Abstract
Background
Five years of tamoxifen reduces breast cancer risk by nearly 50% but is associated with significant side-effects and toxicities. A better understanding of the direct and indirect effects of tamoxifen in benign breast tissue could elucidate new mechanisms of breast carcinogenesis, suggest novel chemoprevention targets, and provide relevant early response biomarkers for Phase II prevention trials.
Methods
Seventy-three women at increased risk for breast cancer were randomized to tamoxifen (20 mg daily) or placebo for three months. Blood and breast tissue samples were collected at baseline and post-treatment. Sixty-nine women completed all study activities (37 tamoxifen and 32 placebo). The selected biomarkers focused on estradiol and IGFs in the blood, DNA methylation and cytology in random periareolar fine needle aspirates, and tissue morphometry, proliferation, apoptosis, and gene expression (microarray and RT-PCR) in the tissue core samples.
Results
Tamoxifen downregulated ets-oncogene transcription factor family members ETV4 and ETV5 and reduced breast epithelial cell proliferation independent of CYP2D6 genotypes or effects on estradiol, ESR1 or IGFs. Reduction in proliferation was correlated with downregulation of ETV4 and DNAJC12. Tamoxifen reduced the expression of ETV4- and ETV5-regulated genes implicated in epithelial-stromal interaction and tissue remodeling. Three months of tamoxifen did not affect breast tissue composition, cytological atypia, preneoplasia or apoptosis.
Conclusions
A plausible mechanism for the chemopreventive effects of tamoxifen is restriction of lobular expansion into stroma through downregulation of ETV4 and ETV5. Multipotential progenitor cap cells of terminal end buds may be the primary target.
Keywords: Tamoxifen, Biomarkers, Gene Expression, Proliferation, DNA Methylation
INTRODUCTION
Estrogen receptor-alpha (ESR1) signaling is a key driver of breast carcinogenesis; consequently, agents which modulate ESR1 activity or deplete its ligand reduce breast cancer incidence. However, estrogen signaling plays a critical role in diverse biological processes accounting for the frequent adverse events that occur when this pathway is pharmacologically perturbed. Tamoxifen is a selective estrogen receptor modifier (SERM) that has been shown to reduce breast cancer incidence by nearly 50% in increased risk women(1), but it also increases the risk for endometrial cancer and venous thromboembolic events. Chemoprevention research has focused primarily on identifying SERMs with better safety profiles, but understanding critical molecular events occurring downstream of ESR1 may permit the development of paradigm-shifting chemoprevention approaches capable of specifically targeting benign breast tissue to reduce breast cancer incidence.
There is increasing interest in the role of insulin-like growth factor (IGF) signaling in breast carcinogenesis and progression. Tamoxifen has previously been shown to reduce circulating IGF-1(2),(3) and IGF-1 has been proposed as a surrogate endpoint biomarker for Phase II chemoprevention trials(4),(5),(6). It is unclear, however, whether systemic modulation of IGF-1 is simply a bystander effect or a key mediator of tamoxifen effects in benign breast tissue.
There is considerable published information concerning the effects of tamoxifen on breast cancer and breast cancer cell lines, but very little for benign breast tissue. Pathways modulated by tamoxifen in benign breast tissue may have relevance for carcinogenesis, may suggest targets for novel prevention approaches, and may provide biomarkers useful as surrogate endpoints in prevention trials.
MATERIALS AND METHODS
Study Design
A Phase II multi-institutional randomized, prospective, double blind placebo-controlled trial was conducted to identify biomarkers that are modulated by tamoxifen but not placebo in women at increased risk for breast cancer (ClinicalTrials.gov NCT00096369). This research was performed in accordance with an assurance filed with and approved by the U.S. Department of Health and Human Services. Institutional Review Board approval was obtained at each site and informed consent was documented in writing for each participant. Women ≥ 35 years of age with a 5-year Gail risk ≥ 1.67% or a personal history of lobular carcinoma in situ (LCIS) were eligible. Exclusion criteria included ever-use of SERMs, use of systemic steroid hormones (including oral contraceptives or hormone replacement therapy) within three months, personal history of invasive or in situ breast cancer, pregnancy or lactation within six months, a history of thromboembolic disease that would preclude the use of tamoxifen, the presence of breast implants, or a bleeding diathesis that would preclude needle sampling of the breast. Eligible and consenting women were centrally randomized (Efron-type biased coin randomization) to tamoxifen or placebo. Stratification variables included 5 year Gail risk < or ≥ 5%, history of high risk preneoplasia including atypical ductal hyperplasia, atypical lobular hyperplasia or LCIS, and menopausal status. Criteria for classifying a woman as postmenopausal included prior bilateral oophorectomy, amenorrhea for > 12 months with an intact uterus and at least one ovary, or amenorrhea and FSH > 20 mIU/ml. Women not meeting any of these criteria were classified as premenopausal. Blood and breast tissue samples were collected at baseline and after three months of tamoxifen or placebo.
The original study design called for enrollment of 130 women to provide 50 evaluable subjects in each treatment group while allowing for a 23% attrition rate prior to the second time point sampling. This sample size was judged sufficient to provide 80% power to recognize 17 – 25% differences in modulation of selected biomarkers in tamoxifen subjects as compared to placebo subjects with two-tailed alpha set at 0.05. The study was activated in 2002 and closed early in 2007 due to poor accrual after enrolling 73 women at five study sites (UT Southwestern Medical Center; UT MD Anderson Cancer Center; Baylor University Medical Center, Dallas; Oklahoma University Health Science Center; and the Cancer Therapy and Research Center, San Antonio). Attrition prior to the second sampling was 5.5% (four subjects).
Biological Samples
Biological samples were collected by venipuncture, bilateral random periareolar fine needle aspiration (RP-FNA), and unilateral breast tissue core biopsy at baseline and after a median of 84 days of treatment with tamoxifen or placebo. For premenopausal women, the baseline and post-treatment samples were obtained on day 28 of the menstrual cycle +/− 2 days. Late luteal phase sampling was specifically chosen to avoid measuring acute estrogenic effects while still capturing events (e.g. apoptosis) in a tissue remodeling phase(7).
Blood was collected by venipuncture into Vacutainer CPT tubes (Becton Dickinson) which were immediately centrifuged to separate plasma and lymphocytes. Aliquots were immediately frozen at −80°C. Bilateral RP-FNA was performed as previously described(8),(9). Direct smears were made for cytological assessment and material from each breast was pooled in PreserveCyt (Cytyk Health Corporation) for subsequent DNA extraction. Fourteen-gauge core needle samples were obtained from the palpably dense tissue in the upper outer quadrant of one breast for each patient. Two cores were immediately snap-frozen in liquid nitrogen for later RNA extraction, and the others were fixed in formalin for histological assessment and immunohistochemistry. Post-treatment core biopsies were performed in the same breast sampled at baseline.
