Abstract
Transcriptional profiling results, using our non-invasive induction assay [short exposure intervals (2–5 h) to sub-lethal amounts of insecticides (<LD3 at 24 h) administered by stress reducing means (contact vs. immersion screen) and with induction assessed in a time frame when tolerance is still present (~LC90 in 2–4 h)], show that ivermectin-induced detoxification genes from body lice are identified by quantitative real-time PCR analyses.
Of the cytochrome P450 monooxygenase and ATP binding cassette transporter genes induced by ivermectin, CYP6CJ1, CYP9AG1, CYP9AG2 and PhABCC4 were respectively most significantly over-expressed, had high basal expression levels and were most closely related to genes from other organisms that metabolized insecticides, including ivermectin.
Injection of dsRNAs against either CYP9AG2 or PhABCC4 into non-induced female lice reduced their respective transcript level and resulted in increase sensitivity to ivermectin, indicating that these two genes are involved in the xenobiotic metabolism of ivermectin and in the production of tolerance.
Keywords: Pediculus humanus humanus, ivermectin, tolerance, transcriptional profiling, RNA interference, P450, ABC transporters
Introduction
Pediculosis is the most prevalent parasitic infestation of humans (Raoult & Roux, 1999) and most people find lice intolerable and repeatedly and prophylactically apply pediculicides (insecticides) without realizing their harm and the rapid onset of resistance that can occur if misused.
Body lice transmit several bacteria (Rickettsia prowazekii, Borrelia recurrentis, and Bartonella quintana) that cause human diseases (epidemic typhus, louse-borne relapsing fever, and trench fever) and these diseases have killed millions (Kelly et al., 2002). Since the advent of antibiotics, outbreaks are sporadic but in 1986 more than 50,000 people were infected with R. prowazekii in Burundi (Weiss, 1988). More recently, outbreaks of bacteremia due to B. quintana have been reported among homeless people in the United States and Europe (Brouqui et al., 1999). Thus, body lice represent a health risk during times of war, famine and social unrest, including homelessness (Piesman & Gage, 2000).
There currently exists a crisis in the chemical management of pediculosis. The pediculicide arsenal is limited to only a few active ingredients and is shrinking due to regulatory restrictions and resistance problems. Insecticide resistance is particularly problematic in the control of human lice for several reasons: (1) they are obligate human blood feeders that are exposed to pediculicides at all stages; (2) they have short generational time and high fecundity; and (3) there are few commercial pediculicidal products, the majority of which share common chemistry (pyrethrins/pyrethroids) and elicit cross-resistance (Yoon et al., 2003). Because of this, louse resistance to most commercial pediculicides has occurred and is increasing (Pray, 2003; Yoon et al., 2003; Gao et al., 2006). Thus, control and resistance problems underscore the need to understand the molecular mechanisms of insecticide resistance in lice.
New pediculicides are a critical need but must have novel chemistry and modes of actions and show no cross-resistance to previously used products. With this in mind, the macrocyclic lactone-type insecticides (i.e., ivermectin) are excellent candidates for development as topical treatments for pediculosis (Strycharz et al., 2008) and have recently proven effective as an oral treatment for hard-to-control louse infestations (Chosidow et al., 2010).
As a natural product, ivermectin is readily detoxified by humans, not absorbed via human skin, and not toxic at topical doses used for louse control. Ivermectin is a mixture of two 16-member macrocyclic lactones (~80% H2B1a and ~20% H2B1b) produced fermentatively by Streptomyces avermitilis. It acts at two ligand-gated channels in the insect nervous system (GABA- and glutamate-chloride channels), resulting in enhanced Cl− flux and paralysis (Prichard & Roulet, 2007). Both mammals and insects form three major oxidative metabolites of ivermectin, 24- and 26-hydroxyavermectin and 3″ O-desmethylavermectin, via Phase I xenobiotic metabolism and dexamethasone induction increases their formation (Yoon et al., 2002). In humans, CYP3A4 metabolizes ivermectin (Zeng et al., 1996) and demethylates erythromycin, a related macrocyclic lactone antibiotic (Wacher et al., 1995). No phase II conjugates of these metabolites have been identified and most absorbed ivermectin is associated with bile apparently due to ATP Binding Cassette (ABC) transporter-dependent efflux. As ivermectin is itself a negatively-charged sugar conjugate, it is likely a direct substrate for ABC transporters (e.g., ABCC-type) and there is accumulating evidence suggesting that ABC transporters are involved in ivermectin efflux, excretion, and possibly in resistance (Pichard & Roulet, 2007; Xu et al., 1998; Lespine et al., 2007; James & Davey, 2009). Thus, Phase I and III xenobiotic metabolism is: (1) involved in the detoxification of ivermectin, and (2) may be involved ultimately in resistance to ivermectin in human lice.
It is well established that xenobiotic-metabolizing enzymes (Feyereisen, 2005) and transporters (James & Davey, 2009) can detoxify or efflux insecticides, respectively, causing resistance. Insecticides also induce the expression of xenobiotic-metabolizing enzymes and transporters, which metabolize them (Willoughby et al., 2006). Resistance occurs when heritable mutations cause either constitutive/inducible over-expression or altered function of these gene products. There are well-documented cases of xenobiotic-inducible genes that are constitutively over-expressed in insecticide-resistant insects (Feyereisen, 2005), indicating that modifications of metabolic pathways are important in the development of tolerance and ultimately resistance. Indeed, one cytochrome P450 monooxygenase (P450) gene (CYP12D1) was induced by DDT in both a DDT-resistant (Brandt et al., 2002; Festucci-Busell et al., 2005) and a susceptible strain (Willoughby et al., 2006) of D. melanogaster. Because DDT induced the expression of CYP12D1 in the susceptible 91-C strain of D. melanogaster and this same P450 gene was found constitutively over-expressed in the DDT-resistant 91-R strain, these findings are in strong support of the contention that some inducible detoxification genes can function as resistant-causing genes once constitutive over-expression occurs. Comparison between cypermethrin-resistant and -susceptible strains of Plutella xylostella revealed that 8 of 11 P450 genes were induced in resistant larvae whereas only a single P450 gene was induced in susceptible larvae under optimal induction conditions (Baek et al., 2010). This finding also suggests that selective P450 gene induction by insecticides in addition to constitutive over expression can confer metabolic resistance.
Identifying insect detoxification genes, based on induced transcript profiles, has therefore been repeatedly suggested as a means of identifying the major metabolic pathways involved in insecticide resistance (Vontas et al., 2005; Willoughby et al., 2006) and initial attempts using transcriptional profiling following insecticide induction in a susceptible strain of D. melanogaster did identified a number of detoxification genes (Willoughby et al., 2006). However, only a limited number of the genes induced appeared to be involved in insecticide metabolism.
To investigate whether detoxification genes can be selectively induced by insecticides, and thereby identify those genes involved in the actual metabolism of the insecticide, the induction scheme should be optimized by including: (1) an assessment of gene transcript levels at a time of peak gene induction that results in insect tolerance; (2) insecticide doses that do not result in physiological stress that can mask the identification of primary detoxification genes due to a large number of genes of secondary importance being co-induced; and (3) application of insecticides in a non-invasive manner, such as contact exposure without solvent carriers. In addition, adequate controls are also required (e.g., application of no insecticide-containing control solutions). Stress associated with starvation can likewise be reduced by using our in vitro rearing system (Yoon et al., 2006). Insecticide induction that leads to tolerance also needs to be fast enough to protect the insect from the rapid onset of toxicity and temporary so the fitness cost associated with the long-term over-expression of detoxification gene products is minimal (Devonshire & Moore, 1982). For these reasons, a non-invasive induction assay (brief exposure to sub-lethal levels of insecticide administered in a stress reducing fashion with a rapid assessment of transcript levels that overlaps with tolerance) was envisaged to optimize the identification of inducible detoxification genes that produce tolerance via metabolism, some of which will result in resistance once inheritable mutations causing constitutive over-expression, more sensitive induction or structural alteration occur.
