Abstract
Shotgun proteomics protocols are widely used for the identification and/or quantitation of proteins in complex biological samples. Described here is a shotgun proteomics protocol that can be used to identify the protein targets of biologically relevant ligands in complex protein mixtures. The protocol combines a quantitative proteomics platform with a covalent modification strategy, termed Stability of Proteins from Rates of Oxidation (SPROX), which utilizes the denaturant dependence of hydrogen peroxide-mediated oxidation of methionine side chains in proteins to assess the thermodynamic properties of proteins and protein-ligand complexes. The quantitative proteomics platform involves the use of isobaric mass tags and a methionine-containing peptide enhancement strategy. The protocol is evaluated in a ligand binding experiment designed to identify the proteins in a yeast cell lysate that bind the well-known enzyme co-factor, β-nicotinamide adenine dinucleotide (NAD+). The protocol is also used to investigate the protein targets of resveratrol, a biologically active ligand with less well-understood protein targets. A known protein target of resveratrol, cytosolic aldehyde dehydrogenase, was identified in addition to six other potential new proteins targets including four that are associated with the protein translation machinery, which has previously been implicated as a target of resveratrol.
Keywords: Protein-ligand binding, resveratrol, NAD+, methionine oxidation, iTRAQ, SPROX
Introduction
The detection and quantitation of protein-ligand binding interactions is important for understanding biological processes. It is also a critical part of many drug discovery efforts, particularly those focused on the development of therapeutic agents with protein targets. Knowledge about the protein targets of therapeutic agents can help define their mode-of-action, and knowledge about a protein’s interaction with specific biological ligands (e.g., enzyme co-factors) can facilitate the functional annotation of proteins. Thus, in order to gain a comprehensive understanding of protein function and protein-drug interactions, it is critical to have the ability to query all the proteins in a proteome for ligand binding and to quantitatively assess their affinity for target ligands.
Few experimental strategies exist for the analysis of protein-ligand binding interactions on the proteomic scale. The most commonly used strategies for the large-scale and high-throughput analysis of protein-ligand interactions, to date, have exploited either the yeast two-hybrid assay1 or affinity-based purification techniques coupled with mass spectrometry.2 While these strategies have proven useful for the large-scale detection of protein-ligand interactions,3–11 the quantitative information that can be extracted from experiments utilizing these two strategies is limited. Applications of the yeast two-hybrid assay and affinity-based purification techniques to non-protein ligands (e.g., small drug molecules) have also been limited due to the inherent difficulties associated with conjugating such ligands to a necessary bait protein in the yeast-based assay or to a necessary solid support in the affinity-based purification assay. More recently, protease protection assays have been developed for the identification of small molecule drug targets on the proteomic scale.12, 13 Protease protection assays are attractive because they are amenable to the detection of both direct and indirect protein targets of drugs. However, one drawback to the use of protease protection assays is that they do not provide quantitative measurements of protein-ligand binding affinities.
Recently, we described an experimental strategy that facilitates both the detection and quantitation of protein-ligand binding interactions on the proteomic scale.14, 15 The strategy involves the use of quantitative proteomics technologies (e.g., isobaric mass tags14 or stable isotopes15) with SPROX (Stability of Proteins from Rates of Oxidation),16 a technique that utilizes the denaturant dependence of hydrogen peroxide-mediated oxidation of methionine side chains in proteins to measure the thermodynamic properties of protein-ligand complexes. One drawback to the bottom-up proteomics protocols interfaced with SPROX, to date,14, 15 is that the number of proteins in a complex mixture that can be analyzed is limited to those proteins that can be identified with methionine-containing peptides in the proteomics experiment. Results from our earlier experiments on the proteins in a yeast cell lysate showed that approximately 20% of the identified peptides contained at least one methionine residue and that these methionine-containing peptides mapped to approximately 33% of the proteins identified in the shotgun proteomics analysis.
