Abstract
Bortezomib induced peripheral neuropathy is a dose-limiting side effect and a major concern in the treatment of multiple myeloma. To identify genetic risk factors associated with the development of this side effect in bortezomib treated multiple myeloma patients, a pharmacogenetic association study was performed using a discovery set (IFM 2005-01; n=238) and a validation set (HOVON65/GMMG-HD4 and a Czech dataset; n=231). After multiplicity correction, none of the 2,149 single nucleotide polymorphisms tested revealed any significant association with bortezomib induced peripheral neuropathy. However, 56 single nucleotide polymorphisms demonstrated an association with bortezomib induced peripheral neuropathy with pointwise, uncorrected significance. Pathway analysis of these polymorphisms demonstrated involvement of neurological disease (FDR <20%). Also a clear enrichment of major bortezomib metabolizing genes was found. Univariate evaluation of these 56 polymorphisms in the validation set demonstrated one single nucleotide polymorphism with pointwise significance: rs619824 in CYP17A1.
Keywords: bortezomib, peripheral neuropathy, risk, single nucleotide polymorphism
Introduction
The introduction of bortezomib (Millennium Pharmaceuticals, Cambridge, MA, USA), an inhibitor of the 26S proteasome, has greatly improved the management of multiple myeloma (MM).1 The dose-limiting toxicity of bortezomib is peripheral neuropathy, which frequently requires a dose reduction or treatment discontinuation.2–4 Bortezomib induced peripheral neuropathy (BiPN) differs from pre-existing peripheral neuropathy associated with 10% of untreated MM patients. BiPN, described in detail by Delforge et al.,4 is predominantly sensory, reversible in most cases, and characterized by distal paresthesias, numbness and neuropathic pain.
A multifactorial pathogenesis for BiPN seems likely, with suggested mechanisms including blockade of nerve-growth-factor-mediated neuronal survival through inhibition of the activation of nuclear factor κB (NFκB),5 damage to mitochondria and the endoplasmic reticulum through activation of apoptosis,6 dysregulation of mitochondrial calcium homoeostasis,7 autoimmune factors, interference with mRNA processing, and translation8 and inflammation.9–10 A number of studies, including a report by our own group, have looked at the pharmacogenetic characterization of BiPN.11–12 In the study carried out by our group, the comparison between early onset (within one treatment cycle) BiPN and late onset (after two or three treatment cycles) BiPN revealed that genes for apoptosis contribute to early onset BiPN, whereas genes that have a role in inflammatory pathways and DNA repair contribute to the development of late onset BiPN, indicating that distinct genetic factors are involved in the development of early onset and late onset forms of this side effect.11 Recently, Favis et al. reported on the association between SNPs and the time to bortezomib induced peripheral neuropathy within the VISTA trial with associated SNPs including a SNP in the gene CTLA4.12
In this study, we further explore the genetic risk factors associated with the development of BiPN in patients with MM who had not been previously treated with bortezomib. A large dataset from the IFM 2005-01 trial was used as discovery set. In addition, a dataset based on the patients from the HOVON-65/GMMG-HD4 trial were used as a validation set.11
Design and Methods
Patients
The study was performed on patients who had been included in two randomized clinical trials, i.e. the Institutional Review Board-approved HOVON-65/GMMG-HD4 (ISRCTN64455289) trial for newly diagnosed patients with MM (n=833), and the IFM 2005-01 trial (NCT00200681; n=493) approved by the Ethics Committee of the University of Nantes, both of which compared standard induction treatment (VAD) with a bortezomib combination prior to high-dose therapy (HDT) and stem cell transplantation (Online Supplementary Figure S1A). In addition, as part of the cooperative program of the International Myeloma Foundation and International Myeloma Working Group, a set of 56 patients (i.e. 56 unique DNA samples), uniformly treated with bortezomib and dexamethasone at relapse, were obtained. In addition, a prospectively collected set of samples (n=56) from the Babak Research Institute (Czech Republic) was included as part of the cooperative program of the International Myeloma Foundation and International Myeloma Working Group. All patients gave written informed consent for this genetic study. Patients with amyloidosis (AL-) or monoclonal gammopathy of undetermined significance (MGUS) were excluded. Adverse events (AEs) were prospectively assessed using standard National Cancer Institute Common Toxicity Criteria for Adverse Events, version 3.0 (CTCAE 3.0). To ensure homogeneity of allelic frequencies, 15 patients of non-European descent were excluded from the study. In total, 238 of 246 patients from IFM 2005-01, 183 of 412 patients from HOVON-65/GMMG-HD4 and 48 of 56 from the Czech Republic who were randomized for treatment with bortezomib were included in the analysis. Samples were divided into a discovery and validation set (Online Supplementary Figure S1B, Online Supplementary Table S1).