In this study, we investigate whether the optimization of dose, the timing of exposure, and the assessment of transcript levels during tolerance can be used to identify detoxification genes that metabolize ivermectin. Because resistance monitoring is an absolute requirement for any sustainable vector control program (David et al., 2005), there is a critical need to efficiently identify detoxification genes that metabolize insecticides during the process of induced tolerance prior to resistance evolving. Some of these genes will certainly be involved in phenotypic resistance that will evolve after pesticide selection and can then be used proactively to monitor for metabolic resistance. This approach is particularly relevant with the advent of pesticides that posses “green chemistries”, such as ivermectin, and prone to rapid detoxification by xenobiotic metabolism.
Results and discussion
Dexamethasone induces tolerance to ivermectin whereas verapamil increases ivermectin toxicity
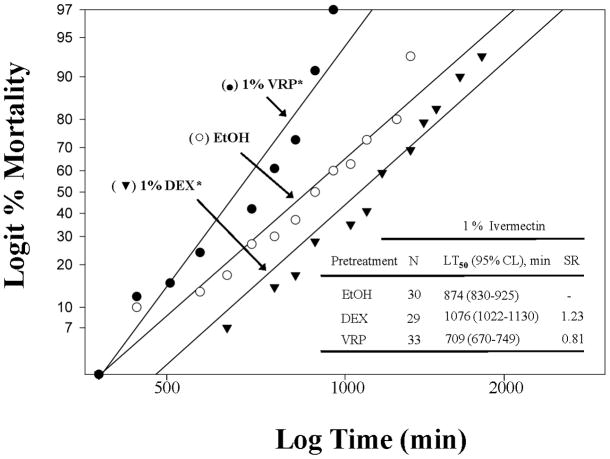
The 1% ivermectin (IVM) mortality response of 1% dexamethasone (DEX)-pretreated lice was significantly slower (~23% slower at the LT50) compared to that of ethanol (EtOH)-pretreated lice (χ2=36, df=2, P<0.001, Fig. 1). This result suggests that P450s, which are induced by dexamethasone, increased oxidative metabolism of ivermectin. Apparently, the increased oxidative metabolism detoxifies ivermectin, leading to a slower rate of mortality as evidenced in the Colorado potato beetle (Yoon et al., 2002).
Figure 1.
Log time versus logit mortality regression analyses of a lethal contact concentration of 1 % (w/v) ivermectin (IVM) using female body lice pretreated by immersion with either a sub-lethal concentration of verapamil (VRP; 1 %, w/v) or dexamethasone (DEX; 1 %, w/v) in ethanol. Asterisks (*) indicate that pretreated regressions are significantly different from ethanol (EtOH)-treated control regressions using the maximum log-likelihood ratio test (P<0.05). Synergistic ratios (SR) are determined by dividing the pretreatment LT50 values (VRP or DEX) by the ethanol-only LT50 value (EtOH).
The 1% ivermectin mortality response of 1% verapamil (VRP)-pretreated lice was significantly faster (~19% faster at the LT50) than that of ethanol-pretreated lice (χ2=50, df=2, P<0.001, Fig. 1). Since verapamil is known to inhibit ABCB-type transporters in mammals, this result suggests that ABCB-type transporters are also likely involved in the increased excretion of ivermectin (MacDonald & Gledhill, 2007). In such a case, the ABCB-type transporters would be overwhelmed by verapamil, which would slow the excretion of ivermectin (Prichard & Roulet, 2007), causing an increased rate of mortality in the verapamil-pretreated lice.
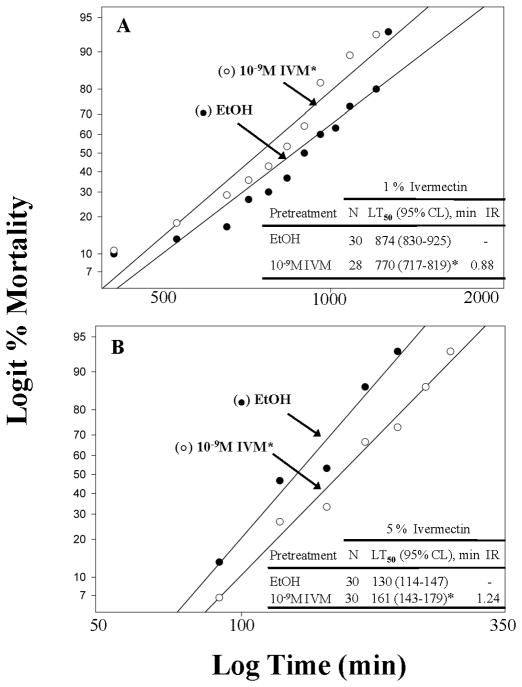
Tolerance to 5% but not 1% ivermectin is induced by a 2 hour pretreatment to a sub-lethal concentration of ivermectin
The 1% ivermectin mortality response of sub-lethal (10−9 M) ivermectin-pretreated lice was significantly faster but not substantially faster (~12% faster at the LT50) than that of ethanol-pretreated lice (χ2=3, df=2, P=0.002), indicating that sub-lethal ivermectin pretreatment for 2 h did not induce substantial oxidative metabolism of ivermectin continuously over the ~25 h mortality bioassay (Fig. 2A). The lack of induced tolerance following pretreatment with sub-lethal amount of ivermectin is likely due to the long assessment time for mortality in the presence of 1% ivermectin (0–25 h, Fig. 1A). Our transcriptional profiling results given below (Figs. 3 and 4) show that the induction of detoxification genes occurs at ~2–4 h following exposure to sub-lethal amounts of ivermectin but returns back to pre-induced levels by ~5 h (data not shown). Recent results have likewise shown that peak induction occurs ~2–4 h after optimal exposure to cypermethrin in the diamondback moth, that induction level was dose dependent, and that induction was only temporary, returning to pre-induced levels by 10 h (Baek et al., 2010). Induction of tolerance was achieved, however, by exposing lice to sub-lethal amounts of ivermectin for 2 h but assessing them in the presence of 5% ivermectin so that the entire mortality assessment time was reduced to ~5 h (Fig. 1B). By doing this, tolerance was assessed at a time when gene induction was apparently still present and had not yet returned to pre-induced levels. Thus, P450s were likely induced, metabolized ivermectin, and resulted in a significant and substantial slowing of mortality (~24% at the LT50, Fig. 2B).
Figure 2.
Comparative tolerance to lethal contact concentrations of ivermectin (1 %, w/v, IVM in panel A or 5 %, w/v, IVM in panel B) using female body lice pretreated by immersion with a sub-lethal concentration of ivermectin (10−9 M) in ethanol. Asterisks (*) indicate that pretreated regressions are significantly different from ethanol (EtOH)-treated control regressions using the maximum log-likelihood ratio test (P<0.05). Induction ratios (IR) for tolerance are determined by dividing the IVM pretreatment LT50 values by the ethanol-only LT50 value (EtOH).