The frequency of methionine residues in naturally occurring protein sequences is relatively low and has been estimated at approximately 2%.17 However, nearly all protein sequences contain at least one methionine residue in addition to the initiator methionine residue, which is generally removed in vivo. For example, it has been noted that 95.8% of E. coli proteins contain at least one methionine residue.18 Thus, a bottom-up proteomics strategy that focused on the identification of methionine-containing peptides would be highly beneficial for SPROX analyses performed on proteins in complex biological mixtures such as cell lysates.
Described here is an experimental protocol that involves the use of a commercially available resin to chemo-selectively isolate the unoxidized methionine-containing peptides generated in a shotgun proteomics-based SPROX experiment. The chemo-selective isolation of methionine-containing peptides in conventional bottom-up proteomics experiments has previously been reported to facilitate protein identifications.19, 20 As part of the work described here, we report the development and initial application of a SPROX protocol that incorporates such a chemo-selective isolation strategy into a quantitative, bottom-up proteomics experiment using isobaric mass tags to make quantitative thermodynamic measurements of protein folding and ligand binding on the proteomic scale. The protocol is developed in the context of two ligand binding experiments, including one experiment to identify the proteins in a yeast cell lysate that bind a well-known and ubiquitous ligand, β-nicotinamide adenine dinucleotide (NAD+), and a second experiment to investigate the protein targets of resveratrol, a biologically active ligand with less well-understood protein targets.
Experimental
Yeast Cell Lysate Preparation
The yeast cell lysate used in the NAD+ binding study was obtained from a Saccharomyces cerevisiae Y258 strain (MATa pep4-3, his4-580, ura 3-53, leu2-3,112) purchased from Open Biosystems (Huntsville, AL) that overexpressed yeast NAD+-dependent glutamate dehydrogenase (YDL215C) under control of the GAL1 promoter in the form of a C-terminal fusion protein. The overexpressed yeast fusion protein contained a 19-kDa purification tag containing an HA epitope, a 3C protease cleavage site, a ZZ domain from Protein A, and a 6xHis-tag. This Y258 strain was grown overnight at 30°C in 2% dextrose SC-Ura, and 2 mL of this solution was used to inoculate 50 ml of 2% raffinose SC-Ura solution, which was then incubated overnight. The solution was diluted into 500 mL of 2% raffinose SC-Ura to give an OD600 of 0.3 and was incubated at 30°C until the OD600 of the solution was 0.8–1.2. A 250-mL volume of 3×YP-Gal (3% yeast extract, 6% peptone, 6% galactose) was added to induce overexpression. After 6 h of incubation, yeast cell pellets were obtained by centrifuging 250 mL of culture at 1000 × g for 10 min. The yeast cell pellets were frozen until further use.
The yeast cell lysate in the resveratrol binding study was obtained from S. cerevisiae strain Y258, also purchased from Open Biosystems. The Y258 strain was grown on YPD media containing 1% yeast extract, 2% peptone, and 2% dextrose. Yeast cells were harvested when the OD600 reached approximately 0.8–1.0, and yeast cell pellets were obtained by centrifuging 250 mL of culture at 1000 × g for 10 min. The yeast cell pellets were frozen until further use.
The yeast cell lysates used in the NAD+ and resveratrol binding studies were each obtained from a 250-mL culture. In each case the yeast cell pellets were lysed in 500 µL of 20 mM phosphate, pH 7.4, containing 1 mM AEBSF, 50 µM bestatin, 15 µM E64, 20 µM leupeptin, and 10 µM peptstatin A (Pierce). Cell lysis was accomplished by vortexing the pellets with acid-washed glass beads (425–600 µm in diameter) (Sigma-Aldrich) for 20 s ten times, with 1 min intervals on ice in between. The samples were centrifuged at 14,000 × g for 5 min to pellet the insoluble material, and the supernatant lysate was used in the SPROX analysis. The total protein concentration in each lysate was ~9 mg/mL, as measured using a Bradford assay.