Genotyping
DNA was extracted from peripheral blood nucleated cells or CD138 negative bone marrow cells. Genotyping was performed using an Affymetrix® Targeted genotyping custom built panel, comprising 3404 SNPs, selected using a hypothesis-driven strategy, targeting genes and SNPs with previously described associations or putative functional effects.13
Statistical analysis
After imputation and applying SNP exclusion criteria (minor allele frequency (MAF) <0.05, Hardy Weinberg equilibrium <1×10−5), a panel containing 2,149 SNPs was analyzed by univariate association analysis using the software package PLINK.14 Categorical comparisons with respect to frequencies were performed with the χ2 or Fisher’s exact test, and continuous variables were analyzed using the Mann-Whitney U test (Online Supplementary Table S1).
SNP association analysis comparing grade 1–4 BiPN with no BiPN patients in the discovery set (IFM 2005-01) was performed as previously described.11
The associated gene sets were subjected to Ingenuity Pathway Analysis (Ingenuity System Inc., USA) using 2,149 SNPs as a reference set. Only the top three associated pathways with a FDR≤20% are reported.
As validation, a Cochran Mantel-Haenszel stratified association test was performed in an independent dataset comprised of patients from the HOVON-65/GMMG-HD4 trial and patients from the Czech Republic to evaluate cross validating SNP associations and Odds Ratios (ORs). Specifically, ORs from significant SNPs (pointwise P<0.05) in the discovery set were selected for validation. A one-sided test for OR was performed to test whether the observed effects in the validation set were associated with the same effect direction as observed in the discovery set.
Based on the numbers of the discovery and validation set, a conservative power calculation for both sets was performed. According to this calculation, ORs need to be higher than 2.28 or lower than 0.44 to be found at a significance level of 0.05 for SNPs with a minor allele frequency (MAF) of 0.5. These ORs diverge as the MAF decreases (Online Supplementary Figures S2 and S3, Table S1 and S2). Please note this is a conservative analysis in which multiplicity correction is performed by Bonferroni correction and no linkage is taken into account.
Results and Discussion
The BiPN rates and clinical characteristics of both the discovery set (n=238) and the validation set (n=231) are shown in the Online Supplementary Table S1. In the discovery set, 27 patients developed BiPN grade 1, 57 grade 2, 11 grade 3, and 4 grade 4. Online Supplementary Figure S4 shows the time to BiPN for each grade separately in patients from the HOVON-65/GMMG-HD4 trial, who are included in the validation set. The median time to BiPN grade 1 was six weeks, and seven weeks to grade 2, 3 or 4. The peripheral neuropathy rates in the VAD treatment arm (i.e. not bortezomib) of the HOVON-65/GMMG-HD4 trial, will not be discussed further here (Online Supplementary Table S3).
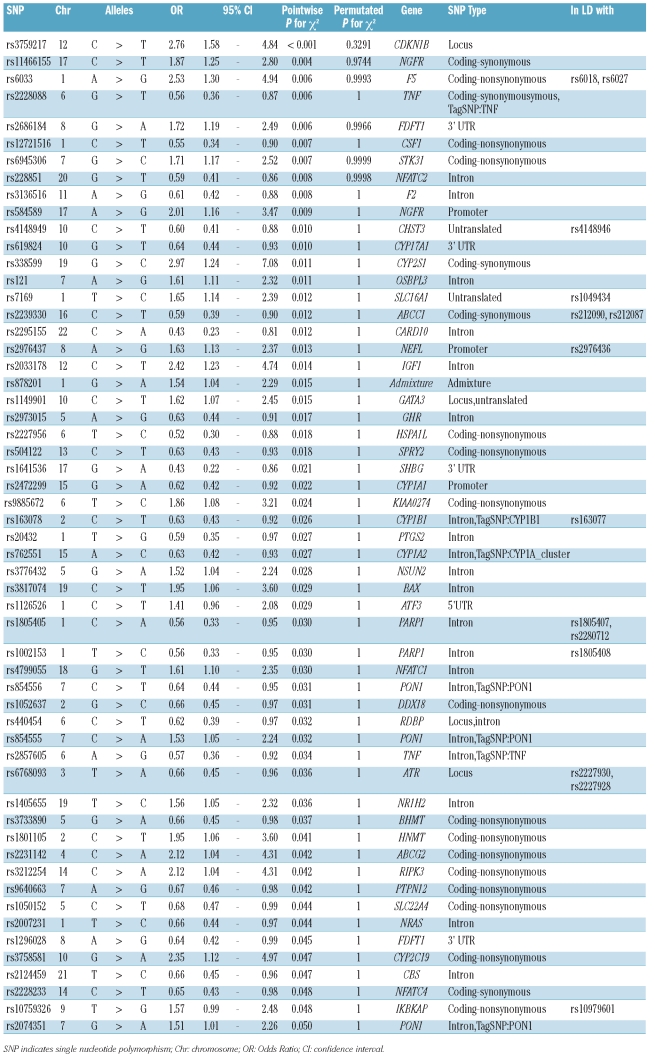
After imputation and applying PLINK exclusion filters, a panel containing 2,149 SNPs was analyzed for association by conducting a χ2 association analysis. None of the SNPs were found to be significantly associated with BiPN using the permutated P value correction for multiple testing in the discovery set (IFM2005-01; Table 1). The highest ranking SNP, with corrected P value of 0.3, is in the locus of the cell cycle gene CDKN1B. This SNP, rs3759217, has been evaluated in a number of cancer studies, but was not reported to be significantly associated with any cancer type.15 Using the pointwise, uncorrected P value, 56 SNPs were found to be associated with BiPN in this set (Table 1).