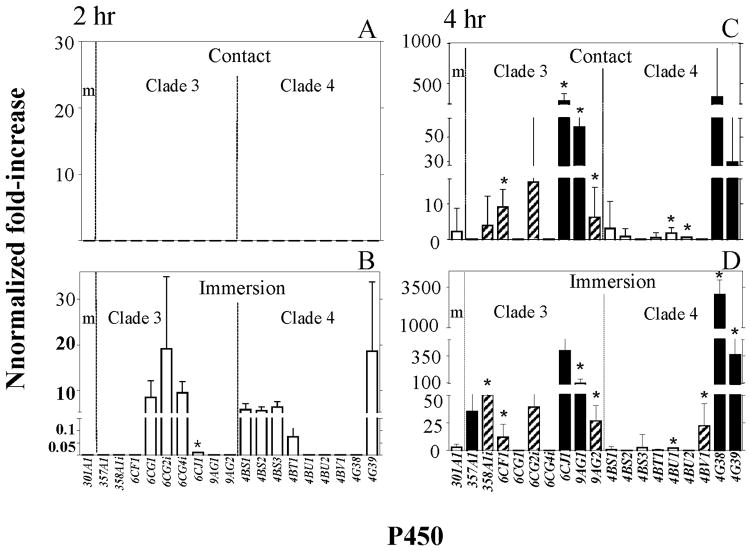
Figure 3.
Relative increases in the transcript levels of cytochrome P450 monooxygenase (P450) genes determined by qPCR following 2 (panels A and B) and 4 (panels C and D) h post ivermectin (IVM) treatments of female lice, where normalized fold-increase = normalized basal transcript level × [relative fold-increase of transcript level following IVM treatment (10−6 M for direct contact (panels A and C); 10−9 M IVM in ethanol for immersion (panels B and D)) − 1]. Asterisks (*) indicate a significant increase in transcript levels over respective controls using Student’s t-test (P<0.05). Solid bars are highly expressed (normalized basal level transcription (NBLT > 220), hatched bars are modestly expressed 30 < NBLT < 220) and open bars are weakly expressed (NBLT < 30) genes.
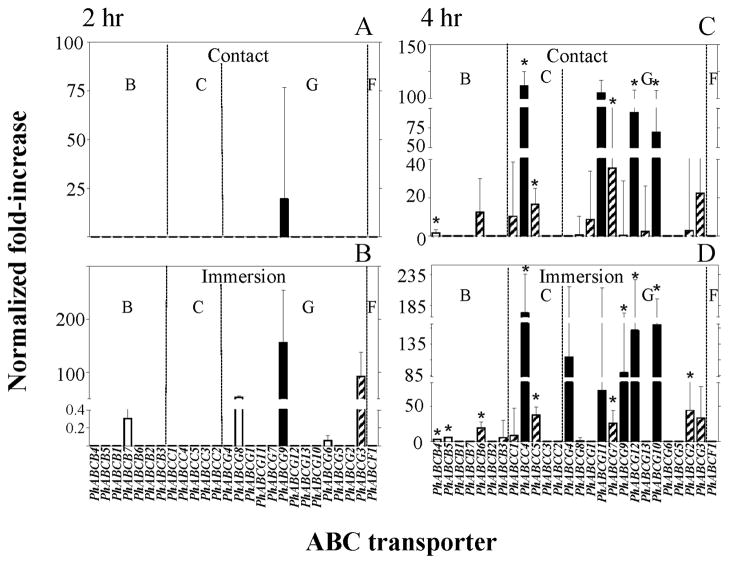
Figure 4.
Relative increases in the transcript levels of ABC transporter genes determined by qPCR following 2 (panels A and B) and 4 (panels C and D) h post ivermectin (IVM) treatments of female lice, where normalized fold-increase = normalized basal transcript level × [relative fold-increase of transcript level following IVM treatment (10−6 M for contact (panels A and C); 10−9 M IVM for immersion (panels B and D)) − 1]. Asterisks (*) indicate a significant increase in transcript levels over respective controls using Student’s t-test (P<0.05). Solid bars are highly expressed (normalized basal level transcription (NBLT > 220), hatched bars are modestly expressed 30 < NBLT < 220) and open bars are weakly expressed (NBLT < 30) genes. The letters B, C, G, and F designate ABC transporter subfamilies.
Transcriptional profiling by quantitative real-time PCR (qPCR) identifies P450 and ABC transporter genes over transcribed following brief exposures to sub-lethal amounts of ivermectin
The transcript levels of the eighteen P450 genes in Clades 3 and 4 and CYP301A1 and the twenty-six ABC transporter genes in the ABCB, ABCC and ABCG subfamilies and PhABCF1 were evaluated by qPCR following brief pre-exposures to sub-lethal amounts of ivermectin. Both exposure methods (topical exposure following immersion for 1 sec and surface contact to filter paper disks) significantly increased the transcript levels of P450 (Fig. 3) and ABC transporter (Fig. 4) genes over the 4 h exposure interval.
With the immersion method of application, the 2 h exposure to ivermectin resulted in increased transcript levels of four P450 genes in Clade 3 (only CYP6CJ1 was significantly increased, 1/9 or 11%) and five P450 genes in Clade 4 (none of which were significantly increased) when compared to the transcript levels of non-ivermectin exposed lice (Fig. 3B). At 4 h following immersion, seven P450 genes in Clade 3 were increased, four significantly (4/9 or 44%), and six P450 genes in Clade 4 were increased, four significantly (4/9 or 44%), when compared to the transcript levels of non-ivermectin exposed lice (Fig. 3D).
With the contact method of application, the 2 h exposure to ivermectin resulted in no increased transcript levels of any of the P450 genes in either Clades 3 or 4 (Fig. 3A). At 4 h following contact, however, six P450 genes were increased in Clade 3, four significantly (4/9 or 44%), and seven P450 genes were increased in Clade 4, two significantly (2/9 or 22%), when compared to the transcript levels of non-ivermectin exposed lice (Fig. 3C).
Overall, ivermectin exposure following immersion (Fig. 3B and D) substantially increased the number of P450 genes in Clades 3 and 4 having increased transcript levels (9 genes, 1 significantly, increased at 2 h and 13 genes, 8 significantly, at 4 h) compared to that following contact exposure (0 genes increased at 2 h and 13 genes, 6 significantly, at 4 h) (Fig. 3A and C). Additionally, the 4 h exposure resulted in substantially more P450 genes having significantly increased transcript levels (8 genes with immersion and 6 genes with contact, Fig. 3D and C) compared to the 2 h exposure (1 gene with immersion and 0 genes with contact, Fig. 3B and A). Of the P450 genes that had significantly increased transcript levels at 4 h post-ivermectin exposure, the transcript levels of CYPCF1, CYP6CJ1, CYP9AG1, CYP9AG2 and CYP4BU1 were significantly increased following ivermectin exposure by both immersion and contact applications, with CYP6CJ1, CYP9AG1 and CYP9AG2 being the most highly expressed (Fig. 3D and C).
Similar results were obtained for the twenty-six ABC transporter genes examined. With the immersion method of application, the 2 h exposure to ivermectin resulted in increased transcript levels of five ABC transporter genes, none of which were significantly increased when compared to the transcript levels of non-ivermectin exposed lice (Fig. 4B). At 4 h following immersion, sixteen ABC transporter genes were increased, among which ten were significantly increased (10/26 or 38%), with three genes significantly increased of the seven ABCB-type (43%), two genes significantly increased of the five ABCC-type (40%), and five genes significantly increased of the thirteen ABCG-type (38%) (Fig. 4D).
