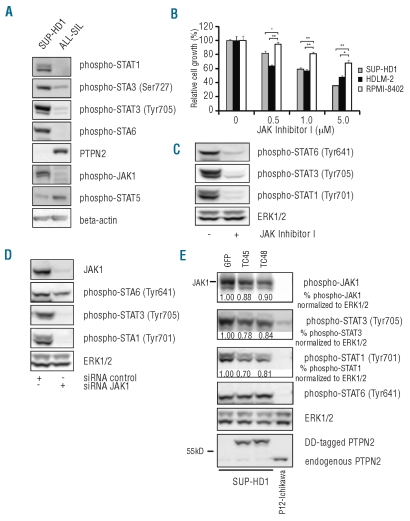
PTPN2 is a negative regulator of JAK/STAT signaling. In agreement with low PTPN2 expression in SUP-HD1 cells, these cells showed constitutive phosphorylation of JAK1 and several members of the STAT family (Figure 2A). Treatment of SUP-HD1 cells with a JAK kinase inhibitor resulted in a clear dephosphorylation of all STATs, and a dose dependent inhibition of the proliferation of the SUP-HD1 cells (Figures 2B and C), confirming that SUP-HD1 cells are largely dependent on JAK/STAT signaling for their proliferation. Similarly, siRNA mediated knockdown of JAK1 expression reduced STAT activation (Figure 2D).
Figure 2.
SUP-HD1 cells show constitutive activation of the JAK/STAT pathway. (A) Identification of constitutively activated JAK/STAT proteins in SUP-HD1 by Western blot analysis of WCL. T-ALL cell line ALL-SIL is known to harbor no genetic abnormality of the PTPN2 gene locus and is shown as control.3 (B) SUP-HD1 and HDLM-2 cells showed sensitivity to JAK inhibitor I. HDLM-2 cells feature a deletion and mutation in the SOCS1 gene and JAK2 amplification causing sensitivity to JAK/STAT inhibition,14,17 whereas RPMI-8402 cells display no activated JAK/STAT pathway and were used as negative control. Y axis represents cell growth relative to respective DMSO treated control recorded 72 h after treatment initiation and bars show average ± s.e.m. n=3; *P<0.05, **P<0.005. (C) Western blot analysis of SUP-HD1 cells upon exposure to JAK inhibitor I (1 μM) demonstrated dephosphorylation of JAK1 downstream targets STAT1, STAT3 and STAT6. ERK1/2 is shown as loading control. (D) Downregulation of JAK1 protein levels using human JAK1 targeting short interfering RNA (siRNA JAK1) resulted in a profound effect on the activation status of STAT family members STAT1 and STAT3 and a slightly minor effect on STAT6 phosphorylation. Equal amount of total protein was assured by detection of ERK1/2. (E) Re-expression of inducible forms of PTPN2 (DD-tagged) resulted in decreased JAK1 phosphorylation, and inhibition of STAT1 and STAT3 activation. GFP fused to the destabilizing domain was used as negative control. Normalized quantification values of phosphorylation levels calculated as relative change compared to GFP-transduced control cells are shown below respective blots. T-ALL cell line P12-Ichikawa was included as control with normal PTPN2 protein expression level. Loading control: ERK1/2.