Table 1.
SNPs associated with BiPN (pointwise P<0.05) using an χ2 association analysis. The genomic inflation factor λ is 1.0201.
The results of the analysis performed in the discovery set (IFM 2005-01 trial) were validated using an independent dataset from the Czech Republic combined with the dataset from the HOVON-65/GMMG-HD4 trial (Online Supplementary Figure S1). A Cochran Mantel-Haenszel stratified association test was performed. Associated SNPs (pointwise P<0.05) in this validation set are shown in Online Supplementary Table S5. To investigate whether associated SNPs (pointwise P<0.05) in the discovery set and available in the validation set (n=51) had the same direction of effect, a one-sided test for ORs was performed in the validation set. This resulted in one pointwise significantly cross validating SNP; rs619824 in CYP17A1 (Online Supplementary Table S6).
CYP17A1, cytochrome P-450c17α, is involved in steroid hormone biosynthesis, and has both steroid 17α-hydroxylase activity and 17,20-lyase activity.16 Steroids have been shown to affect nerve cells, and have even been suggested for use as a therapeutic option to prevent the development of neuropathy.17 Treatment with progesterone has been reported to increase the expression of myelin protein zero in both rat sciatic nerve and Schwann cells.17 Due to the paucity of cross validated SNPs, we have examined the SNPs with a significant pointwise P value in the discovery set (Table 1). Foremost, we have performed a pathway analysis based on this set of SNPs. This analysis showed enrichment of genes involved in cardiovascular disease (11 genes), genetic disorder (22 genes) and neurological disease (21 genes). The latter include the genes NEFL, PON1, PTGS2 and ABCG2, which have been reported frequently in relation to neurological disease such as Alzheimer’s.
Previous studies showed that bortezomib is primarily metabolized by cytochrome P450 isoforms CYP3A4, CYP2C19, CYP1A2, with a minor contribution of CYP2D6 and CYP2C9.18 The results show an enrichment of the major bortezomib metabolizing genes within the top 56 SNPs (P=0.0013).
Previously, genes involved in inflammation were found to be associated with late onset BiPN.11 Indeed, one of the most associated SNPs, rs3136516 (pointwise P=0.008) was an intronic SNP located in prothrombin (coagulation factor II; F2), which has been reported in relation to the neuro-toxic cascade leading to neurodegenerative diseases.19 Two SNPs that lie within or in close proximity to the TNFα gene (rs2857605 and rs2228088; Online Supplementary Figure S5) were associated with BiPN. TNFα has been implicated in the pathogenesis of several neurodegenerative diseases, including multiple sclerosis, Alzheimer’s disease, and human immunodeficiency virus-related encephalopathy.20 Additionally, the TNFα system is activated in diabetic polyneuropathy, which leads to increased microvascular permeability, hypercoagulability and even direct nerve damage. Improvement of diabetic polyneuropathy following suppression of TNFα has been shown in several animal models.21 Furthermore, neuropathic pain, one of the determinants of the CTCAE-neuropathy score, and thus of BiPN severity, is mediated through TNF-mediated induction of stress-activated kinases like p38 MAPK.22
The NFκB pathway is central to the immune response and two associated SNPs are located in the IKBKAP gene; rs10979601 and rs10759326. This is a particularly relevant association because hereditary sensory and autonomic neuropathy type III, or familial dysautonomia (FD), can be caused by mutations in the IKBKAP gene, leading to poor development, reduced survival, and progressive degeneration of the sensory and autonomic nervous system.23
Mutations in neurofilament light polypeptide (NEFL) cause Charcot-Marie-Tooth Neuropathy Type 2E/1F, the most common inherited peripheral neuropathy.24 Two promoter SNPs (rs2976437 and rs2976436) in NEFL were associated with BiPN. Two SNPs were located in the nerve growth factor receptor (NGFR; rs11466155 and rs584589), a gene particularly important with respect to neurological functions. The NFGR signals via NFκB activation and binds neutrophin precursors that stimulate neuronal cell survival and differentiation. These results support the finding in our previous study that late onset BiPN is associated with genes involved in the development and function of the nervous system.11 In a recent paper, the time to BiPN was found to be associated with the occurrence of the SNP rs4553808 in the gene CTLA4.12 Comparison with that study is not feasible, due to the fact that the SNP set tested had only minimal overlap with our SNP set (2% overlap).