With the contact method of application, the 2 h exposure to ivermectin resulted in only one ABC transporter gene over transcribed but this level was not significantly different from that of the non-ivermectin exposed control lice (Fig. 4A). At 4 h following contact, however, fifteen ABC transporter genes were increased, among which six were increased significantly (6/26 or 23%), with one gene significantly increased of the seven ABCB-type (14%), two genes significantly increased of the five ABCC-type (40%), and three genes significantly increased of the thirteen ABCG-type (23%) (Fig. 4C).
Overall, ivermectin exposure following immersion substantially increased the number of ABC transporter genes having increased transcript levels (5 genes increased at 2 h and 16 genes, 10 significantly, at 4 h) (Fig. 4B and D) compared to the contact exposure (1 gene at 2 h and 15 genes, 6 significantly, at 4 h) (Fig. 4A and C). Additionally, the 4 h ivermectin exposure resulted in substantially more ABC transporter genes having significantly increased transcript levels (10 genes with immersion and 6 genes with contact) (Fig. 4D and C) compared to the 2 h exposure (0 genes with immersion and 0 genes with contact) (Fig. 4B and A). Of the ABC transporter genes that had significantly increased transcript levels at 4 h post ivermectin exposure, the transcript levels of PhABCB4, PhABCC4, PhABCC5, PhABCG7, PhABCG10 and PhABCG12 were significantly increased by both immersion and contact applications, with PhABCC4 being the most highly expressed (Fig. 4D and C).
Because the transcript levels of the Clade 3 P450 genes CYP6CJ1, CYP9AG1 and CYP9AG2, and the ABC transporter gene PhABCC4 were significantly increased following ivermectin application by both the immersion and contact methods and all achieved substantial levels of expression, they appear to be primary candidates for mediating phase I (oxidative) and phase III (efflux) metabolism of ivermectin, respectively. Also the manner in which an insecticide is applied appears to be as important as the amount applied or the duration of exposure. Ivermectin applied in ethanol solutions by immersion resulted in substantially more genes being over-transcribed as compared to the contact (ivermectin-treated filter paper) application following both the 2 and 4 h exposures. Interestingly, some of the genes that were significantly over-transcribed following immersion were not significantly over transcribed following contact exposure (CYP358A1i, CYP4BV1, CYP4G38, CYP4G39, PhABCB5, PhABCB6, PhABCG2 and PhABCGG9), indicating that these genes may be responding to higher levels of ivermectin due to ethanol acting as a carrier solvent. Focusing the selection of candidate genes only on those that are over-transcribed follow both application methods is therefore an efficient screen to determine the most sensitive detoxification genes to ivermectin induction.
Phylogenetic relatedness of P450 and ABC transporter genes over transcribed following ivermectin exposure
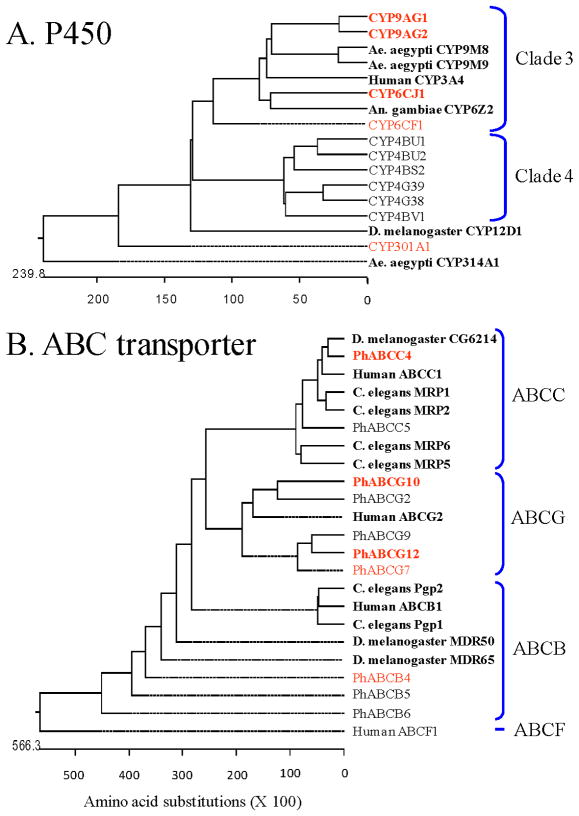
The phylogenetic relatedness of eleven of the nineteen P450 (Fig. 5A) and ten of the twenty-six ABC transporter (Fig. 5B) genes, which were significantly over-transcribed following the 4 h exposure to ivermectin by both immersion and contact application methods, was determined by deduced amino acid sequence comparisons to P450 and ABC transporter genes from various organisms that have previously been implicated in induction, binding and/or metabolism of insecticides, including ivermectin.
Figure 5.
Phylogenetic trees of cytochrome P450 monooxygenases (P450) and ABC transporters from deduced amino acid sequences of genes from female body lice that were significantly induced following a brief exposure to a sub-lethal amount of ivermectin. Louse genes over-transcribed following both immersion and contact exposures and that are highly expressed are given in bold red font. Louse genes over-transcribed following both immersion and contact exposures that are modestly expressed are given in normal red font. Louse genes that were over-transcribed by only one of the two exposure means and/or were either modestly or weakly expressed are given in normal black font. Non-louse genes used for comparisons are given in bold black font with the organism’s name.
CYP6CJ1, CYP9AG1 and CYP9AG2 are Clade 3 genes and are related to the human CYP3A4 gene, a P450 known to metabolize a diversity of insecticides, including ivermectin, and erythromycin, a close structural analog of ivermectin (Wacher et al., 1995) (Fig. 6A). CYP6CJ1 showed the greatest level of ivermectin induction by qPCR analysis and was most closely related to An. gambiae CYP6Z2, which binds a broad range of xenobiotics, including ivermectin (McLaughlin et al., 2008). CYP9AG2 was induced by ivermectin in both the 2 and 4 h qPCR analyses and was most closely related to Ae. aegyptii CYP9M8 and CYP9M9, both of which were induced by sub-lethal permethrin treatment (Poupardin et al., 2008). CYP301A1 was weakly induced by the 4 h ivermectin exposure and was most closely related to D. melanogaster CYP12D, which was induced by DDT (Brandt et al., 2002; Willoughby et al., 2006) and over-expressed in a DDT-resistant strain (Festucci-Buselli et al., 2005).
Figure 6.
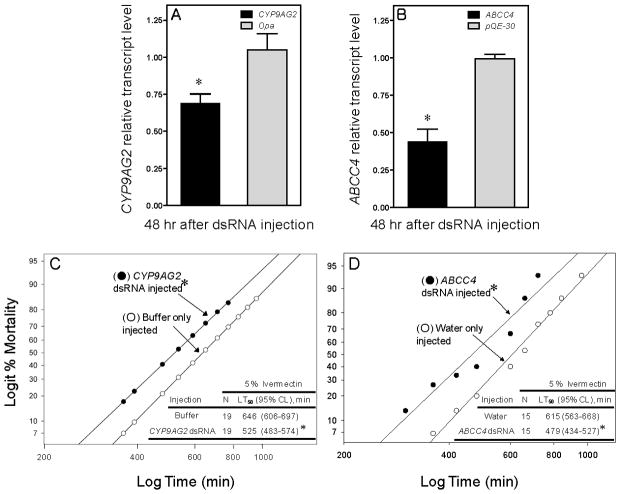
Relative transcript levels (panel A and B) and mortality responses (panel C and D) of body louse females to a lethal contact amount of ivermectin (5% IVM) following injection of dsRNA targeting either louse CYP9AG2 or ABCC4. Lice were also injected with either dsRNA of the odd-paired gene, opa, (GeneBank accession # S78339) for P450 silencing or with dsRNA of the E.coli plasmid, pQE30, for ABC transporter silencing as sham injected controls. Asterisks (*) in panels A and B indicate that CYP9AG2 and ABCC4 dsRNA significantly suppress the levels of CYP9AG2 and ABCC4 transcripts, respectively (Student’s t-test, P<0.05). In panel C, the bioassay was started 48 hr after CYP9AG2 dsRNA injection. In panel D, the bioassay was started 12 hr after ABCC4 dsRNA injection. Asterisks (*) in panels C and D indicate that the mortality responses of lice injected with dsRNAs were significantly different from their respective controls (buffer or water only injected, maximum log-likelihood ratio test, P<0.05).