To evaluate whether PTPN2 indeed modulates JAK/STAT signaling in SUP-HD1, we re-expressed the PTPN2 isoforms using an inducible retroviral vector system.3 Re-expression of the TC45 or TC48 isoform of PTPN2 diminished JAK1 activation as observed by a slight reduction in JAK1 phosphorylation itself and a more clear effect on the downstream targets STAT1 and STAT3 (Figure 2E). No effect was observed on STAT6 phosphorylation. These data demonstrate that PTPN2 modulates JAK/STAT signaling in the cells, but also suggest that additional signaling proteins upstream of the JAK/STAT pathway are activated that are not negatively regulated by PTPN2. Indeed, JAK2 is known to be over-expressed in SUP-HD1 cells due to high-level 9p24 amplification, but JAK2 is not a direct substrate of PTPN2.14
In conclusion, we demonstrate that SUP-HD1 cells harbor bi-allelic inactivation of PTPN2, which causes activation of JAK1 and several STAT proteins, required for proliferation of the SUP-HD1 cells. Our data also suggest that additional genetic lesions present in SUP-HD1 cells cooperate with loss of PTPN2 to activate the JAK/STAT signaling pathways.14
Sequence analysis of PTPN2 in HL and T-NHL
STAT activation has been described in HL and partially attributed to the presence of JAK2 amplification and mutations in SOCS genes.16,17 Since PTPN2 represents a phosphatase for both JAKs and STATs, and since PTPN2 was found to be mutated in HL cell line, we investigated whether mutation of PTPN2 could be responsible for the deregulation of the JAK/STAT signaling cascade in HL.8,18 We selected a cohort of HL samples (10 nodular lymphocyte-predominant HL, 27 nodular sclerosis cHL, 6 mixed-cellularity cHL, 5 lymphocyte-rich cHL, and 2 lymphocyte-depleted cHL) for a comprehensive sequence analysis of the coding region of PTPN2 using the 454 amplicon deep sequencing technology (Roche 454 GS-FLX). This technology was chosen because HL samples usually carry only a very low percentage of clonal Hodgkin’s and Reed Sternberg cells; typically less than 1%. Even though in this way we achieved sufficient coverage of all exons to be able to detect mutations in such a low percentage of cells (Online Supplementary Table S1), we identified only a single missense mutation located in exon 2 of PTPN2 (c.89A>G). The percentage of readings carrying the mutation was in the expected range: 54 of 3,659 readings, corresponding to 3% of diploid cells carrying a heterozygous mutation. Readings carrying the mutation were equally present in both forward and reverse readings. The base pair alteration A>G replaced a histidine at position 30 by an arginine (p.His30Arg). Even though substituted amino acids are very different, no functional consequence was found in our in vitro assays. Transient expressed His30Arg mutant (239T cells) effectively dephosphorylated a co-transfected constitutively active JAK1 mutant (p.Ala634Asp), suggesting that this variation in PTPN2 is unlikely to be an inactivating mutation (Online Supplementary Figure S1).
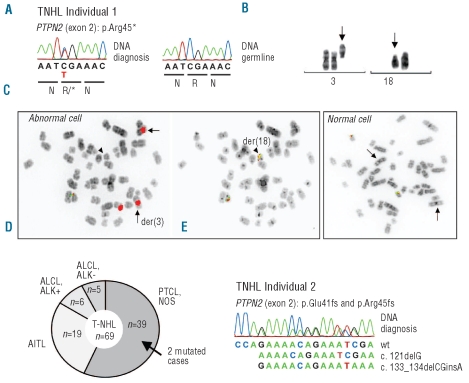
Surprisingly, we also identified a nonsense mutation in approximately 50% of the cells in one of the HL samples. This sample carried two cytogenetically unrelated abnormal clones of distinct disease origins, T-NHL (peripheral T-cell lymphoma, not otherwise specified (PTCL, NOS)) and classical HL (mixed-cellularity cHL). Genetic characterization of this case identified complete inactivation of the PTPN2 gene in the T-cell lymphoma clone. The 454 sequence analysis showed a nonsense mutation located in exon 2 (c.133G>A, p.Arg45*) of PTPN2 in about 50% of the cells, which was subsequently confirmed by Sanger sequencing (Figure 3A). Moreover, fluorescence in situ hybridization (FISH) analysis showed loss of the second copy due to deletion of the short arm of chromosome 18 (18p) in the T-NHL clone (Figure 3B and C). These data document a bi-allelic inactivation of PTPN2 in the T-NHL clone in this patient.
Figure 3.