Treatment
Subjects were provided with a one month supply of identically labeled tamoxifen or placebo. The tamoxifen dose was 20 mg orally each day. Compliance was measured by pill counts every four weeks.
Biomarker Selection
The biomarker panel included markers known to be modulated by tamoxifen (e.g. IGF-1), recently proposed markers of breast cancer risk (e.g. RP-FNA cytology(10), number of acini per lobule(11),(12), and DNA methylation(8),(9)), and gene expression markers with the potential to generate new hypotheses concerning the effects of tamoxifen on benign breast tissue.
Plasma Markers
To avoid repeated freezing and thawing, all of the assays for a given sample were usually run on the same day. Baseline and post-treatment samples for a given individual were always run on the same plate. Plasma markers included estradiol (RIA, DSL-39100, Diagnostic Systems Laboratories Inc.), albumin (QuantiChrom BCG Albumin Assay Kit, BioAssay Systems), sex hormone binding globulin (SHBG, ELISA, DSL-10-7400), prolactin (ELISA, DSL-10-4500), IGF-1 (ELISA, DSL-10-5600), IGF-2 (ELISA, DSL-10-2600), IGFBP-1 (ELISA, DSL-10-7800), and IGFBP-3 (ELISA, DSL-10-6600). Free estradiol was calculated from total estradiol, albumin and SHBG as previously described(13).
Tissue Core Markers
Histological assessment of hematoxyllin and eosin-stained slides was performed by a single breast pathologist (VS), specifically evaluating the characteristics of the epithelium: normal, non-proliferative fibrocystic change, or proliferative fibrocystic change. Specific histological patterns were recorded, including cystic changes, apocrine metaplasia, adenosis, sclerosing adenosis, papilloma/papillomatosis, epithelial hyperplasia (mild, moderate, or florid), atypical ductal hyperplasia (ADH), atypical lobular hyperplasia (ALH), columnar cell change, or LCIS.
Markers measured by immunohistochemistry by the CLIA/CAP certified laboratory, OncoDiagnostics (Dallas, TX), included: proliferation (MIB-1 DAKO), epidermal growth factor receptor (EGFR, ZYMED 31G7), BCL2 (DAKO 124), and estrogen receptor-alpha (ESR1, DAKO 1D5). The pattern of ETV4 (SC-113, Santa Cruz Biotechnology) and ETV5 (SC-22807, Santa Cruz Biotechnology) protein expression was assessed in four core samples from two premenopausal participants. Proliferation was quantified by manual cell counting as described in the Data Analysis section. Estrogen receptor and EGFR were assessed using a DAKO autostainer and the percentage of positive cells quantified using an Automated Cellular Imaging System (ACIS). BCL2 was scored manually as 0, 1+, 2+, or 3+. Apoptosis was measured by TUNEL assay in the laboratory of Weiya Xia at M.D. Anderson Cancer Center.
Breast tissue morphometry was quantified by computer-assisted tissue component analysis (NIH Image, Scion Corp). The fractional area corresponding to epithelial structures, fibrous stroma, and adipose was measured for one entire H&E stained section for each sample. This included cuts through multiple cores. On average, 37.1 mm2 were assessed (range = 5.8 – 74.5, Standard Deviation = 13.9). The number of acini in each lobule was manually counted and recorded, permitting calculation of the mean acini/lobule ratio and total acini.
Fine Needle Aspiration Markers
Each Papanicolaou-stained smear was classified by a breast cytopathologist (RA) as acellular, normal epithelium, hyperplasia, or atypia. Each sample was also assigned a Masood score(14) based on cell arrangement, cellular pleomorphism, paucity of myoepithelial cells, anisonucleosis, prominence of nucleoli, and chromatin clumping. Epithelial cells were manually counted for each slide to provide a direct measure of sample cellularity.
Tumor suppressor gene methylation was measured by Quantitative Multiplex Methylation-specific PCR (QM-PCR)(15). Markers evaluated included cyclin D2, APC, HIN-1, CST6, RASSF1A, and RAR-β2. The primers, probes, and performance characteristics of this assay have been described previously(16).
CYP2D6 Genotypes
CYP2D6 genotyping was performed in the laboratory of Dr. David Flockhart on DNA extracted from peripheral blood mononuclear cells isolated from whole blood of the tamoxifen subjects using methods that have previously been described(17). The observed alleles included *1, *2, *4, *5, *9, *11, *12, and *41. Patients with one or more *4 or *5 alleles were classified as poor metabolizers.
Gene Expression
RNA was extracted from 113 snap frozen breast tissue core samples (Qiazol and the RNeasy Micro Kit, Qiagen), amplified once, (TargetAmp Biotin-aRNA Amplification Kit, Epicentre Biotechnologies) and then hybridized to the Illumina whole genome 48k chip (Human-6 v2). An RNA Integrity Number (RIN) was measured for each sample using an Agilent 2100 Bioanalyzer. The RINs ranged from 6.6 to 9.1 with an average of 7.9. Core biopsy samples with few or no epithelial structures were excluded; consequently, the microarray analysis described in the results section is based on 70 arrays (35 two-time point pairs). Reasons for excluding arrays included < 5 lobules in both paired core samples (19), < 5 lobules in one of the paired samples (5), no paired sample (11), duplicate sample (2), chip did not pass quality criterion of mean correlation ≥ 0.94 compared to all other chips (5), and correlation < 0.94 in the paired sample (1). The expression data is available at NCBI GEO GSE293338.
RNA was also extracted from microdissected lobules from 5 tamoxifen-treated subjects (2 premenopausal and 3 postmenopausal), amplified twice, and then hybridized to Illumina whole genome 48k chips (NCBI GEO GSE29338). Data from these doubly amplified samples did not meet the quality criteria established for the whole tissue cores and was judged unreliable for whole genome assessment of tamoxifen-modulated gene expression. Instead, these data were used to confirm the epithelial cell relevance of observations from the whole tissue cores. Mean Log2 ratios (post-treatment/baseline) ≥ 1.5-fold up or down were considered significant in the microdissected lobules.
Reverse transcriptase PCR (RT-PCR) was used to confirm microarray observations for selected genes. cDNA was prepared from RNA extracted from snap frozen core samples using the SuperScript VILO cDNA Synthesis kit (Invitrogen). RT-PCR reactions were performed in Sybr premix Ex Taq II (TAKARA BIO, Inc.) using 5 ng of starting template on a Chromo4 thermocycler (BIO-RAD). The RT-PCR primer sequences are provided in Supplemental Table 3. Normalized Relative Quantities (NRQ) were calculated according to the method of Hellesman(18) using PCR efficiencies calculated from standard curves as 10(−1/slope). After extensive testing for stability in benign breast tissue, three reference genes were chosen: HPRT1, ACTB, and RPL13A. Two calibrators were included on every plate: cDNA prepared from a universal human reference RNA (Stratagene, Agilent Technologies), and cDNA prepared from a standard RNA solution prepared locally from four pooled benign breast tissue samples.