PhABCC4 was the most highly expressed ABC transporter gene and was most closely related to C. elegans multi-drug resistant protein 1 (MRP1), D. melanogaster CG6214 and human ABCC1/MRP1 (Fig. 5B), with human MRP1 known to efflux ivermectin (Lespine et al., 2006). PhABCB4 was significantly over-expressed as determined by qPCR but its basal level of expression was low. Nevertheless, it was closely related to C. elegans Pgp1, D. melanogaster MDR50 and human ABCB1. Of these, Pgp1 is over-expressed in ivermectin-resistant C. elegans (James & Davey, 2009) and known to efflux ivermectin (Lespine et al., 2006). Human ABCB1 likewise actively effluxes ivermectin and erythromycin (Choudhuri & Klaassen et al., 2006) and MDR50 was over-expressed in DDT-resistant D. melanogaster (Strycharz et al., 2008). Less is known of the functional roles of PhABCG10 and PhABCG12 and their interaction with ivermectin. However, some G subfamily ABC transporters exhibit broad substrate specificity for xenobiotic compounds and are associated with MDR in humans (Sarkadi et al., 2006). Interestingly, all thirteen ABCG-type transporters for P. h. humanus contain only a single ATP binding domain (ABD) domain without the transmembrane domain (TM) domain, indicative of a role in drug/xenobiotic sequestration in the cytosol (Lee et al., 2010).
In summary, multiple P450 (e.g., CYP6CJ1, CYP9AG1 and CYP9AG2) and ABC transporter (e.g., PhABCC4, PhABCG10 and PhABCG12) genes have been identified by the non-invasive induction assay as putative candidate genes involved in ivermectin metabolism. P450-dependent oxidation of ivermectin is well established both in mammals and insects, resulting in the formation of three major oxidative metabolites. Additionally, we have shown in the present study that ivermectin tolerance is produced by pre-exposing lice to dexamethasone, an inducer of P450s. Both erythromycin and ivermectin are effuxed by ABC transporters, the drug-drug interaction of erythromycin and ivermectin are well documented, and we have shown in the present study that verapamil, a substrate for ABCB-type transporters, synergizes the toxicity of ivermectin (Fig. 1). Thus, it is highly likely that phase I (oxidation by P450s) and phase III (efflux and excretion of ivermectin by ABC transporters) metabolism may play a major role in tolerance and perhaps ultimately in resistance to ivermectin.
Knockdown of CYP9AG2 P450 and ABCC4 transporter gene expression by RNA interference (RNAi) and subsequent increase in the sensitivity of lice to ivermectin
Two louse genes, CYP9AG2 and ABCC4, were selected as the most relevant P450 and ABC transporter genes, respectively, for in vivo functional validation by RNAi knockdown experiments. Selection of these two genes was based on: (1) basal expression level (the level of transcription under untreated conditions were significantly higher than others); (2) extent of induction by ivermectin using qPCR analyses; and (3) phylogenetic relatedness to other P450 and ABC transporter genes shown previously to be involved in ivermectin metabolism and resistance.
When compared to female lice injected with the sham control odd-paired gene (opa) dsRNA, lice injected with CYP9AG2 dsRNA exhibited a 35% reduction in the transcript level of CYP9AG2 at 48 post-injection (Fig. 6A). When compared to lice injected with sham control pQE-30 dsRNA, lice injected with ABCC4 dsRNA exhibited a 56% reduction in the transcript levels of ABCC4 at 48 h post-injection (Fig. 6B). No apparent physiological alterations were noticed in lice injected with CYP9AG2 or ABCC4 dsRNAs compared to sham controls. These findings demonstrate that louse P450 and ABC transporter gene transcripts can be reduced by RNAi using a dsRNA approach.
The injection of either of these dsRNAs into non-ivermectin-induced female lice increased their sensitivity to ivermectin by ~ 20–30% compared to buffer/water injected lice as judged by their respective LT50 values (Fig. 6C and D). These results are supportive of the contention that both CYP9AG2 and ABCC4 are involved in the xenobiotic metabolism of ivermectin and likely involved in the production of tolerance and perhaps ultimately in resistance to ivermectin.
Conclusions
The above transcriptional profiling results show that ivermectin-induced detoxification genes from body lice can be identified by qPCR analyses using our non-invasive induction assay. CYP6CJ1, CYP9AG1, CYP9AG2 and PhABCC4 were most significantly induced, had high basal expression levels and were most closely related to genes from other organisms that metabolized insecticides, including ivermectin. P450-dependent oxidation of ivermectin is well established both in mammals and insects, resulting in the formation of three major oxidative metabolites. Additionally, pre-exposing lice to dexamethasone, an inducer of P450s, produces tolerance. Both erythromycin and ivermectin are effuxed by ABC transporters, the drug-drug interaction of erythromycin and ivermectin are well documented, and we have shown in the present study that verapamil, a substrate for ABCB-type transporters, synergizes the toxicity of ivermectin.
Injection of dsRNAs against either CYP9AG2 or PhABCC4 into non-induced female lice reduced their respective transcript level and resulted in increase sensitivity to ivermectin, indicating that these two genes are involved in the xenobiotic metabolism of ivermectin and in the production of tolerance. Heterologous expression of both genes, however, will be necessary to establish the actual metabolism of ivermectin and the identification of the metabolites formed.
As mentioned previously, Willoughby et al., (2006) investigated the induction response to six insecticides using a custom designed microarray in Drosophila melanogaster following a short exposure (4 h) to high lethal concentrations of insecticides. Third instars were exposed to five insecticides via their food source. Lufenuron and dicyclanil were used at concentrations significantly higher than those required to arrest development at a life stage transition (500 and 1000 times, respectively) and diazinon, nitenpyram and spinosad were used at concentrations that exceeded their LC99. Four-day old male flies were also exposure to DDT and nitenpyram by direct contact to the surface of a glass vial coated at concentrations that were lethal after 12 h of exposure. None of the five insecticides in food induced the expression of any P450, GST or esterase gene in third instar larvae. Only DDT induced a gene induction response in adult males following direct contact and resulted in the over transcription of only 1 of 89 P450 and 1 of 37 GST genes. Nitenpyram is usually used as a systemic insecticide against plant sucking insects or as an oral treatment for ectoparasites, such as ticks and fleas on cats and dogs. It did not induce gene expression following a contact exposure. These previously published results seem to be in disagreement with our current findings. However, there are significant differences between the two approaches used. Most notable is that we used a substantially lower concentration of ivermectin (LC3 at 24 h) that was applied by rapid emersion into an ethanol solution (~1 sec) followed by a rapid solvent evaporation (~5 sec) and a 2 h induction process. These approaches were adapted because we wanted to insure ivermectin exposure and uptake and look at induction events that occurred very rapidly, result in no mortality, little or no morbidity and cause little or no apparent stress. Most importantly, we ascertained that our induction process overlapped with the actual occurrence of tolerance to ivermectin. In doing so, we determined that tolerance to ivermectin occurred by 1 h post exposure and lasted approximately 5 h. Thus, when we measured the transcript levels of selected P450 and ABC transporter genes using the more sensitive quantitative real-time PCR technique at 2 and 4 h post-exposure, it overlapped with a tolerance event most likely caused by the increased metabolic detoxification of ivermectin. In the scheme used by Willoughby et al., (2006), the production of insecticide tolerance was not measured and time-course experiment for induction was determined only for phenobarbital and not with any of the insecticides tested.