PTPN2 is mutated in T-NHL (PTCL, NOS subtype) (A) Sanger sequence analysis confirmed the presence of the hemizygous PTPN2 nonsense mutation (p.Arg45*). Germline DNA was analyzed to ensure the somatic origin of the mutation. (B) Cytogenetic findings in T-NHL subject 1. A partial karyotype is shown demonstrating deletion of 18p (arrowhead) and presence of 3 centromeric regions of chromosome 3 (arrows) in the same clone. (C) Metaphase FISH analysis confirmed loss of one copy of PTPN2 on der(18) in the T-NHL clone of subject 1. Abnormal cells (left panel) had three centromere 3 signals instead of the two signals expected in a normal metaphase nucleus (left panel, CEP3, red), and only one chromosome 18 with signals for PTPN2 instead of two signals present in normal cells (left panel, green/red). Arrowhead highlights der(18) with loss of PTPN2. Der(18) was identified using break apart BCL2 probes locating to 18q21 (right panel, arrowhead). FISH was performed with gene locus specific dual-color FOSMID probes for PTPN2,3 with centromeric satellite DNA probes for chromosome 3 (CEP3) and with BCL2 Dual-Color break apart probes for BCL2/18q21 (RP11-215A20, SpectrumGreen; RP11-495C15, SpectrumOrange). (D) Schematic overview of the T-NHL cohort (n=69) analyzed in this study. Both subjects harboring mutations of PTPN2 belong to the PTCL, NOS subgroup. PTCL: peripheral T-cell lymphoma; NOS: not otherwise specified; AILT: angioimmunoblastic T-cell lymphoma; ALCL: anaplastic large-cell lymphoma; ALK+: ALK-positive; ALK-: ALK-negative. (E) Sequence of part of exon 2 of PTPN2 (T-NHL individual 2). Double peaks resulted from the presence of two mutations on the different alleles (confirmed by cloning of PCR products and subsequent sequence analysis). The wild-type coding sequence is shown below the electropherogram.
To determine whether PTPN2 inactivation is a recurrent event in T-NHL, we sequenced PTPN2 in lymph node biopsies obtained from 69 T-NHL (Figure 3D and Online Supplementary Table S2). In one additional sample obtained from a patient with PTCL, NOS was identified with bi-allelic inactivation of PTPN2. This lymphoma was characterized by the presence of two frameshift truncating mutations in exon 2 (p.Glu41fs and p.Arg45fs, Figure 3E) resulting in bi-allelic inactivation of PTPN2. Another T-NHL sample had a missense mutation in one allele of PTPN2 (c.1111A>G, p.Arg371Gly in exon 9 of PTPN2), but functional assays showed normal phosphatase activity of the PTPN2 Arg371Gly protein (Online Supplementary Figure S1). Based on this, we conclude that the p.Arg371Gly variation is either a passenger mutation or a rare polymorphism. No other variations were identified, but several samples had low tumor cell content, making detection of the mutations more difficult with Sanger sequencing (Online Supplementary Table S2). In total, 2 of 69 T-NHL had PTPN2 mutations, with both cases belonging to the PTCL, NOS subgroup (2 of 39 (5%) PTCL, NOS cases) (Figure 3D).
In conclusion, we have previously identified PTPN2 as a tumor suppressor gene in T-ALL, where PTPN2 is deleted in 6% of cases.3 In this study, we identified inactivation of PTPN2 by nonsense mutations in 5% of PTCL/NOS cases. These data further demonstrate the tumor suppressor activity of PTPN2 in T-cell malignancies.
Acknowledgments
The authors would like to thank Ralf Küppers, Institute of Cell Biology (Tumor Research), University of Duisburg-Essen, for providing genomic DNA of Hodgkin’s lymphoma cell lines.
Footnotes
Funding: this work was supported by the K.U.Leuven (grant GOA/11/010 to JC and IW; grant PF/10/016 SymBioSys to JC and SA), the FWO-Vlaanderen (G.0287.07, JC), the Foundation against Cancer (SCIE2006-34, JC), and the European Research Council (ERC-starting grant to JC).
