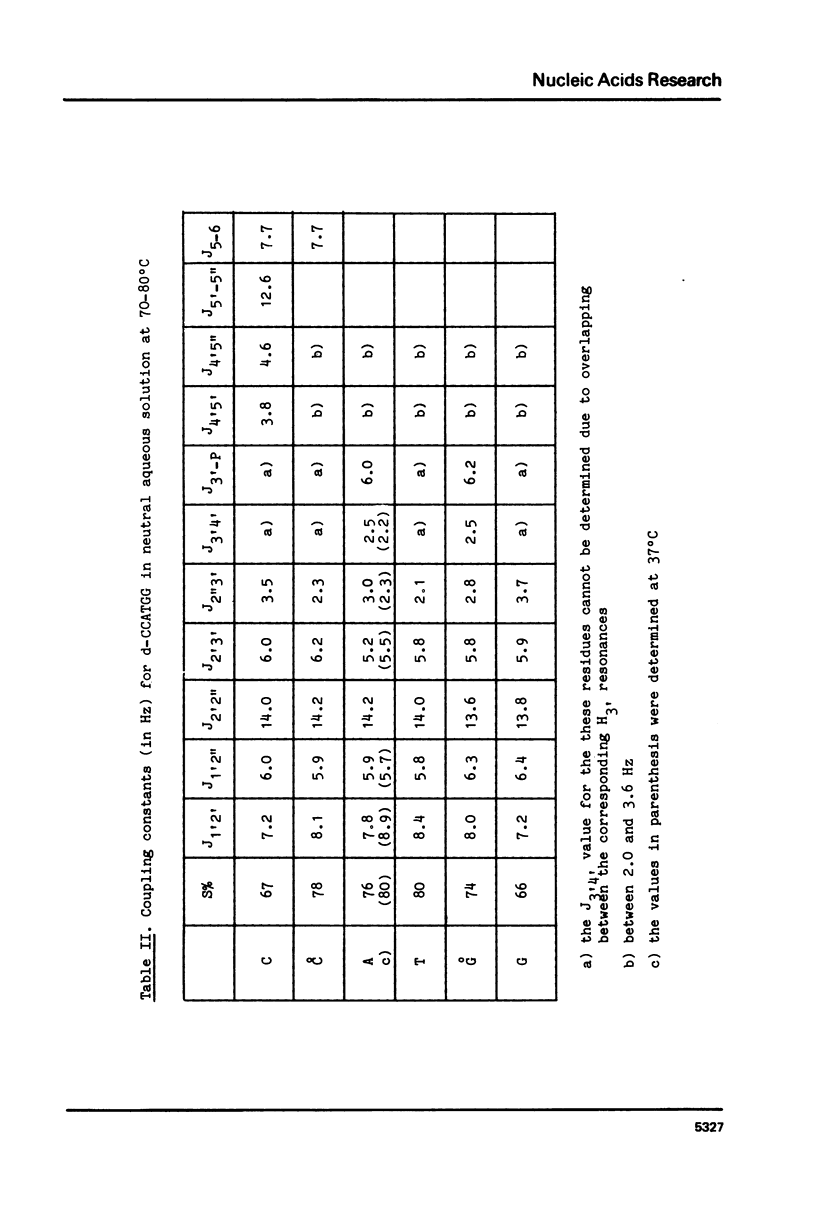
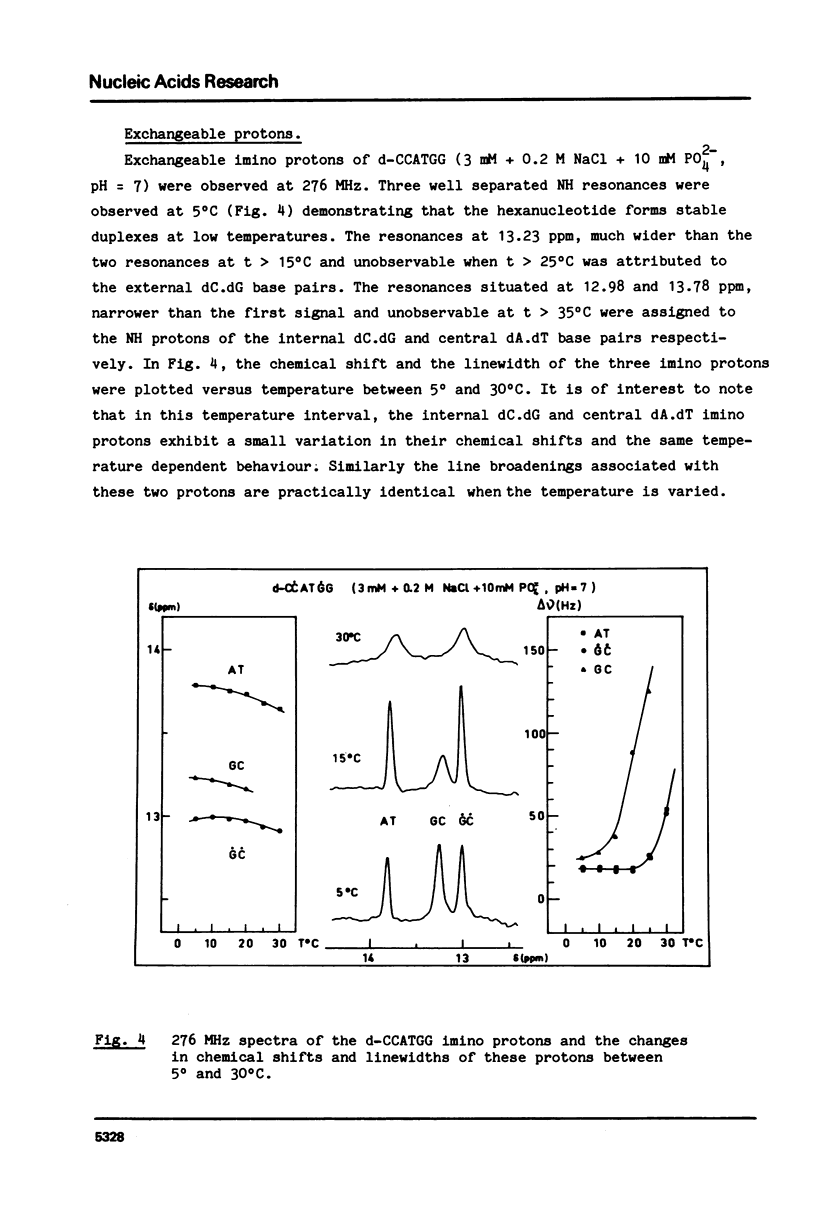
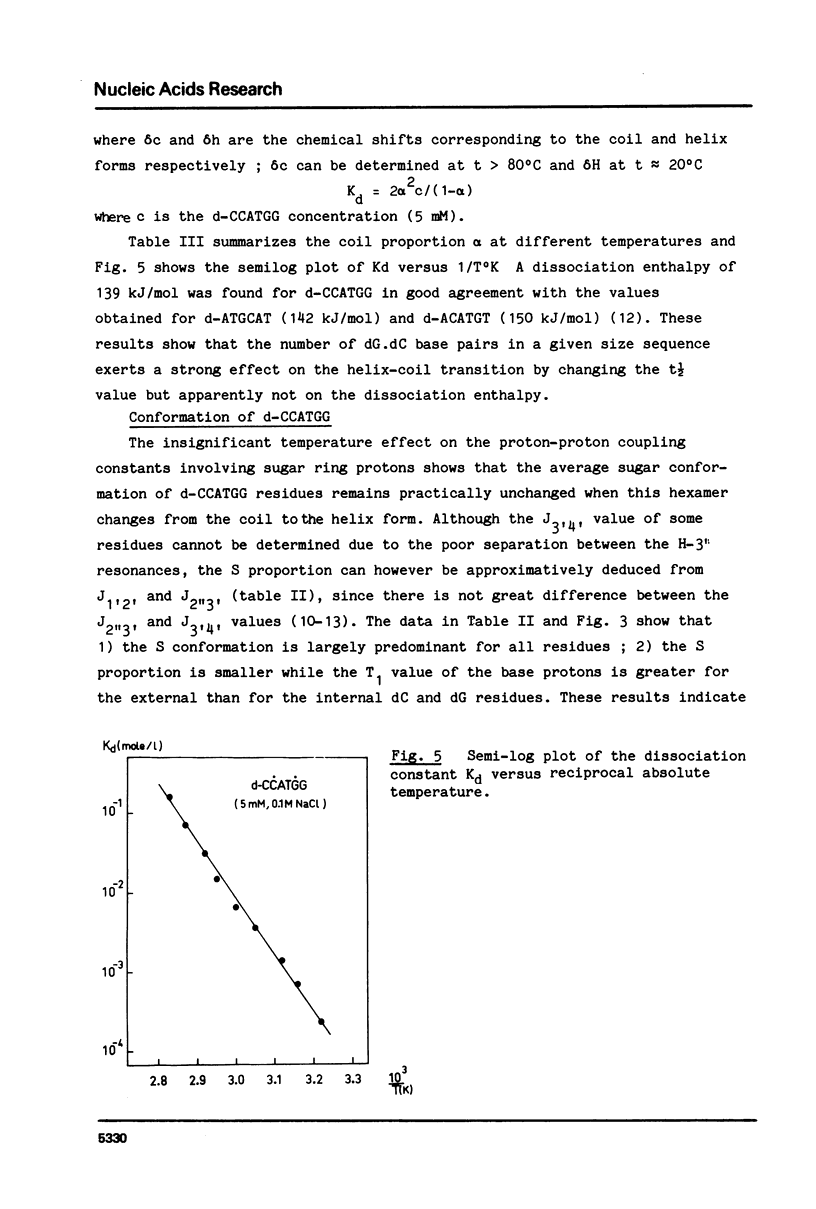
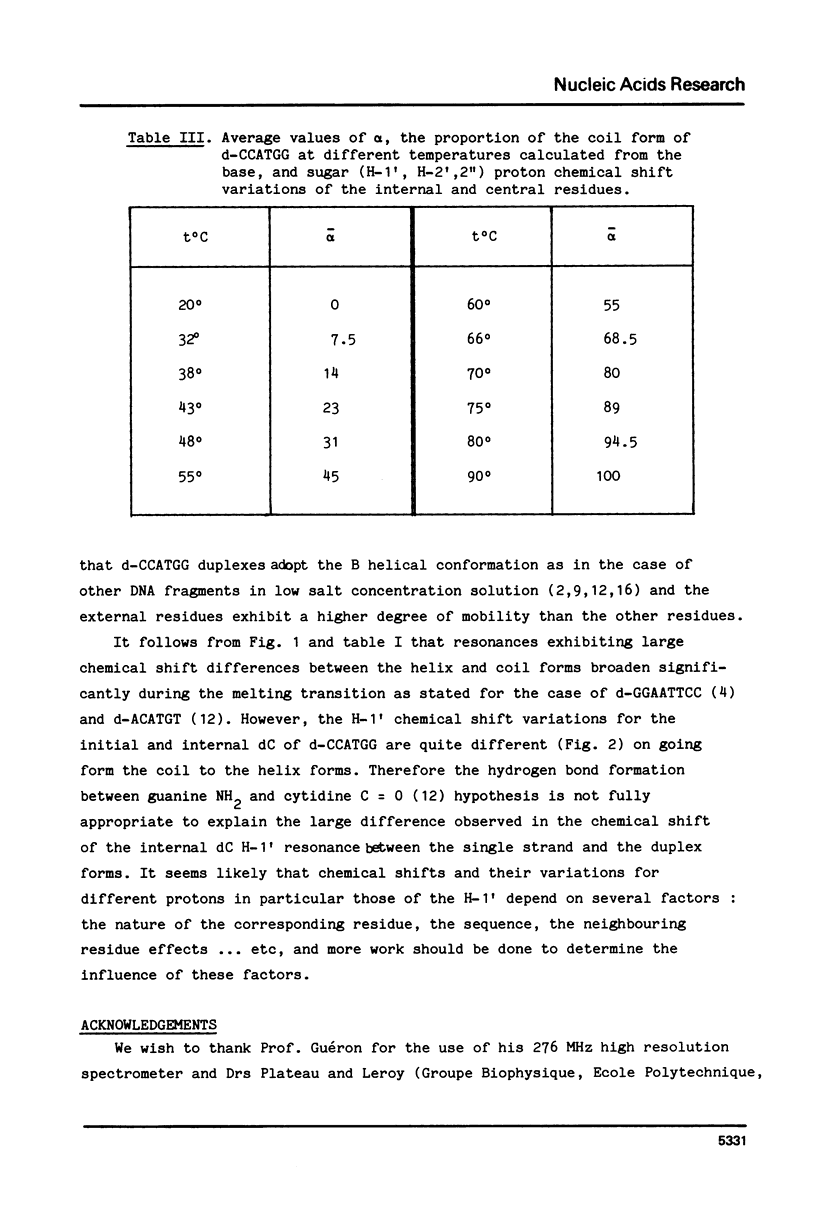
Abstract
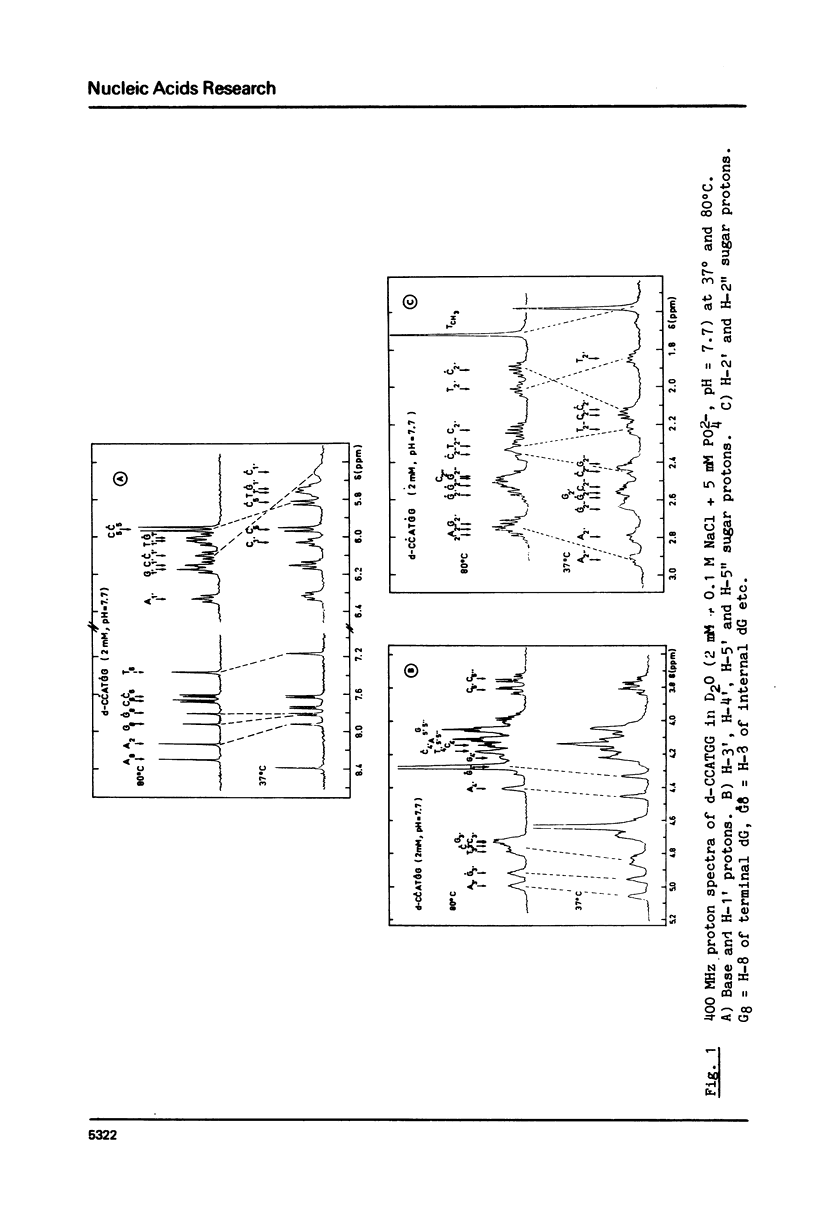
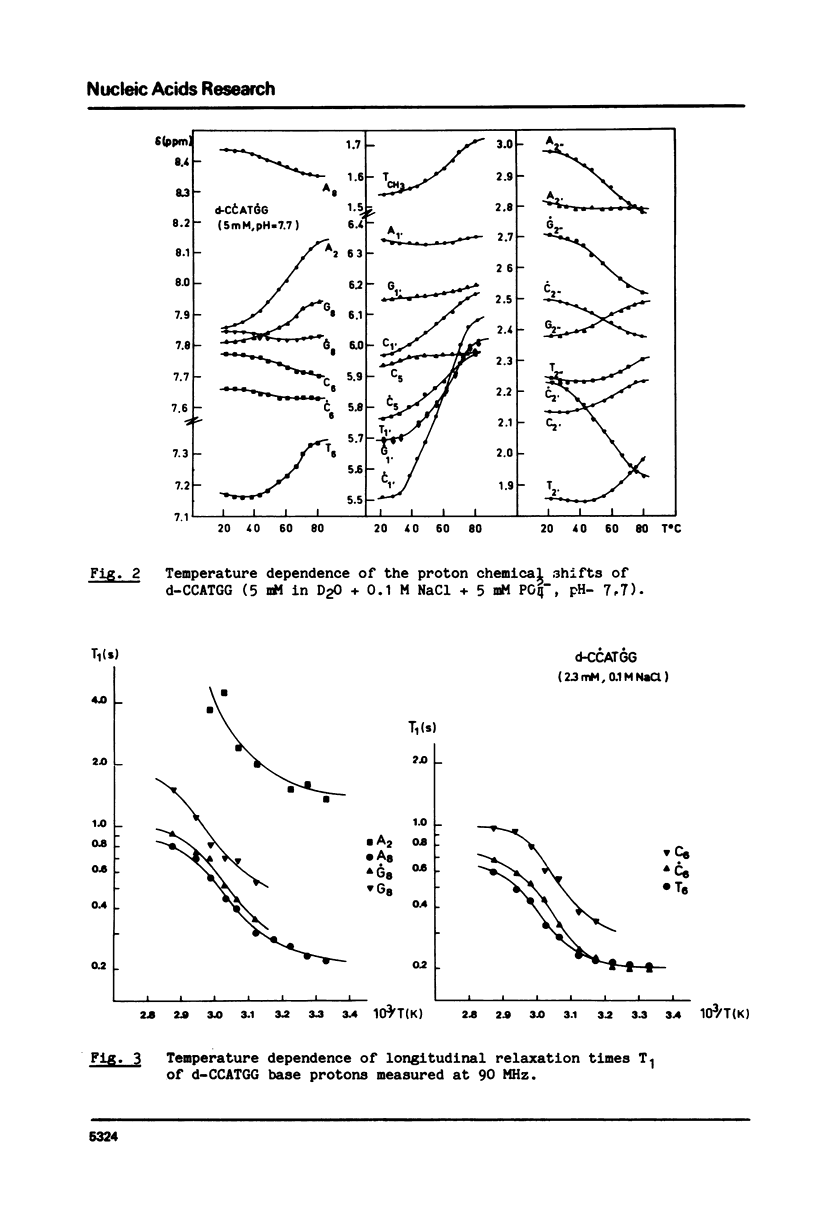
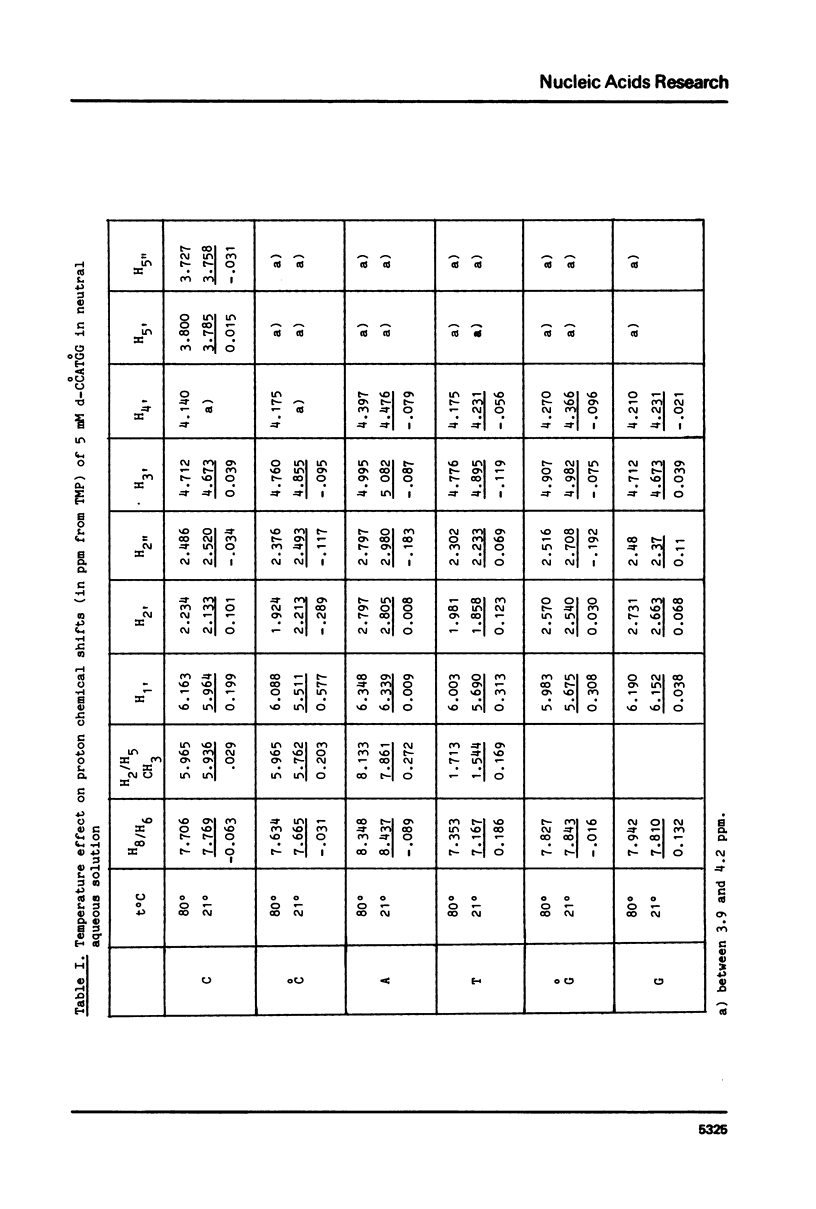
The helix-coil transition and conformation of d-CCATGG were investigated using 1H-NMR spectroscopy at various frequencies (90, 276, 400 MHz). The changes in the chemical shifts and linewidths of imino protons between 5 degrees and 35 degrees C show that the d-CCATGG fraying process consists of two stages: the external dC.dG base pairs open at first, th internal dC.dG and central dA.dT base pairs then open simultaneously at higher temperatures similar to the case of