Data Analysis
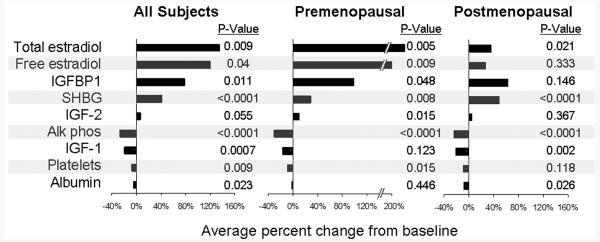
For each plasma marker, the mean Log2 ratio (post-treatment/baseline) for tamoxifen subjects was compared with placebo subjects using two-tailed T-tests. For display purposes (Figure 1), the difference (δ) between post-treatment and baseline values was calculated for each sample pair and then the direction and degree of modulation expressed as (mean δ tamoxifen – mean δ placebo)/ mean baseline all samples.
Figure 1.
Plasma Markers Modulated by Tamoxifen but not Placebo. Horizontal bars show the direction and extent of modulation associated with tamoxifen as compared to placebo.
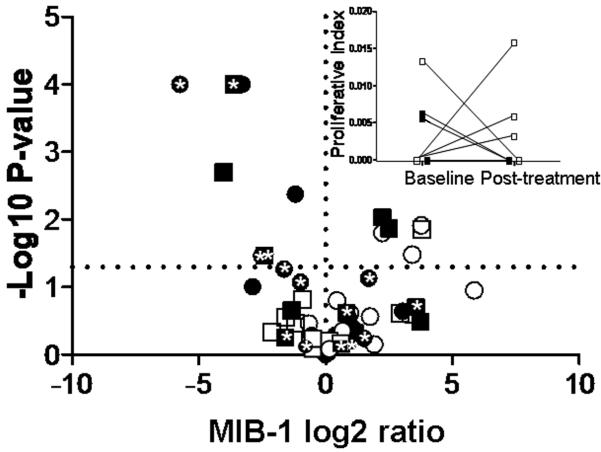
Proliferation rates were calculated separately for every lobule in each core biopsy section by manually counting all MIB-1 positive cells and the total number of cells in the lobule. This permitted a statistical comparison between mean baseline and post-treatment proliferation rates for each individual as shown in the volcano plot in Figure 5. In addition, a summary proliferative index was calculated for each section as the total number of MIB-1 positive cells in all lobules divided by the total number of epithelial cells. The median number of epithelial cells counted per subject was 1,958 (range = 50 – 11,020).
Figure 5.
Volcano plot showing modulation of proliferation for each subject. The X-axis is the MIB-1 Log2 Ratio (post-treatment/baseline) and the Y-axis is the −Log10 for the P-value for each individual. The horizontal dashed line corresponds to P = 0.05. Negative Log2 Ratio values correspond to treatment-related reductions in proliferation and positive values to increased proliferation. Black symbols are tamoxifen subjects and white symbols placebo subjects. Circles are premenopausal subjects and squares postmenopausal subjects. * designates tamoxifen subjects with a CYP2D6 *4 or *5 poor metabolizer allele. The inset shows mean baseline and post-treatment proliferation indices for the 9 postmenopausal subjects with a proliferative index of zero at one or both time points. These subjects cannot be rendered on the volcano plot which requires Log2 transformations. Both baseline and post-treatment proliferative indices were zero for three of the tamoxifen subjects.
Gene expression analysis was performed on 70 breast tissue core biopsies (representing 35 baseline and post-treatment pairs with ≥ 5 lobules in both the baseline and post-treatment core sample) using GeneSpring 11.0.1 as follows. Flags were set to present for entities with detection P-values ≤ 0.2 and absent for entities with detection P-values > 0.4. Raw signals with values < 1 were reset to 1. Quantile normalization was used and baseline transformation performed based on the median expression level for all entities. Of the 48,701 entities included on the Illumina array, flags were present or marginal for 75% of the samples for 27,219 entities and these were retained. The initial analysis combined data from both pre- and postmenopausal women. Genes showing significant modulation in the tamoxifen subjects, based on Benjamin-Hockberg false discovery rate-corrected P-values < 0.05 (paired T-test), were retained (232 genes). To identify genes significantly modulated in the tamoxifen, but not the placebo subjects, mean post-treatment/baseline Log2 ratios and standard deviations were calculated for the tamoxifen (μT and SD_T) and the placebo (μP and SD_P) subjects for each gene according to the class prediction method of Slonim(19). 50 genes were identified with (μT − μP)/(SD_T + SD_P) ≥ 0.5. In a second analysis, pre-menopausal subjects (10 tamoxifen pairs and 8 placebo pairs) were analyzed separately from postmenopausal subjects (11 tamoxifen pairs and 8 placebo pairs). With the reduced samples sizes in this subgroup analysis, no significant Benjamin-Hockberg-corrected P-values were obtained. Criteria for classifying a gene as modulated by tamoxifen but not placebo was relaxed to include, 1) mean fold change in tamoxifen subjects > 1.5 with P < 0.001, and 2) (μT − μP)/(SD_T + SD_P) ≥ 0.5.
Normalized Relative Quantities (NRQ) calculated from the RT-PCR data were compared between post-treatment and baseline samples using paired T-tests. RT-PCR was run on the same 19 tamoxifen pairs and 16 placebo pairs that had been assessed by microarray.
RESULTS
In order to identify biomarkers modulated by tamoxifen but not placebo, 73 women were randomized to 20 mg of tamoxifen each day (N = 40) or placebo (N = 33). Blood and breast tissue samples were collected at baseline and after three months of treatment. Four of these women (5.5%) withdrew from the study prior to the second sampling. One placebo subject was found to have a mammographically occult infiltrating lobular carcinoma on screening MRI obtained as part of her regular clinical management. Three tamoxifen subjects withdrew because of symptoms they attributed to the study medication. A total of 69 women completed all study-related activities including 37 in the tamoxifen group and 32 in the placebo group. Stratification was judged successful based on the similarity of the treatment groups for age, menopausal status, and breast cancer risk (Table 1). Medication compliance, based on pill counts, was > 90% for all but two placebo subjects. Additional evidence for compliance is provided by the observation that 63% of the tamoxifen subjects reported hot flashes as compared to 33% of the placebo subjects (P = 0.024).
Table 1.