We evaluated genetic risk factors associated with BiPN in MM patients who had not been previously treated with bortezomib in the largest study to date using a hypothesis-driven approach. This method is limited by the possibility of population heterogeneity. However, a limited set of patients with different genetic backgrounds were selected out, as described in Design and Methods and reported previously.11 Further limitations are: i) the inability of assessing SNPs outside the candidate panel; and ii) the possibility of finding false-positive associations as a result of multiple testing. To address both issues, we are currently performing a genome-wide scan that will clarify and possibly confirm the associations reported in this study. The power analysis indicated in this study has sufficient power to detect associations with an OR of less than 0.44 or an OR of more than 2.28 and diverging with MAF. It is unlikely that smaller effects can be found. Using the custom BOAC SNP array in a discovery set of 238 patients, no SNP was found to be significantly associated to BiPN at the corrected P<0.05 significance level. However, based on the highest-ranking SNPs found using the uncorrected P value in the discovery set, pathway analysis did demonstrate clear enrichment of neurological disease SNPs.
Acknowledgments
We would like to thank participants of the HOVON-65/GMMG-HD4 and IFM 2005-01 trials. We thank the International Myeloma Work Group for providing patient samples. We would like to thank Brian Durie from the International Myeloma Foundation (IMF) for his contribution. Funding: this work was supported via the Biomedical Research Centre at the Royal Marsden Hospital, by the International Myeloma Foundation (IMF), the Dutch Cancer Foundation, Skyline Diagnostics, the German Federal Ministry of Education and Research, MSMT of the Czech Republic (MSM 0021622434, LC 06027) and Erasmus MC.
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352(24):2487–98. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 2.Richardson PG, Briemberg H, Jagannath S, Wen PY, Barlogie B, Berenson J, et al. Frequency, characteristics, and reversibility of peripheral neuropathy during treatment of advanced multiple myeloma with bortezomib. J Clin Oncol. 2006;24(19):3113–20. doi: 10.1200/JCO.2005.04.7779. [DOI] [PubMed] [Google Scholar]
- 3.Richardson PG, Xie W, Mitsiades C, Chanan-Khan AA, Lonial S, Hassoun H, et al. Single-agent bortezomib in previously untreated multiple myeloma: efficacy, characterization of peripheral neuropathy, and molecular correlations with response and neuropathy. J Clin Oncol. 2009;27(21):3518–25. doi: 10.1200/JCO.2008.18.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delforge M, Blade J, Dimopoulos MA, Facon T, Kropff M, Ludwig H, et al. Treatment-related peripheral neuropathy in multiple myeloma: the challenge continues. Lancet Oncol. 2010;11(11):1086–95. doi: 10.1016/S1470-2045(10)70068-1. [DOI] [PubMed] [Google Scholar]
- 5.Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348(26):2609–17. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 6.Cavaletti G, Gilardini A, Canta A, Rigamonti L, Rodriguez-Menendez V, Ceresa C, et al. Bortezomib-induced peripheral neurotoxicity: a neurophysiological and pathological study in the rat. Exp Neurol. 2007;204(1):317–25. doi: 10.1016/j.expneurol.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Landowski TH, Megli CJ, Nullmeyer KD, Lynch RM, Dorr RT. Mitochondrial-mediated disregulation of Ca2+ is a critical determinant of Velcade (PS-341/bortezomib) cytotoxicity in myeloma cell lines. Cancer Res. 2005;65(9):3828–36. doi: 10.1158/0008-5472.CAN-04-3684. [DOI] [PubMed] [Google Scholar]