d-ACATGT. The midpoint temperatures, the helix and coil proportions and the dissociation constant were determined from the sigma = f(t degree) curves of the base and sugar protons. The results indicate that the midpoint temperature increases with the number of the dG.dC base pair in a given size sequence, while the dissociation enthalpy appears to be independent. The difference between the T1 value of a base proton of the external and internal residues of the same nature is found to be a good criterion for base proton assignment. The high predominance of the S conformation for all residues shows that d-CCATGG duplexes adopt the B-helical conformation.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arter D. B., Walker G. C., Uhlenbeck O. C., Schmidt P. G. PMR or the self-complementary oligoribonucleotide CpCpGpG. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1089–1094. doi: 10.1016/s0006-291x(74)80395-5. [DOI] [PubMed] [Google Scholar]
- Borer P. N., Kan L. S., Ts'o P. O. Conformation and interaction of short nucleic acid double-stranded helices. I. Proton magnetic resonance studies on the nonexchangeable protons of ribosyl ApApGpCpUpU. Biochemistry. 1975 Nov 4;14(22):4847–4863. doi: 10.1021/bi00693a012. [DOI] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R., Hillen W., Wells R. D. A 300 MHz and 600 MHz proton NMR study of a 12 base pair restriction fragment: investigation of structure by relaxation measurements. Nucleic Acids Res. 1980 Dec 11;8(23):5795–5812. doi: 10.1093/nar/8.23.