Characteristics of the Study Sample
| Characteristic | Placebo | Tamoxifen | P-value |
|---|---|---|---|
| Total Number | 33 | 40 | NA |
| Number dropped | 1 | 3 | NA |
| FOR THOSE COMPLETING BOTH SAMPLINGS | |||
| Age (median and range) | 50.2 (41.6-67.5) | 50.2 (37.0-86.1) | 0.938 |
| Race | |||
| Caucasian, N (%) | 27 (84.4%) | 32 (86.5%) | 1.000 |
| African-American, N (%) |
2 (6.2%) | 2 (5.4%) | |
| Asian, N (%) | 0 (0%) | 1 (2.7%) | |
| Ethnicity | 0.657 | ||
| Hispanic, N (%) | 3 (9.4%) | 2 (5.4%) | |
| Menopausal Status | 0.607 | ||
| Premenopausal | 15 (46.9%) | 14 (37.8%) | |
| Postmenopausal | 17 (53.1%) | 23 (62.2%) | |
| LCIS | 0.696 | ||
| No | 28 (87.5 %) | 34 (91.9%) | |
| Yes | 4 (12.5%) | 3 (8.1%) | |
| > 90% compliant | 30 (93.8%) | 37 (100%) | 0.211 |
| 5-year Gail Risk | 0.436 | ||
| <1.7 | 1 (3.1%) | 2 (5.4%) | |
| 1.7 – 3.3 | 17 (53.1%) | 24 (64.9%) | |
| 3.4 – 4.9 | 12 (37.5%) | 9 (24.3%) | |
| ≥5.0 | 2 (6.3%) | 2 (5.4%) | |
| BMI Kg/M2 (mean) | 29.3 | 27.0 | 0.104 |
Blood Markers
Tamoxifen significantly increased total estradiol in both pre- and postmenopausal women, but this effect was most pronounced for premenopausal women (Figure 1). Increases in total estradiol translated into increased free estradiol for premenopausal women, but the large increases in SHBG seen in postmenopausal women largely offset the modest increase in total estradiol. In general, tamoxifen reduced IGF-1 and increased IGF-2 and IGFBP1, but only marginally affected IGFBP3 (7% increase, P = 0.126).
Gene Expression Modulated by Tamoxifen but not Placebo
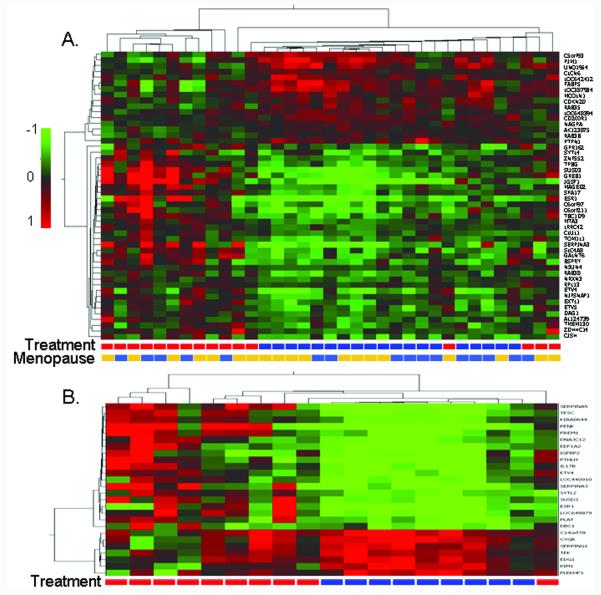
The initial microarray analysis, which included samples from both premenopausal and postmenopausal women, identified 50 genes significantly modulated by tamoxifen but not placebo (Supplemental Table 1). Unsupervised hierarchical Pearson-centered clustering using a centroid linkage rule based on Log2 ratios (post-treatment/baseline) for these 50 genes clearly identified a highly related tamoxifen cluster largely distinct from the placebo-treated subjects (Figure 2A). Twenty-one of these genes were significantly modulated in microdissected lobules from tamoxifen-treated subjects (marked with * in Supplemental Table 1), and in each case the direction of modulation was identical to that observed in the whole breast cores. Known estrogen response genes such as ESR1, GREB1, and SERPINA3 were significantly downregulated; but, notably, the ets-oncogene family transcription factors ETV4 and ETV5 were among the most significantly downregulated genes.
Figure 2.
A) Hierarchical clustering for genes whose expression was significantly modulated by tamoxifen but not placebo in breast core biopsies. Premenopausal and postmenopausal samples were combined in this analysis which used Benjamin-Hockberg-corrected P < 0.05 as the initial selection criteria. Treatment:  placebo,
placebo,  tamoxifen; Menopause is menopausal status:
tamoxifen; Menopause is menopausal status:  premenopausal,
premenopausal,  postmenopausal. B) Hierarchical clustering for genes whose expression was significantly modulated by tamoxifen but not placebo in breast core biopsies for premenopausal subjects only. Group:
postmenopausal. B) Hierarchical clustering for genes whose expression was significantly modulated by tamoxifen but not placebo in breast core biopsies for premenopausal subjects only. Group:  placebo,
placebo,  tamoxifen. The color scale is based on the Log2 ratio (post-treatment/baseline).
tamoxifen. The color scale is based on the Log2 ratio (post-treatment/baseline).
In the second analysis, pre-menopausal subjects were analyzed separately from postmenopausal subjects. This identified 26 genes significantly modulated by tamoxifen but not placebo in premenopausal women (Supplemental Table 2), but no genes met the criteria described in the Methods section for postmenopausal women (ESR1 was downregulated 1.94-fold, P = 0.005). Hierarchical cluster analysis based on the 26 genes identified in premenopausal women clearly identified a highly related premenopausal tamoxifen cluster largely distinct from the premenopausal placebo-treated subjects (Figure 2B). Fifteen of these genes were significantly modulated in microdissected lobules from tamoxifen-treated subjects (marked with * in Supplemental Table 2), and in each case the direction of modulation was identical to that observed in the whole breast cores. Known estrogen response genes such as ESR1 and SERPINA3 were significantly downregulated; but, notably the ets-oncogene family transcription factor ETV4 was also significantly downregulated.
Among the 69 unique genes modulated by tamoxifen but not placebo in the combined and premenopausal analyses 18 are known estrogen response genes (designated by bold text in Supplemental Tables 1 and 2). Tamoxifen downregulated the expression of each of these genes except CD200R1, TEK, and RAB35 which were upregulated. Pathway analysis based on these 69 genes identified ESR1 and the RAB-interacting protein TBC1D9 as major interaction nodes, and identified estrogen receptor signaling and ceramide metabolism as key processes (Supplemental Figure 1).
Gene expression array data were confirmed by RT-PCR for selected genes thought to be particularly relevant to the main effects of tamoxifen in benign breast tissue (Figure 3). In general, there was excellent correlation between microarray and RT-PCR expression values (Supplemental Table 4). As with the array data, significant modulation was generally only observed for premenopausal subjects. Among premenopausal subjects, tamoxifen, but not placebo, significantly downregulated the ets-oncogene transcription factors ETV4 and ETV5, as well as a gene thought to function as an ESR1 chaperone, DNAJC12. Of note, modulation of ETV4, ETV5, and DNAJC12 was entirely independent of tamoxifen-induced changes in plasma estradiol or IGFs but increasing plasma IGFBP3 was correlated with reduced expression of ETV4 (Spearman r = −0.616, P = 0.005). Baseline ETV4 expression was highly correlated with ETV5 expression (Spearman r = 0.707, P <0.0001) and treatment-associated changes in ETV4 expression were highly correlated with changes in ETV5 expression (Spearman r = 0.672, P = 0.002). The microarray data from microdissected lobules show that ETV4 and ETV5 are expressed by mammary epithelium and this expression is downregulated by tamoxifen. Additional evidence for epithelial cell expression is provided by the observations that baseline expression of both ETV4 and ETV5 is directly correlated with the fractional epithelial area of the core samples (Spearman r = 0.501 and 0.465, P = 0.002 and 0.005, respectively), and ETV4 and ETV5 protein expression, assessed by IHC in benign breast tissue core samples, is largely limited to epithelial structures (Figure 4).