- 8.Casafont I, Berciano MT, Lafarga M. Bortezomib induces the formation of nuclear poly(A) RNA granules enriched in Sam68 and PABPN1 in sensory ganglia neurons. Neurotox Res. 2010;17(2):167–78. doi: 10.1007/s12640-009-9086-1. [DOI] [PubMed] [Google Scholar]
- 9.Ravaglia S, Corso A, Piccolo G, Lozza A, Alfonsi E, Mangiacavalli S, et al. Immune-mediated neuropathies in myeloma patients treated with bortezomib. Clin Neurophysiol. 2008;119(11):2507–12. doi: 10.1016/j.clinph.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Cavaletti G, Marmiroli P. Chemotherapy-induced peripheral neurotoxicity. Nat Rev Neurol. 2010;6(12):657–66. doi: 10.1038/nrneurol.2010.160. [DOI] [PubMed] [Google Scholar]
- 11.Broyl A, Corthals SL, Jongen JL, van der Holt B, Kuiper R, de Knegt Y, et al. Mechanisms of peripheral neuropathy associated with bortezomib and vincristine in patients with newly diagnosed multiple myeloma: a prospective analysis of data from the HOVON-65/GMMG-HD4 trial. Lancet Oncol. 2010;11(11):1057–65. doi: 10.1016/S1470-2045(10)70206-0. [DOI] [PubMed] [Google Scholar]
- 12.Favis R, Sun Y, van de Velde H, Broderick E, Levey L, Meyers M, et al. Genetic variation associated with bortezomib-induced peripheral neuropathy. Pharmacogenet Genomics. 2011;21(3):121–9. doi: 10.1097/FPC.0b013e3283436b45. [DOI] [PubMed] [Google Scholar]
- 13.Van Ness B, Ramos C, Haznadar M, Hoering A, Haessler J, Crowley J, et al. Genomic variation in myeloma: design, content, and initial application of the Bank On A Cure SNP Panel to detect associations with progression-free survival. BMC Med. 2008;6:26. doi: 10.1186/1741-7015-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gayther SA, Song H, Ramus SJ, Kjaer SK, Whittemore AS, Quaye L, et al. Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007;67(7):3027–35. doi: 10.1158/0008-5472.CAN-06-3261. [DOI] [PubMed] [Google Scholar]
- 16.Sharp L, Cardy AH, Cotton SC, Little J. CYP17 gene polymorphisms: prevalence and associations with hormone levels and related factors. a HuGE review. Am J Epidemiol. 2004;160(8):729–40. doi: 10.1093/aje/kwh287. [DOI] [PubMed] [Google Scholar]
- 17.Roglio I, Giatti S, Pesaresi M, Bianchi R, Cavaletti G, Lauria G, et al. Neuroactive steroids and peripheral neuropathy. Brain Res Rev. 2008;57(2):460–9. doi: 10.1016/j.brainresrev.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Curran MP, McKeage K. Bortezomib: a review of its use in patients with multiple myeloma. Drugs. 2009;69(7):859–88. doi: 10.2165/00003495-200969070-00006. [DOI] [PubMed] [Google Scholar]
- 19.Mhatre M, Nguyen A, Kashani S, Pham T, Adesina A, Grammas P. Thrombin, a mediator of neurotoxicity and memory impairment. Neurobiol Aging. 2004;25(6):783–93. doi: 10.1016/j.neurobiolaging.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Sriram K, O’Callaghan JP. Divergent roles for tumor necrosis factor-alpha in the brain. J Neuroimmune Pharmacol. 2007;2(2):140–53. doi: 10.1007/s11481-007-9070-6. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Clemente JM, Mauricio D, Richart C, Broch M, Caixas A, Megia A, et al. Diabetic neuropathy is associated with activation of the TNF-alpha system in subjects with type 1 diabetes mellitus. Clin Endocrinol (Oxf) 2005;63(5):525–9. doi: 10.1111/j.1365-2265.2005.02376.x. [DOI] [PubMed] [Google Scholar]
- 22.Boyle DL, Jones TL, Hammaker D, Svensson CI, Rosengren S, Albani S, et al. Regulation of peripheral inflammation by spinal p38 MAP kinase in rats. PLoS Med. 2006;3(9):e338. doi: 10.1371/journal.pmed.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, et al. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet. 2001;68(3):753–8. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordanova A, De Jonghe P, Boerkoel CF, Takashima H, De Vriendt E, Ceuterick C, et al. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain. 2003;126(Pt 3):590–7. doi: 10.1093/brain/awg059. [DOI] [PubMed] [Google Scholar]