It is our contention, therefore, that resistance to ivermectin in body lice will occur, in part, by a combination of oxidative metabolism and efflux via ABC transporters driven by the over-expression of some of the genes identified above once either constitutive over-expression or a more sensitive induction mechanism is selected for in the field. However, once identified as above, these inducible detoxification genes may be used in proactive resistance monitoring schemes and in the construction of metabolic maps using a variety of insecticides to establish cross- and negative cross-expression patterns during the acquisition of tolerance following induction. Such information is critical in establishing effective mixtures to be used in proactive resistance management of pediculosis.
Experimental procedures
Body louse strain and maintenance
Female lice were obtained from an inbred colony derived from the USDA/Culpepper strain of human body lice, Pediculus humanus humanus, which has been maintained on rabbits since 1999 at the University of Massachusetts-Amherst (Yoon et al., 2006).
Induction of tolerance with dexamethasone or ivermectin and synergism with verapamil
Female lice were immersed into sub-lethal concentrations of either dexamethasone (DEX, 1% w/v, <LD1 at 24 h), an inducer of P450(s), or ivermectin (IVM, 10−9 M, <LD3 at 24 h), both in ethanol, for ~1 sec, air-dried (~ 5 sec), and incubated (30°C, 70–80% RH) for 2 h on the in vitro rearing system (Yoon et al., 2006). Besides inducing both mammalian (Huss et al., 1995) and insect (Yoon et al., 2002) P450s, dexamethasone has been shown to be a substrate/competitive inhibitor of p-glycoproteins (MDR1a, Schinkel et al., 1995) and an eicosanoid biosynthesis inhibitor in insects (Carton et al., 2002). Control lice received only ethanol. For mortality bioassays following inducer pretreatments, ivermectin-impregnated filter paper disks were prepared by dipping them into 1% or 5% ivermectin in ethanol (w/v) for 10 sec and air dried (Gao et al., 2006). Disks were placed into Petri dishes and dexamethasone/ivermectin- or ethanol-treated lice transferred separately onto the ivermectin-impregnated disks. Log time versus logit percent mortality regressions were generated to determine LT50 values and the maximum log-likelihood ratio test used to determine equality (slope and intercept) of regression lines (P<0.05, POLO PC, LeOra Software, 1987).
Verapamil (VRP), a competitive substrate/inhibitor of ABCB-type transporters (Agarwala et al., 2004), was dissolved in ethanol to a final concentration of 1% w/v (<10% mortality at 24 h). Verapamil is also a well-known L-type calcium channel blocker (Szakacs et al., 2008) but has been shown to increase the toxicity of DDT in a highly DDT-resistant 91-R strain of D. melanogaster (Strychaz et al., 2010) and has recently been used to inhibit ABC transporters in honey bees, a process that greatly increases the toxicity of organophosphorous, pyrethroid and neonicotinoid insecticides (Hawthorne, 2011). Female lice were pretreated by immersing them into the verapamil solution for ~1 sec, air-dried ~5 sec, and incubated (30°C, 70–80% RH) for 2 h on the in vitro rearing system. Control lice were immersed into ethanol only. Ivermectin mortality bioassays were performed and analyzed as described above.
qPCR analysis of 19 P450 and 26 ABC transporter genes following brief exposure to a sub-lethal concentration of Ivermectin
We selected 19 P450 genes (all 18 annotated Clades 3 and 4 P450s and mitochondrial Clade CYP301A1) and 26 ABC transporter genes (all annotated subfamilies ABCB, ABCC and ABCG transporter genes and PhABCF1) from the body louse genome to determine their transcript levels using qPCR following 2 and 4 h exposures to sub-lethal amounts of ivermectin. Ivermectin was applied either by “immersion” of female lice into a sub-lethal 10−9 M ivermectin-ethanol solution for ~1 sec with ~5 sec air drying (<LC3 at 24 h) or allowing them to come into “contact” with a filter paper disk that had been impregnated with ivermectin by dipping it into 10−6 M ivermectin in ethanol and air-drying completely. Control lice where treated exactly as ivermectin-treated lice only without ivermectin, respectively. Contact exposure to this amount of ivermectin resulted in < 3% mortality at 24 h. The ethanol immersion scenario included a solvent (ethanol)-induced stress whereas the contact scenario excluded this stress and was designed to simulate the actual situation of pediculicide treatment to the human scalp.
Primers for qPCR were designed from individual P450- or ABC transporter-specific regions after aligning nucleotide sequences of all P450 or ABC transporter genes identified from the TIGR database (see Supplemental Tables 1 and 2, respectively). Total RNA was extracted with Trizol (MRC, Cincinnati, OH) from ivermectin-treated and untreated lice, treated with DNAase l and cDNA template for qPCR synthesized from 5 μg total RNA with SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA). qPCR was performed in a 20 μl reaction mixture containing 5 μl of 1/50-diluted cDNA, 5 pmol forward and reverse primers and 2× DyNAmo HS SYBR Green qPCR master mix (Finnzyme, Espoo, Finland) using Chromo4 thermal cycler (Bio-Rad, Hercules, CA). The reaction mixture was incubated at 95°C for 15 min for activation of DNA polymerase and qPCR conducted by 35 cycles of 95°C/10 sec, 56°C/15 sec, and 72°C/20 sec. The amplification efficiency of the qPCR was adjusted close to 2.0 using following equation, E = 10−1/slope, where the slope was derived from the plot of amplification critical time (Ct value) versus serially diluted template cDNA concentration. The specificity and authenticity of each PCR product was checked by melting curve analysis, where the melting curve was generated every 0.2°C from 55°C to 95°C. Elongation factor 1-α (EF1-α) gene was used as a reference gene to normalize the transcription levels of target P450 and ABC transporter genes. EF1-α showed the highest amplification stability across treatments (S.D. of Ct values < 0.5) among five housekeeping genes tested (EF1-α, two beta-tubulin1 genes, and two beta-tubulin2C genes) (data not shown).
The relative fold-increases of normalized induced transcript levels versus normalized non-induced transcript levels were determined using the following equation:
These values were converted to “normalized fold-increase values” used in Figs. 3 and 4 by the following equation:
where the normalized basal transcript level is determined by dividing the Ct value of non-induced samples normalized to the Ct values of EF1-α by either the normalized Ct value for CYP4BU2 (the highest P450 gene Ct value) or the normalized Ct value of PhABCG6 (the highest ABC transporter gene Ct value).
All qPCR were conducted with at least three biological replicates, with each having three technical replicates.