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancelot G., Mayer R., Thuong N. T., Chassignol M., Hélène C. Synthesis and structural studies of a self-complementary decadeoxynucleotide d(AATTGCAATT). II.-Proton magnetic resonance studies. Biochimie. 1981 Oct;63(10):785–790. doi: 10.1016/s0300-9084(81)80038-7. [DOI] [PubMed] [Google Scholar]
- Mellema J. R., Haasnoot C. A., Van Boom J. H., Altona C. Complete assignment and conformational analysis of a deoxyribotetranucleotide. d(TAAT). A 360 and 500 Mhz NMR study. Biochim Biophys Acta. 1981 Sep 28;655(2):256–264. doi: 10.1016/0005-2787(81)90016-2. [DOI] [PubMed] [Google Scholar]
- Neumann J. M., Huynh-Dinh T., Kan S. K., Genissel B., Igolen J., Tran-Dinh S. DNA fragment conformations. 1H-NMR and relaxation studies of 2'-deoxyadenylyl(3'-5')thymidylyl-(3'-5')deoxyguanosylyl(3'-5')thymidine, d(A-T-G-T), in neutral aqueous solution. Eur J Biochem. 1982 Jan;121(2):317–323. doi: 10.1111/j.1432-1033.1982.tb05788.x. [DOI] [PubMed] [Google Scholar]
- Pardi A., Martin F. H., Tinoco I., Jr Comparative study of ribonucleotide, deoxyribonucleotide, and hybrid oligonucleotide helices by nuclear magnetic resonance. Biochemistry. 1981 Jul 7;20(14):3986–3996. doi: 10.1021/bi00517a007. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. L. Helix-coil transition of the self-complementary dG-dG-dA-dA-dT-dT-dC-dC duplex. Eur J Biochem. 1979 May 15;96(2):267–276. doi: 10.1111/j.1432-1033.1979.tb13037.x. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Hilbers C. W. Proton nuclear magnetic resonance investigations of fraying in double-stranded d-ApTpGpCpApT in H2O solution. Biochemistry. 