Figure 3.
RT-PCR confirmation of gene expression array data. NRQ is Normalized Relative Quantity. The mean Log2 ratio (post-treatment/baseline) is displayed for each gene to show the direction and degree of modulation by ■ tamoxifen or □ placebo. Mean post-treatment NRQ was compared with baseline NRQ using paired T-tests, *** P ≤ 0.001, ** P ≤ 0.01, and *P ≤ 0.05.
Figure 4.
Immunohistochemical assessment of ETV4 (A – D) and ETV5 (E – H) protein expression in normal testis (A and E), invasive breast cancer (B and F), and benign breast tissue core biopsies (C, D, G, and H). ETV4 was not expressed in human testis, but extensive nuclear staining was observed in breast cancer. ETV4 protein expression was primarily confined to the cytoplasm of luminal mammary epithelial cells (C, arrow), but rare epithelial structures with strong nuclear staining were also observed (D, arrow). ETV5 showed strong nuclear staining in a subpopulation of cells from testis (E), breast cancer (F), and benign breast epithelial structures (G and H).
Breast Epithelial Proliferation
The median proliferative index for breast epithelial cells at baseline was 1.5% for premenopausal women and 0.6% for postmenopausal women. On average, three months of tamoxifen was associated with a 51% reduction in proliferation (P = 0.05) and placebo a 50% increase in proliferation (P = 0.15).
The volcano plot in Figure 5 shows the MIB-1 Log2 ratio (post-treatment/baseline) versus the −Log10 of the P-value for each individual. It is clear from this figure that tamoxifen, but not placebo, exerts a significant anti-proliferative effect in some, but not all women, regardless of menopausal status. Of note, changes in ETV4 and DNAJC12 expression during treatment were directly correlated with changes in proliferation (i.e. reduced gene expression was associated with reduced proliferation ETV4 RT-PCR r = 0.493, P = 0.03 and DNAJC12 RT-PCR r = 0.642, P = 0.003).
CYP2D6 Genotypes
Certain CYP2D6 genotypes, notably the *4 and *5 alleles, have been associated with reduced conversion of tamoxifen to the active metabolite endoxifen. One (2.7%) of the 37 tamoxifen subjects was homozygous *5, and 18 (49%) were heterozygous for *4 or *5. The homozygous *5 subject was a postmenopausal woman with a proliferative index of 0 at baseline and post-treatment. The mean difference between post-treatment and baseline MIB-1 among tamoxifen-treated women was similar for *4 or *5 heterozygotes as compared to the homozygous wild-type subjects (P = 0.47). Figure 5 shows that the CYP2D6 *4 or *5 heterozygotes are evenly distributed among subjects manifesting and not manifesting a significant anti-proliferative response to tamoxifen. The effects of homozygous poor metabolizer genotypes are not known.
Apoptosis
Baseline mean apoptosis rate, as measured by TUNEL in breast tissue cores, was 4.9% for premenopausal subjects and 4.5% for postmenopausal subjects. ETV5 expression was modestly inversely correlated with apoptosis in baseline samples (Spearman r = −0.382, 95% CI −0.640 - −0.045, P = 0.024). However, tamoxifen did not modulate apoptosis as measured by TUNEL at three months.
Estrogen Receptor
Though ESR1 measured by IHC was reduced a median of 27% with tamoxifen, and increased a median of 11% with placebo, this was not statistically significant (P = 0.058). However, tamoxifen was associated with a median reduction in ESR1 gene expression of 49% as measured by microarray (P <0.0001) but only an 8% reduction in placebo subjects (P = 0.447). This result was confirmed by RT-PCR which showed a median reduction of 49% for tamoxifen-treated subjects (P = 0.005) and a 4% decrease for placebo-treated subjects (P = 0.98). In breast cancer, ETV4(20) and ETV5(21) expression have been inversely correlated with ESR1 expression. We observed an inverse correlation between baseline ETV5 expression in benign breast tissue and ESR1 protein expression measured by IHC (Spearman r = −0.398, 95% CI = −0.652 - −0.065, P = 0.018).
Breast Tissue Histology and Cytology
Lobular architecture, specifically the number of acini per lobule, has previously been linked to breast cancer risk (11),(12). The mean acini per lobule ratio, assessed at baseline, was directly correlated with ETV5 expression measured by RT-PCR (Spearman r = 0.481, 95% CI = 0.166 – 0.707, P = 0.003) and inversely correlated with RAB35 (P = 0.025), PIM1 (P = 0.004), and TEK (P = 0.001). Cytologic atypia in RP-FNA has also previously been linked to breast cancer risk(10). Baseline ETV5 expression measured by RT-PCR was directly correlated with RP-FNA Masood scores (Spearman r = 0.388, 95% CI = 0.351 – 0.655, P = 0.03). However, three months of tamoxifen did not convincingly modulate RP-FNA cellularity, RP-FNA cytological classification (Masood score or categorical classifications), core biopsy histological classification, breast tissue composition (epithelial, fibrous stroma, or adipose area), number of acini per lobule or total acini.
Tumor Suppressor Gene Methylation
Methylation of APC and RASSF1A in benign RP-FNA samples has previously been associated with breast cancer risk(9). ETV4 gene expression, measured at baseline by RT-PCR in breast tissue core samples, correlated directly with the sum of APC and RASSF1A methylation measured at baseline in RP-FNA samples (Spearman r = 0.442, 95% CI = 0.117 - 0.681, P = 0.008). Tamoxifen did not modulate methylation of CCND2, HIN1, CST6, or RAR-β2, but the mean difference for the sum of APC and RASSF1A methylation fractions between post-treatment and baseline samples was +7.1% for placebo subjects and −3.5% for tamoxifen subjects (P = 0.021).
Other Markers
Tamoxifen did not modulate breast tissue EGFR expression as measured by IHC or plasma prolactin. The proportion of BCL2 3+ cases increased from 40.0% to 85.7% for premenopausal tamoxifen subjects (Fisher’s exact test p = 0.029), and from 31.3% to 60.0% for premenopausal placebo subjects (p = 0.212), but BCL2 was not modulated in postmenopausal subjects.