The lysate used in the NAD+ binding study was divided into two 225-µL aliquots. A total of 25 µL of 50 mM NAD+ (Sigma) was added to one aliquot, and 25 µL 20 mM phosphate was added to the other. The two lysate samples were incubated for 2 h, before dilution into the SPROX buffers. The lysate used in the resveratrol binding study was divided into two 150-µL aliquots, and 16.67 µL 10 mM resveratrol (Sigma-Aldrich) in DMSO was added to one, while 16.67 µL DMSO was added to the other. The lysates were allowed to equilibrate in the presence or absence of the small molecule ligands for 30 min before dilution into the SPROX buffers.
SPROX Analysis
A total of 20 µL of the above yeast cell lysate samples (i.e., the yeast cell lysates in the presence and in the absence of the NAD+ and resveratrol ligands) were combined with 75 µL (25 µL for the resveratrol binding experiment) of each denaturant-containing SPROX buffer stock solution. The denaturant-containing SPROX buffer stock solutions for the NAD+ binding study were comprised of 20 mM sodium phosphate buffer pH 7.4 and different concentrations of urea, which were 0, 1.4, 2.7, 3.6, 4.4, 5.3, 6.5, or 8 M. Urea was chosen as the denaturant in the NAD+ binding study as opposed to GdmCl because it is better suited to the analysis of protein-ligand complexes involving electrostatic interactions. We have previously found that such electrostatic interactions can be significantly weakened in the presence of GdmCl.21 The denaturant-containing SPROX buffer stock solutions for the resveratrol binding study were comprised of 20 mM sodium phosphate buffer pH 7.4 and different concentrations of GdmCl, which were 1.0, 2.0, 2.3, 3.3, 4.0, 5.0, 6.0, and 7.0 M. After a 30 min equilibration, the oxidation reaction in each denaturant-containing SPROX buffer was initiated by adding 5 µL of 9.8 M H2O2 (Sigma-Aldrich). The final urea concentrations in the oxidation reactions performed in the NAD+ binding experiment were 0, 1.0, 2.0, 2.7, 3.3, 4.0, 4.9, and 6.0 M; and the final GdmCl concentrations in the oxidation reactions performed in the SPROX buffers used in the resveratrol binding experiment were 0.5, 1.0, 1.3, 1.7, 2.0, 2.5, 3.0, and 3.5 M. After 6 min (3 min for the resveratrol binding experiment) the oxidation reaction in each SPROX buffer was quenched with 950 µL (1 mL for the resveratrol binding experiment) of 300 mM free L-methionine (Sigma-Aldrich).
Ultimately, the protein material in each denaturant-containing SPROX buffer was precipitated with the addition of 200 µL ice-cold TCA solution that was 1 g/mL. Following an overnight incubation on ice, the samples were centrifuged at 8,000 × g for 30 min at 4°C to pellet the protein. The supernatants were removed, and the protein pellets were washed 3 times with 300 µL of ice-cold ethanol.
The NAD+ binding study involved a single SPROX analysis in which the yeast cell lysate was analyzed in the presence and in the absence of the NAD+. The resveratrol binding study involved two identical SPROX analyses in which the yeast cell lysate was analyzed in the presence and in the absence of resveratrol.
Proteomic Sample Preparation
The protein pellets obtained from each denaturant-containing SPROX buffer were redissolved in 30 µL of 0.5 M triethylammonium bicarbonate (Sigma), pH 8.5, and 1.5 µL 2% (w/w) SDS (Sigma), with aid of vortexing and sonication. After dissolution, 5 µL of each sample was removed for a Bradford assay to determine how much protein was recovered following the TCA precipitation. The protein concentrations recorded in the samples were typically ~1–2 µg/µL. The remainder of the sample was reduced with TCEP, alkylated with MMTS, and digested with trypsin all according to the iTRAQ labeling protocol provided by the manufacturer. The peptide samples were then labeled with iTRAQ 8-plex isobaric mass tags (AB Sciex). Each set of samples (i.e., with and without ligand) was labeled with 0.5 units of the iTRAQ tags dissolved in 50 µL isopropanol. After addition of the iTRAQ tags, the pH of the samples was tested to confirm it was above pH 8. The iTRAQ labeling reaction proceeded for 2 h at room temperature, and the samples were then stored at −20°C until they were analyzed by LCMS/ MS. Samples that were not subjected to the methionine-containing peptide enhancement were desalted using C18 resin (The Nest Group) just prior to LC-MS/MS analysis.