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Tartaglia M, Martinelli S, Cazzaniga G, Cordeddu V, Iavarone I, Spinelli M, et al. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 2004;104(2):307–13. doi: 10.1182/blood-2003-11-3876. [DOI] [PubMed] [Google Scholar]
- 2.Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34(2):148–50. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 3.Kleppe M, Lahortiga I, El Chaar T, De Keersmaecker K, Mentens N, Graux C, et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42(6):530–5. doi: 10.1038/ng.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosinger B, Jr, Tillmann U, Westphal H, Tremblay ML. Cloning and characterization of a mouse cDNA encoding a cytoplasmic protein-tyrosine-phosphatase. Proc Natl Acad Sci USA. 1992;89(2):499–503. doi: 10.1073/pnas.89.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen JN, Jansen PG, Echwald SM, Mortensen OH, Fukada T, Del Vecchio R, et al. A genomic perspective on protein tyrosine phosphatases: gene structure, pseudogenes, and genetic disease linkage. FASEB J. 2004;18(1):8–30. doi: 10.1096/fj.02-1212rev. [DOI] [PubMed] [Google Scholar]
- 6.Doody KM, Bourdeau A, Tremblay ML. T-cell protein tyrosine phosphatase is a key regulator in immune cell signaling: lessons from the knockout mouse model and implications in human disease. Immunol Rev. 2009;228(1):325–41. doi: 10.1111/j.1600-065X.2008.00743.x. [DOI] [PubMed] [Google Scholar]
- 7.Tiganis T, Kemp BE, Tonks NK. The protein-tyrosine phosphatase TCPTP regulates epidermal growth factor receptor-mediated and phosphatidylinositol 3-kinase-dependent signaling. J Biol Chem. 1999;274(39):27768–75. doi: 10.1074/jbc.274.39.27768. [DOI] [PubMed] [Google Scholar]
- 8.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, McGlade CJ. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr Biol. 2002;12(6):446–53. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 9.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Fourth Edition. Lyon: World Health Organization; 2008. [Google Scholar]
- 10.Kleppe M, Mentens N, Tousseyn T, Wlodarska I, Cools J. MOHITO, a novel mouse cytokine-dependent T-cell line, enables studies of oncogenic signaling in the T-cell context. Haematologica. 2011;96(5):779–83. doi: 10.3324/haematol.2010.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cool DE, Tonks NK, Charbonneau H, Walsh KA, Fischer EH, Krebs EG. cDNA isolated from a human T-cell library encodes a member of the protein-tyrosine-phosphatase family. Proc Natl Acad Sci USA. 1989;86(14):5257–61. doi: 10.1073/pnas.86.14.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116(17):3268–77. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diehl V, Kirchner HH, Schaadt M, Fonatsch C, Stein H, Gerdes J, et al. Hodgkin’s disease: establishment and characterization of four in vitro cell lines. J Cancer Res Clin Oncol. 1981;101(1):111–24. doi: 10.1007/BF00405072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joos S, Kupper M, Ohl S, von Bonin F, Mechtersheimer G, Bentz M, et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res. 2000;60(3):549–52. [PubMed] [Google Scholar]
- 17.Weniger MA, Melzner I, Menz CK, Wegener S, Bucur AJ, Dorsch K, et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phos-pho-STAT5 accumulation. Oncogene. 2006;25(18):2679–84. doi: 10.1038/sj.onc.1209151. [DOI] [PubMed] [Google Scholar]
- 18.Lu X, Chen J, Sasmono RT, Hsi ED, Sarosiek KA, Tiganis T, et al. T-cell protein tyrosine phosphatase, distinctively expressed in activated-B-cell-like diffuse large B-cell lymphomas, is the nuclear phosphatase of STAT6. Mol Cell Biol. 2007;27(6):2166–79. doi: 10.1128/MCB.01234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]