DISCUSSION
Tamoxifen downregulates mRNA expression of ets-oncogene family members, ETV4 and ETV5, independent of changes in plasma estradiol, IGF1, IGF2, or IGFBP1 suggesting a direct transcriptional effect. ETV4 is a known estrogen response gene(22). ETV4 and ETV5 belong to the PEA3 subfamily of ets-oncogene family transcription factors. In rodent models, these transcription factors are essential for maintaining stem cell niches(23),(24),(25) and regulating branching morphogenesis of epithelial structures(26),(27), During pubertal murine mammary gland development, and subsequent lobular expansion in early pregnancy, ETV4 and ETV5 maintain the self-renewal capacity of the multipotent progenitor cap cells that guide extension of tubules through the stroma, and also regulate alveolar differentiation at the terminal end buds(28),(27),(29). The precise role of ETV4 and ETV5 in the human mammary gland is not known.
Known transcriptional targets of ETV4 and ETV5 include proteases (20),(30),(31), genes involved in epithelial stromal interaction, such as PTHLH(32) and UPAR(31), and IGFBPs(31). Our gene expression data are consistent with ETV4/5-mediated effects as tamoxifen significantly modulated the expression of genes related to epithelial-stromal interaction and tissue remodeling (PLAT, SERPINA3, SERPINA5, SERPING1, DBC1, EXTL1, and PTHLH). In addition, ets-oncogene family transcription factors have been shown to interact with chromatin remodeling complexes(33),(34) suggesting a possible mechanism for the decrease in tumor suppressor gene methylation observed with tamoxifen.
Tamoxifen has been shown to reduce the incidence of estrogen receptor positive, but not estrogen receptor negative breast cancer(1). ETV4 and ETV5 are most highly expressed in murine mammary tissue at menarche and early in pregnancy, periods of intense sex hormone-mediated tissue remodeling. Timing of these events is a critical component of the Gail model which performs best for the prediction of estrogen receptor positive breast cancer(35). A main effect of tamoxifen may be constraint of lobular development and maintenance through downregulation of ETV4 and ETV5. Persistence of large, acini-rich lobules has previously been linked to increased breast cancer risk(11),(12), but the hormone receptor status of the associated breast cancers in these studies is not known. A synthesis of the animal data(29), and the data from the current study, suggests that the etiology of estrogen receptor positive breast cancer may be related to ETV4/5-mediated effects on lineage specific differentiation of the multipotential progenitor cap cells of the terminal end buds.
Tamoxifen also significantly modulated the expression of genes belonging to, or interacting with, Ras superfamily G proteins involved in intracellular vesicle trafficking (RAB3B, RAB38, RAB35, and SYTL2). These genes are not known to be regulated by ETV4 or ETV5. Of note, tamoxifen significantly upregulated TEK, a tyrosine kinase associated with angiogenesis(36) and poor prognosis breast cancer(37),(38), and also marginally upregulated PIM1, a serine/threonine protein kinase recognized as a proto-oncogene(39),(40). Tamoxifen also reduced the expression of several known estrogen response genes including GREB1, which is thought to mediate estrogen-induced proliferation(41),(42),(43).
Our approach to gene expression analysis imposes certain limitations in interpretation. Analysis of the gene expression microarray data was directed at identifying the most consistent general effects of tamoxifen in benign breast tissue and will have excluded genes with large expression changes in just a few individuals. Hierarchical clustering, as shown in Figure 2, was used to confirm that the variability in up or downregulation of the selected genes was greater between treatment groups then within groups. In addition, the stringent statistical criteria used for gene selection will have excluded many important genes with more modest modulation. For example, downregulation of the calcium/calmodulin-dependent protein kinase (CAMK) CAMK2G has previously been reported in tamoxifen-treated premenopausal women(44). Our data did not confirm this observation, but did show modulation of a variety of other CAMK-related genes, none of which met our statistical criteria for inclusion in the final lists.
Tamoxifen-mediated reductions in ETV4/5 gene expression would be expected to interfere with lobular development and maintenance. However, three months of tamoxifen did not alter the morphological features of benign breast tissue or impact the rate of cytological atypia. Notably, in one study, 12 months but not six months of tamoxifen significantly reduced RP-FNA Masood cytology scores for 17 treated women as compared to 16 untreated controls(45), suggesting that longer treatment periods are required to translate early molecular events into morphological changes observable under the microscope.
Tamoxifen has been associated with reduced proliferation in benign breast epithelial cells in some studies(46),(47),(48), but not others(49). The three positive studies are from the same institution and proliferation was only measured at a single post-treatment time point in premenopausal women undergoing fibroadenoma excision. Of note, mean proliferation in the control groups was highly variable for these three studies (50.3%, 9.5% and 2.04%). Proliferation is difficult to quantify by MIB-1 IHC in benign breast tissue because of the enormous variability within the same section (e.g. in our most variable case, proliferative indices ranged from 0.006 – 0.75 for one section with 26 lobules). To account for this, we calculated a proliferative index for every available lobule in each core biopsy section and then compared baseline and post-treatment averages for each subject as shown in the volcano plot in Figure 5. This convincingly demonstrates an anti-proliferative effect for tamoxifen in benign breast tissue for some women, but not others. Downregulation of ETV4 and DNAJC12 expression during treatment were directly correlated with reductions in proliferation.
In summary, tamoxifen significantly downregulated the expression of ets-oncogene family members ETV4 and ETV5 which are known to play a central role in stem/progenitor cell renewal and differentiation during initial mammary gland development and subsequent remodeling. The reduced proliferation and changes in gene expression we observed are consistent with ETV4/5-mediated effects. Tissue remodeling following ETV4/5 downregulation is also a likely mechanism for the reduction in mammographic density observed with tamoxifen(4),(50). Further investigations into the role of ETV4 and ETV5 in breast carcinogenesis, specifically the role in maintaining stem/progenitor cell populations, are warranted as targeting this pathway may provide an approach for reducing breast cancer risk while avoiding the toxicity associated with systemic modulation of estrogen response pathways.
Supplementary Material
ACKOWLEDGEMENTS
Simmons Comprehensive Cancer Center Genomics Core Laboratory.
GRANT SUPPORT
This work was supported by the National Cancer Institute at the National Institutes of Health (Contract Number N01-CN-95139) and the Simmons Comprehensive Cancer Center NCI Cancer Center Support Grant (1P30 CA142543-01).