Phylogenetic relatedness of ivermectin-induced P450 and ABC transporter genes
The rooted phylogenetic trees were generated using ClustalW method (Thompson et al., 1994) using deduced amino acid sequences of P450 and ABC transporter genes from body louse females (Kirkness et al., 2010).
Knockdown of CYP9AG2 P450 and ABCC4 transporter gene expression by RNA interference (RNAi) and subsequent increase in the sensitivity of lice to ivermectin
A 652 (242-89) bp cDNA fragment of the P450 gene, CYP9AG2, was PCR amplified using the primer set (TAA TAC GAC TCA CTA TAG GGT ACG GGA TCC GGA ATT GAT A; TAA TAC GAC TCA CTA TAG GGC CAC GTC CGT ATT CTT CGT T), and a 484 (3024-3507) bp cDNA fragment of the ABC transporter gene, ABCC4, was amplified using the primer set (TAA TAC GAC TCA CTA TAG GGA GAA GAT GTC GAT ACG CTT GAT AGC; TAA TAC GAC TCA CTA TAG GGA GAG GCA TAC GTC ACG GAT AAA CCA AC). The T7 polymerase promoter element (TAA TAC GAC TCA CTA TAG GG) was attached to the 5′ ends of both forward and reverse primers. The dsRNA fragments were generated from the PCR product by using the MEGAscript T7 transcription kit (Ambion, TX). The dsRNA was alcohol-precipitated and resuspended in H2O. An aliquot of dsRNA (~112 ng for CYP9AG2 and ~35 ng for ABCC4) was ventrally injected into female lice (in between the second and third posterior abdominal segments) using a nanoinjector (Nanoliter 2000, World Precision Instruments, Sarasota, FL). The optimum concentrations resulting in maximum levels of target gene silencing were determined empirically through preliminary experiments by determining concentrations and volumes of dsRNA that cause no mortality during the 48 hr post-injection period. Lice were maintained on the in vitro rearing system as before. Total RNA was extracted from live lice at 48 h post-injection time points using TRI reagent (MRC). Total RNA was treated with DNase I and processed to cDNA using Superscript III (Invitrogen). Lice were also injected with either dsRNA of the odd-paired gene, opa (GenBank accession # S78339) for P450 silencing or dsRNA of the E. coli plasmid, pQE30, for ABC transporter silencing as sham-injected controls. qPCR was conducted as described previously to evaluate the degree of target gene silencing using the elongation factor 1-α (EF1α) gene as an internal reference. For qPCR of CYP9AG2, a new primer set was used (TAT CGT TGC TCT CGT TTC CGC CAT; TTC AGC GGG ATC CGA TGT ATG GAA (153 (13-166 bp).
Supplementary Material
P450 gene-specific oligonucleotide primer sets for the determination of relative transcript levels by qPCR. A primer set for the amplification of a specific region within the elongation factor 1-α (EF 1-α) gene was used for normalization of the P450 transcript levels.
ABC transporter gene-specific oligonucleotide primer sets for the determination of relative transcription levels by qPCR. A primer set for the amplification of a specific region within the elongation factor 1-α (EF 1-α) gene was used for normalization of the ABC transporter transcript levels.
Acknowledgments
This work was supported by a grant from the NIH/NIAID (5 R01 AI045062-06) to JMC and SHL and by C.W. Kearns, C.L. Metcalf and W.P. Flint Endowment funds to BRP. We thank the Body Louse Genome Consortium for their efforts in the annotation the P.h. humanus genome (Kirkness et al., 2010).
References
- Agarwala S, Chen W, Cook TJ. Effect of chlorpyrifos on efflux transporter gene expression and function in Caco-2 cells. Toxicol In Vitro. 2004;18:403–409. doi: 10.1016/j.tiv.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Baek JH, Clark JM, Lee SH. Cross-strain comparison of cypermethrin-induced cytochrome P450 transcription under different induction conditions in diamondback moth. Pestic Biochem Physiol. 2010;96:43–50. [Google Scholar]
- Brandt A, Scharf M, Pedra JHF, Holmes G, Dean A, Kreitman M, Pittendrigh BR. Differential expression and induction of two Drosophila cytochrome P450 genes near the Rst (2) locus. Insect Mol Biol. 2002;11:337–341. doi: 10.1046/j.1365-2583.2002.00344.x. [DOI] [PubMed] [Google Scholar]
- Brouqui P, Lascola B, Roux V, Raoult D. Chronic Bartonella quintana bacteremia in homeless patients. The New England J of Med. 1999;340:184–189. doi: 10.1056/NEJM199901213400303. [DOI] [PubMed] [Google Scholar]
- Carton Y, Frey F, Stanley DW, Vass E, Nappi A. Dexamethasone inhibition of the cellular response of Drosophila melanogaster against a parasitoid. J Parasit. 2002;88:405–407. doi: 10.1645/0022-3395(2002)088[0405:DIOTCI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Chosidow O, Giraudeau B, Cottrell J, Ozri A, Hofmann R, Mann SG, Burgess I. Oral ivermectin versus malathion lotion for difficult-to-treat head lice. New Engl J Med. 2010;362:896–905. doi: 10.1056/NEJMoa0905471. [DOI] [PubMed] [Google Scholar]
- Choudhuri S, Klaassen CD. Structure, function, expression, genomic organization, and a single nucleotide polymorphism of human ABCB1 (MDR1), ABCC (MRP) and ABCG2 (BCRP) efflux transporters. Int J of Toxicol. 2006;25:231–259. doi: 10.1080/10915810600746023. [DOI] [PubMed] [Google Scholar]
- David JP, Strode C, Vontas J, Nikou D, Vaughan A, Pignatelli Louis C, Hemingway J, Ranson H. The Anopheles gambiae detoxification chip: A highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc Natl Acad Sci USA. 2005;102:4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devonshire AL, Moores GD. A carboxylesterase with broad substrate specificity causes organophosphorous, carbamate, and pyrethroid resistance in peach-potato aphid. Pestic Biochem Physiol. 1982;18:235–46. [Google Scholar]
- Festucci-Buselli RA, Carvalho-Dias AS, de Oliveira-Andrade M, Caixeta-Nunes C, Li HM, Stuart JJ, Muir W, Scharf ME, Pittendrigh BR. Expression of Cyp6g1 and Cyp12d1 in DDT resistant and susceptible strains of Drosophila melanogaster. Insect Mol Biol. 2005;14:69–77. doi: 10.1111/j.1365-2583.2005.00532.x. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. Insect cytochrome P450. In: Gilbrert LI, Latrou K, Gill SS, editors. Comp Mol Insect Sci. Elsevier; Oxford, UK: 2005. pp. 1–77. [Google Scholar]
- Gao JR, Yoon KS, Frisbie RK, Coles GC, Clark JM. Esterase-mediated malathion resistance in the human head louse, Pediculus capitis (Anoplura: Pediculidae) Pestic Biochem Physiol. 2006;85:28–37. [Google Scholar]