Chemo-Selective Isolation of Methionine-Containing Peptides
A 25–30 µL aliquot from each iTRAQ-labeled sample (corresponding to approximately 25 µg of total protein measured in the Bradford Assay described above) was used for the methionine enhancement protocol. This amount was removed from each set of iTRAQ-labeled samples and combined with the other 7 labeled samples within each set of samples. The pooled samples containing the peptide samples from all eight of the iTRAQ labeling reactions were reduced in volume to approximately 50–75 µL using a SpeedVac concentrator, and approximately 25–30 µL glacial acetic acid was added to lower the pH of the sample to between 2–3.
Pi3 Methionine Selective Resin (The Nest Group) was used according to the manufacturer’s recommended protocol to select for iTRAQ-labeled, unoxidized methionine-containing peptides. Following this selection, the iTRAQ-labeled, unoxidized methionine-containing peptide samples were desalted using 50 µg C18 resin (The Nest Group). The iTRAQ-labeled, unoxidized methionine-containing peptide samples were eluted from the resin with 60–70% ACN/0.1% TFA. The ACN was evaporated using a SpeedVac concentrator, and 0.1% TFA in water was added to bring the total volume to about 150 µL.
Quantitative Proteomic Sample Analysis
Samples were analyzed using an Agilent 6520 Q-TOF mass spectrometer system equipped with a Chip Cube Interface. The HPLC Chip used in this experiment contained a 150 mm × 75 µm column with Zorbax 300SB-C18 5 µm packing. The tryptic peptides were eluted using a linear gradient consisting of an increase from 5% to 15% Buffer B in 2.5 min, 15% to 45% Buffer B in 78 min, and then 45% to 100% Buffer B over 10 min. Buffer A was 0.1% FA in water and Buffer B was 0.1% FA in ACN. The flow rate was 0.4 µL/min. The capillary voltage was 1850 V. The drying gas was 350°C at a flow rate of 6 L/min. The skimmer and fragmentor were set to 65 V and 175 V, respectively. The inclusion window width for precursor ions was 4 m/z units. The scan rate was 3 scans per second in the mass spectra and 2 scans per second in the product ion mass spectra, and there were 4 precursors selected for fragmentation per cycle. The collision energy was 3.9 V/100 Da with a 2.9 V offset for the NAD+ binding study and 3.7 V/100 Da for the resveratrol binding study.
Multiple LC-MS.MS runs were performed for each set of samples. In the NAD+ binding experiment, the methionine-containing peptide enhanced samples from the SPROX analyses with and without ligand and the non-enhanced samples from the SPROX analyses with and without ligand were each subjected to 3 LC-MS/MS. In the resveratrol binding experiment, the non-enhanced samples from the duplicate SPROX analyses with and without ligand were subjected to a total of 4 and 3 LC-MS/MS runs, respectively; and the enhanced samples from the duplicate SPROX analyses with and without ligand were subject to a total of 15 and 11 LC-MS/MS runs, respectively.