Financial Support: Euhus, NIH/NCI N01-CN-95139
REFERENCES
- 1.Fisher B, Constantino JP, Wickerham L, Redmond CK, Kavanah M, Cronin WM, et al. Tamoxifen for prevention of breast cancer: Report of the National Surgical Adjuvant Bowel and Breast Project P-1 study. J Natl Cancer Inst. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 2.Bonanni B, Johansson H, Gandini S, Guerrieri-Gonzaga AAR, Sandri MT, Cazzaniga M, et al. Effect of low dose tamoxifen on the insulin-like growth factor system in healthy women. Breast Cancer Research & Treatment. 2001;69:21–7. doi: 10.1023/a:1012241505717. [DOI] [PubMed] [Google Scholar]
- 3.Helle SI, Holly JM, Tally M, Hall K, Vander Stappen J, Lonning PE. Influence of treatment with tamoxifen and change in tumor burden on the IGF-system in breast cancer patients. Int J Cancer. 1996;69:335–9. doi: 10.1002/(SICI)1097-0215(19960822)69:4<335::AID-IJC17>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 4.Decensi A, Robertson C, Guerrieri-Gonzaga A, Serrano D, Cazzaniga M, Mora S, et al. Randomized double-blind 2 × 2 trial of low-dose tamoxifen and fenretinide for breast cancer prevention in high-risk premenopausal women. J Clin Oncol. 2009;27:3749–56. doi: 10.1200/JCO.2008.19.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decensi A, Formelli F, Torrisi R, Costa A. Breast cancer chemoprevention: studies with 4-HPR alone and in combination with tamoxifen using circulating growth factors as potential surrogate endpoints. J Cell Biochem Suppl. 1993;17G:226–33. doi: 10.1002/jcb.240531142. [DOI] [PubMed] [Google Scholar]
- 6.Fabian CJ. Surrogate response biomarkers in prevention research: do they point the way or lead us astray? J Clin Oncol. 2009;27:3734–6. doi: 10.1200/JCO.2009.22.9211. [DOI] [PubMed] [Google Scholar]
- 7.Navarrete MA, Maier CM, Falzoni R, Quadros LG, Lima GR, Baracat EC, et al. Assessment of the proliferative, apoptotic and cellular renovation indices of the human mammary epithelium during the follicular and luteal phases of the menstrual cycle. Breast Cancer Res. 2005;7:R306–13. doi: 10.1186/bcr994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis CM, Cler LR, Bu DW, Zochbauer-Muller S, Milchgrub S, Naftalis EZ, et al. Promoter hypermethylation in benign breast epithelium in relation to predicted breast cancer risk. Clin Cancer Res. 2005;11:166–72. [PubMed] [Google Scholar]
- 9.Euhus DM, Bu D, Milchgrub S, Xie XJ, Bian A, Leitch AM, et al. DNA methylation in benign breast epithelium in relation to age and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:1051–9. doi: 10.1158/1055-9965.EPI-07-2582. [DOI] [PubMed] [Google Scholar]
- 10.Fabian CJ, Kimler BF, Zalles CM, Klemp JR, Kamel S, Zeiger S, et al. Short-term breast cancer prediction by random periareolar fine-needle aspiration cytology and the Gail risk model. J Natl Cancer Inst. 2000;92:1217–27. doi: 10.1093/jnci/92.15.1217. [DOI] [PubMed] [Google Scholar]
- 11.Russo J, Romero AL, Russo IH. Architectural pattern of the normal and cancerous breast under the influence of parity. Cancer Epidemiol Biomark Prev. 1994;3:219–24. [PubMed] [Google Scholar]
- 12.McKian KP, Reynolds CA, Visscher DW, Nassar A, Radisky DC, Vierkant RA, et al. Novel breast tissue feature strongly associated with risk of breast cancer. J Clin Oncol. 2009;27:5893–8. doi: 10.1200/JCO.2008.21.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endogenous Hormones Breast Cancer Collaborative Group Free Estradiol and Breast Cancer Risk in Postmenopausal Women. Cancer Epidemiology Biomarkers & Prevention. 2003;12:1457–61. [PubMed] [Google Scholar]
- 14.Masood S, Fryberg ER, McLellan GL, Dee S, Bullard JB. Cytologic differentiation between proliferative and non-proliferative breast disease in mammographically guided fine-needle aspirates. Diag Cytopathol. 1991;7:581–90. doi: 10.1002/dc.2840070607. [DOI] [PubMed] [Google Scholar]
- 15.Fackler MJ, McVeigh M, Mehrotra J, Blum MA, Lange J, Lapides A, et al. Quantitative Multiplex Methylation-Specific PCR Assay for the Detection of Promoter Hypermethylation in Multiple Genes in Breast Cancer. Cancer Res. 2004;64:4442–52. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 16.Euhus DM, Bu D, Ashfaq R, Xie XJ, Bian A, Leitch AM, et al. Atypia and DNA methylation in nipple duct lavage in relation to predicted breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:1812–21. doi: 10.1158/1055-9965.EPI-06-1034. [DOI] [PubMed] [Google Scholar]
- 17.Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slonim DK, Tamayo P, Mesirov JP, Golub TR, Lander ES. Class Prediction and Discovery Using Gene Expression Data; Fourth International Conference on Computational Molecular Biology; Tokyo, Japan. 2000. [Google Scholar]
- 20.Bieche I, Tozlu S, Girault I, Onody P, Driouch K, Vidaud M, et al. Expression of PEA3/E1AF/ETV4, an Ets-related transcription factor, in breast tumors: positive links to MMP2, NRG1 and CGB expression. Carcinogenesis. 2004;25:405–11. doi: 10.1093/carcin/bgh024. [DOI] [PubMed] [Google Scholar]
- 21.Chotteau-Lelievre A, Revillion F, Lhotellier V, Hornez L, Desbiens X, Cabaret V, et al. Prognostic value of ERM gene expression in human primary breast cancers. Clin Cancer Res. 2004;10:7297–303. doi: 10.1158/1078-0432.CCR-04-0593. [DOI] [PubMed] [Google Scholar]
- 22.Ishida S, Higashino F, Aoyagi M, Takahashi A, Suzuki T, Shindoh M, et al. Genomic structure and promoter activity of the E1AF gene, a member of the ETS oncogene family. Biochem Biophys Res Commun. 2006;339:325–30. doi: 10.1016/j.bbrc.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Tyagi G, Carnes K, Morrow C, Kostereva NV, Ekman GC, Meling DD, et al. Loss of Etv5 decreases proliferation and RET levels in neonatal mouse testicular germ cells and causes an abnormal first wave of spermatogenesis. Biol Reprod. 2009;81:258–66. doi: 10.1095/biolreprod.108.075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubo H, Shimizu M, Taya Y, Kawamoto T, Michida M, Kaneko E, et al. Identification of mesenchymal stem cell (MSC)-transcription factors by microarray and knockdown analyses, and signature molecule-marked MSC in bone marrow by immunohistochemistry. Genes Cells. 2009;14:407–24. doi: 10.1111/j.1365-2443.2009.01281.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–4. doi: 10.1038/nature03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chotteau-Lelievre A, Montesano R, Soriano J, Soulie P, Desbiens X, deLaunoit Y. PEA3 transcription factors are expressed in tissues undergoing branching morphogenesis and promote formation of duct-like structures by mammary epithelial cells in vitro. Dev Biol. 2003;259:241–57. doi: 10.1016/s0012-1606(03)00182-9. [DOI] [PubMed] [Google Scholar]