- Hawthorne D. Killing them with kindness? In-hive medications may inhibit xenobiotic efflux transporters and endanger honey bees. PLoS ONE. 2011 doi: 10.1371/journal.pone.0026796. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Wang SI, Astrom A, McQuiddy P, Kasper CB. Dexamethasone responsiveness of a major glucocorticoid-inducible CYP3A gene is mediated by elements unrelated to a glucocorticoid receptor binding motif. Proc Natl Acad Sci USA. 1995;93:4666–75. doi: 10.1073/pnas.93.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CE, Davey MW. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. I J Parsitol. 2009;39:213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Kelly DJ, Richards AL, Temenak J, Strickman D, Dasch GA. The past and present threat of rickettsial diseases to military medicine and international public health. Clin Infect Dis. 2002;34 (suppl 4):S245–S169. doi: 10.1086/339908. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, et al. Genome sequences of the human body louse and its primary endosymbiont provides insights into the permanent parasitic lifestyle. Proc Natl Acad Sci USA. 2010;107:12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Min JS, Yoon KS, Strycharz JP, Johnson R, Mittapalli O, Margam VM, Sun W, Berenbaum MR, Pittendrigh BR, Clark JM. Decreased detoxification genes and genome size makes the human body louse an efficient model to study xenobiotic metabolism. Insect Mol Biology. 2010;19:599–615. doi: 10.1111/j.1365-2583.2010.01024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespine A, Dupuy J, Orlowski S, Nagy T, Glavinas H, Krajcsi P, Alvinerie M. Interaction of ivermectin with multidrug resistance proteins (MRP1, 2 and 3) Chemico-Biological Interactions. 2006;159:169–179. doi: 10.1016/j.cbi.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Lespine A, Martin S, Dupuy J, Roulet A, Pineau T, Orlowski S, Alvinerie M. Interaction of macrocyclic lactones with P-glycoprotein: Structure–affinity relationship. European Journal of Pharmaceutical Sciences. 2007;30:84–94. doi: 10.1016/j.ejps.2006.10.004. [DOI] [PubMed] [Google Scholar]
- MacDonald M, Gledhill A. Potential impact of ABCB1 (p-glycoprotein) polymorphisms on avermectin toxicity in humans. Arch Toxicol. 2007;81:553–563. doi: 10.1007/s00204-007-0193-6. [DOI] [PubMed] [Google Scholar]
- McLaughlin LA, Niazi U, Bibby J, David J-P, Vontas J, Hemingway J, Ranson H, Sutcliffe M, Paine MJI. Characterization of inhibitors and substrates of Anopheles gambiae CYP6Z2. Insect Mol Biol. 2008;17:125–135. doi: 10.1111/j.1365-2583.2007.00788.x. [DOI] [PubMed] [Google Scholar]
- Piesman P, Gage L. Bacterial and rickettsial diseases. In: Eldridge BF, Edman JD, editors. Medical Entomology. Kluwer Academic Publ; Boston, MA: 2000. pp. 337–413. [Google Scholar]
- Poupardin R, Reynaud S, Strode C, Ranson H, Vontas J, David JP. Cross-induction of detoxification genes by environmental xenobiotics and insecticides in the mosquito Aedes aegyptii: Impact on larval tolerance to chemical insecticides. Insect Biochem Mol Biol. 2008;38:540–551. doi: 10.1016/j.ibmb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Pray WS. Pediculicide resistance in head lice: A survey. Hosp Pharm. 2003;38:241–246. [Google Scholar]
- Prichard RK, Roulet A. ABC transporters and β-tubulin in macrocyclic lactone resistance: prospects for marker development. Parasitology. 2007;134:1123–1132. doi: 10.1017/S0031182007000091. [DOI] [PubMed] [Google Scholar]
- Raoult D, Roux V. The body louse as a vector of reemerging human disease. Clin Infect Dis. 1999;29:888–911. doi: 10.1086/520454. [DOI] [PubMed] [Google Scholar]
- Sarkadi S, Homolya L, Szakacs G, Varadi A. Human multidrug resistance ABCB and ABCG transporters: Participation in a chemoimmunity defense system. Physiol Rev. 2006;86:1179–1236. doi: 10.1152/physrev.00037.2005. [DOI] [PubMed] [Google Scholar]
- Strycharz JP, Yoon KS, Clark JM. A New Ivermectin Formulation Topically Kills Permethrin-resistant Human Head Lice, Pediculus humanus capitis (Phthiraptera: Pediculidae) J Med Entomol. 2008;45:75–81. doi: 10.1603/0022-2585(2008)45[75:aniftk]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Strycharz JP, Lee SH, Sun W, Pittendrigh BR, Clark JM. Picogram: Abstract Book. 182. Vol. 78. ACS/AGRO; 2010. RNAi knockdown of ABC transporters causes decreased tolerance in the highly DDT-resistant 91-R strain of Drosophila melanogaster; p. 161 .p. 116. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vontas J, Blass C, Koutsos AC, David JP, Kafatos FC, Louis C, Hemingway J, Christophides GK, Ranson H. Gene expression in insecticide resistant and susceptible Anopheles gambiae strains constitutively or after insecticide exposure. Insect Mol Biol. 2005;14:509–521. doi: 10.1111/j.1365-2583.2005.00582.x. [DOI] [PubMed] [Google Scholar]
- Wacher VJ, Wu CY, Benet LZ. Overlapping substrate specificity and tissue distribution of cytochrome P45 3a and P-glycoprotein: Implications for drug delivery and activity in cancer chemotherapy. Mol Carcinog. 1995;13:129–134. doi: 10.1002/mc.2940130302. [DOI] [PubMed] [Google Scholar]
- Weiss K. The role of rickettsioses in history. In: Walker DH, editor. Biology of Rickettsial Diseases. Vol. 1. CRC; Boca Raton, FL: 1988. pp. 1–14. [Google Scholar]
- Willoughby L, Chung H, Lumb C, Robin C, Batterham P, Daborn PJ. A comparison of Drosophila melanogaster detoxification gene induction responses for six insecticides, caffeine and phenobarbital. Insect Biochem and Mol Biol. 2006;36:934–942. doi: 10.1016/j.ibmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Yoon KS, Gao JR, Lee SH, Clark JM, Brown L, Taplin D. Permethrin-resistant human head lice, Pediculus capitis, and their treatment. Arch Dermatol. 2003;139:994–1000. doi: 10.1001/archderm.139.8.994. [DOI] [PubMed] [Google Scholar]
- Yoon KS, Nelson JO, Clark JM. Selective induction of abamectin metabolism by dexamethasone, 3-methyl cholanthrene, and phenobarbital in Colorado potato beetle, Leptinotarsus decemlineata (Say) Pestic Biochem Physiol. 2002;73:74–86. [Google Scholar]
- Yoon KS, Strycharz JP, Gao JR, Takano-Lee M, Edman JD, Clark JM. An Improved In Vitro Rearing System for the Human Head Louse allows the Determination of Resistance to Formulated Pediculicides. Pestic Biochem Physiol. 2006;86:195–202. [Google Scholar]
- Xu M, Molento M, Blackhall W, Ribeiro P, Beech R, Prichard R. Ivermectin resistance to nematodes may be caused by alteration of P-glycoprotein homolog. Mol Biochem Parasitol. 1998;91:327–335. doi: 10.1016/s0166-6851(97)00215-6. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Andrew NW, Woda JM, Halley BA, Crouch LS, Wang RW. Role of cytochrome P450 isoforms in the metabolism of abamectin and ivermectin in rats. J Agric Food Chem. 1996;44:3374–3378. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P450 gene-specific oligonucleotide primer sets for the determination of relative transcript levels by qPCR. A primer set for the amplification of a specific region within the elongation factor 1-α (EF 1-α) gene was used for normalization of the P450 transcript levels.
ABC transporter gene-specific oligonucleotide primer sets for the determination of relative transcription levels by qPCR. A primer set for the amplification of a specific region within the elongation factor 1-α (EF 1-α) gene was used for normalization of the ABC transporter transcript levels.