1975 Jun 17;14(12):2651–2656. doi: 10.1021/bi00683a014. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Broka C., Rice J. A., Itakura K., Breslauer K. J. Premelting and melting transitions in the d(CGCGAATTCGCG) self-complementary duplex in solution. Biochemistry. 1982 Feb 2;21(3):428–436. doi: 10.1021/bi00532a002. [DOI] [PubMed] [Google Scholar]
- Patel D. J. Peptide antibiotic-oligonucleotide interactions. Nuclear magnetic resonance investigations of complex formation between actinomycin D and d-ApTpGpCpApT in aqueous solution. Biochemistry. 1974 May 21;13(11):2396–2402. doi: 10.1021/bi00708a025. [DOI] [PubMed] [Google Scholar]
- Patel D. J. d-CpCpGpG and d-GpGpCpC self-complementary duplexes: Nmr studies of the helix-coil transition. Biopolymers. 1977 Aug;16(8):1635–1656. doi: 10.1002/bip.1977.360160804. [DOI] [PubMed] [Google Scholar]
- Phillips D. R., Roberts G. C. Proton nuclear magnetic resonance study of the self-complementary hexanucleotide d(pTpA)3 and its interaction with daunomycin. Biochemistry. 1980 Oct 14;19(21):4795–4801. doi: 10.1021/bi00562a013. [DOI] [PubMed] [Google Scholar]
- Tran-Dinh S., Neumann J. M., Huynh-Dinh T., Genissel B., Igolen J., Simonnot G. DNA fragment conformations. 1H-NMR studies of helix-coil transition, conformations and dynamic structures of the self-complementary deoxyhexanucleotide d(A-C-A-T-G-T) in aqueous solution. Eur J Biochem. 1982 Jun;124(3):415–425. [PubMed] [Google Scholar]