- 27.Kurpios NA, Sabolic NA, Shepherd TG, Fidalgo GM, Hassell JA. Function of PEA3 Ets transcription factors in mammary gland development and oncogenesis. J Mammary Gland Biol Neoplasia. 2003;8:177–90. doi: 10.1023/a:1025948823955. [DOI] [PubMed] [Google Scholar]
- 28.Chotteau-Lelievre A, Desbiens X, Pelczar H, Defossez PA, de Launoit Y. Differential expression patterns of the PEA3 group transcription factors through murine embryonic development. Oncogene. 1997;15:937–52. doi: 10.1038/sj.onc.1201261. [DOI] [PubMed] [Google Scholar]
- 29.Kurpios NA, MacNeil L, Shepherd TG, Gludish DW, Giacomelli AO, Hassell JA. The Pea3 Ets transcription factor regulates differentiation of multipotent progenitor cells during mammary gland development. Dev Biol. 2009;325:106–21. doi: 10.1016/j.ydbio.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 30.Kaya M, Yoshida K, Higashino F, Mitaka T, Ishii S, Fujinaga K. A single ets-related transcription factor, E1AF, confers invasive phenotype on human cancer cells. Oncogene. 1996;12:221–7. [PubMed] [Google Scholar]
- 31.Firlej V, Ladam F, Brysbaert G, Dumont P, Fuks F, de Launoit Y, et al. Reduced tumorigenesis in mouse mammary cancer cells following inhibition of Pea3- or Erm-dependent transcription. J Cell Sci. 2008;121:3393–402. doi: 10.1242/jcs.027201. [DOI] [PubMed] [Google Scholar]
- 32.Richard V, Rosol TJ, Foley J. PTHrP gene expression in cancer: do all paths lead to Ets? Crit Rev Eukaryot Gene Expr. 2005;15:115–32. doi: 10.1615/critreveukaryotgeneexpr.v15.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker KM, Wei G, Schaffner AE, Ostrowski MC. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. Journal of Biological Chemistry. 2003;278:17876–84. doi: 10.1074/jbc.M209480200. [DOI] [PubMed] [Google Scholar]
- 34.Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, Martins-Green M. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. Embo J. 2003;22:281–91. doi: 10.1093/emboj/cdg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chlebowski RT, Anderson GL, Lane DS, Aragaki AK, Rohan T, Yasmeen S, et al. Predicting risk of breast cancer in postmenopausal women by hormone receptor status. J Natl Cancer Inst. 2007;99:1695–705. doi: 10.1093/jnci/djm224. [DOI] [PubMed] [Google Scholar]
- 36.Konstantinou K, Yamamoto K, Ishibashi F, Mizoguchi Y, Kurata M, Nakagawa Y, et al. Angiogenic mediators of the angiopoietin system are highly expressed by CD10-positive lymphoma cells in angioimmunoblastic T-cell lymphoma. Br J Haematol. 2009;144:696–704. doi: 10.1111/j.1365-2141.2008.07534.x. [DOI] [PubMed] [Google Scholar]
- 37.Dales JP, Garcia S, Carpentier S, Andrac L, Ramuz O, Lavaut MN, et al. Prediction of metastasis risk (11 year follow-up) using VEGF-R1, VEGF-R2, Tie-2/Tek and CD105 expression in breast cancer (n=905) Br J Cancer. 2004;90:1216–21. doi: 10.1038/sj.bjc.6601452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meunier-Carpentier S, Dales JP, Djemli A, Garcia S, Bonnier P, Andrac-Meyer L, et al. Comparison of the prognosis indication of VEGFR-1 and VEGFR-2 and Tie2 receptor expression in breast carcinoma. Int J Oncol. 2005;26:977–84. [PubMed] [Google Scholar]
- 39.Bachmann M, Hennemann H, Xing PX, Hoffmann I, Moroy T. The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint. Journal of Biological Chemistry. 2004;279:48319–28. doi: 10.1074/jbc.M404440200. [DOI] [PubMed] [Google Scholar]
- 40.Roh M, Franco OE, Hayward SW, van der Meer R, Abdulkadir SA. A role for polyploidy in the tumorigenicity of Pim-1-expressing human prostate and mammary epithelial cells. PLoS ONE. 2008;3:e2572. doi: 10.1371/journal.pone.0002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Nawaz Z, Slingerland JM. Long-range activation of GREB1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Molecular Endocrinology. 2007;21:2651–62. doi: 10.1210/me.2007-0082. [DOI] [PubMed] [Google Scholar]
- 42.Lin CY, Strom A, Vega VB, Kong SL, Yeo AL, Thomsen JS, et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rae J, Johnson M, Scheys J, Cordero K, Larios J, Lippman M. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–9. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 44.Da Silva ID, Dias-Netto E, Villanova FE, Yamamoto L, Baracat EC, Lima GR, et al. Tamoxifen down-regulates CaMKII messenger RNA levels in normal human breast tissue. Clin Exp Obstet Gynecol. 2004;31:204–8. [PubMed] [Google Scholar]
- 45.Baker JC, Ostrander JH, Lem S, Broadwater G, Bean GR, D’Amato NC, et al. ESR1 Promoter Hypermethylation Does Not Predict Atypia in RPFNA nor Persistent Atypia after 12 Months Tamoxifen Chemoprevention. Cancer Epidemiol Biomark Prev. 2008;17:1884–90. doi: 10.1158/1055-9965.EPI-07-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernardes JR, Jr., Nonogaki S, Seixas MT, Rodrigues de Lima G, Baracat EC, Gebrim LH. Effect of a half dose of tamoxifen on proliferative activity in normal breast tissue. Int J Gynaecol Obstet. 1999;67:33–8. doi: 10.1016/s0020-7292(99)00092-2. [DOI] [PubMed] [Google Scholar]
- 47.de Sousa JA, de Seixas MT, de Lima GR, Baracat EC, Gebrim LH. Evaluation of monoclonal antibody MIB-1 in the mammary epithelium adjacent to fibroadenomas in premenopausal women treated with tamoxifen. Breast J. 2001;7:392–7. doi: 10.1046/j.1524-4741.2001.07603.x. [DOI] [PubMed] [Google Scholar]
- 48.de Lima GR, Facina G, Shida JY, Chein MB, Tanaka P, Dardes RC, et al. Effects of low dose tamoxifen on normal breast tissue from premenopausal women. Eur J Cancer. 2003;39:891–8. doi: 10.1016/s0959-8049(02)00530-0. [DOI] [PubMed] [Google Scholar]
- 49.Walker KJ, Price-Thomas JM, Candlish W, Nicholson RI. Influence of the antioestrogen tamoxifen on normal breast tissue. Br J Cancer. 1991;64:764–8. doi: 10.1038/bjc.1991.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meggiorini ML, Labi L, Vestri AR, Porfiri LM, Savelli S, De Felice C. Tamoxifen in women with breast cancer and mammographic density. Eur J Gynaecol Oncol. 2008;29:598–601. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.