Quantitative Proteomic Data Analysis
Peptides were identified using Agilent’s Spectrum Mill MS Proteomics Workbench software, Rev A03.03.084 SR4. Search parameters included alkylation of cysteines with MMTS as a fixed modification, both the N-terminus and lysine residues labeled with iTRAQ 8-plex reagent as a fixed modification, deamidation of glutamines and asparagines as a variable modification, and oxidation of methionines as a variable modification. The precursor and product ion mass tolerances were set to 20 ppm, the protein cleavage chemistry was set for trypsin with 3 maximum missed cleavages, and the maximum ambiguous precursor charge was set to 7. The peptide fragment products and peptide precursor masses were searched against the NCBInr database for S. cerevisiae. Searches performed on the methionine peptide enhanced samples from the resveratrol binding experiment were searched only against a database of tryptic Met peptides. Searches were also performed against a decoy database, which was the entire S. cerevisiae database of reversed protein sequences, using the same search parameters to determine false discovery rates. Spectrum Mill was also used to extract the iTRAQ intensities of the identified peptides. Only the identified peptides yielding product ion spectra with intense iTRAQ reporter ion intensities (i.e., raw reporter ion intensities that averaged >125 counts/reporter ion) were used in further analyses. The extracted iTRAQ reporter ion intensities were normalized as we have previously described.14 Detailed information about the normalization procedure is also provided in the Supporting Information.
Transition midpoints were assigned by visual inspection of the normalized iTRAQ intensity values and by using a set of rules that assumed the data had a specific structure (i.e., a single unfolding/folding transition with a pre- and post-transition baseline). Prior to visual inspection, the distributions of the normalized iTRAQ reporter ion intensities obtained at the highest and lowest denaturant concentrations in each ligand binding experiment (see Figure S1) were used to determine the normalized reporter ion intensity value that best separated the pre- and post-transition baselines in the SPROX data. This normalized reporter ion intensity was 1.0 in both the NAD+ and resveratrol binding experiments (see Figure S1). The transition midpoint was assigned to be the chemical denaturant concentration at which the normalized iTRAQ intensity values transitioned from the pre- to the post-transition baseline. If there was a normalized iTRAQ reporter ion intensity value of 1.0 ± 0.1 at the transition then the denaturant concentration corresponding to that iTRAQ reporter ion was taken as the midpoint. Otherwise the denaturant concentrations corresponding to the iTRAQ reporter ions flanking the transition were averaged and this average value was assigned as the transition midpoint.
In our visual inspection of the normalized iTRAQ intensity values, unoxidized methionine-containing peptides with SPROX data in which more than one data point was not > 1.0 or <1.0 in the pre- and post-transition baselines, respectively, were classified as uninterpretable and not assigned a transition midpoint. In cases where only a single normalized reporter ion intensity was inconsistent with the expected pre- and post-transition baseline values, the outlying value was removed from the data set and the remaining seven values were used in the visual inspection to assign the transition midpoint.
Quantitation of Ligand Binding
The transition midpoint value shifts obtained by visual inspection of the chemical denaturation data generated in the presence and in the absence of ligand were used to quantify protein ligand binding affinities. This was accomplished by first calculating a binding free energy using equation 1, which has been previously described.16
| Equation 1 |
In equation 1, ΔΔGf is the binding free energy, m is the δΔGf/δ[Denaturant] (where ΔGf is the protein folding free energy), and is the transition midpoint shift observed upon ligand binding. The m-values used in this work were estimated to be 2.6 and 1.4 kcal mol−1 M−1 for GdmCl and urea, respectively. These values were estimated by assuming an average protein domain size of 100 amino acids and an average contribution to the m-value per residue of 0.014 and 0.026 kcal mol−1 M−1 for urea and for GdmCl (respectively). These per residue values were empirically derived by Myers et al. 22
Equation 2 from reference23 was used to calculate the Kd values in this work.
| Equation 2 |
In equation 2, [L] is the molar concentration of the free ligand, n is the number of independent binding sites, R is the universal gas constant, and T is the temperature in Kelvin. In both the NAD+ and the resveratrol binding studies the total ligand concentration (0.4 and 2 mM for resveratrol and NAD+, respectively) was much greater than any individual target protein concentration, therefore the total ligand concentration was used for the free ligand concentration. In all cases, it was also assumed that n = 1.
Results and Discussion
General Strategy
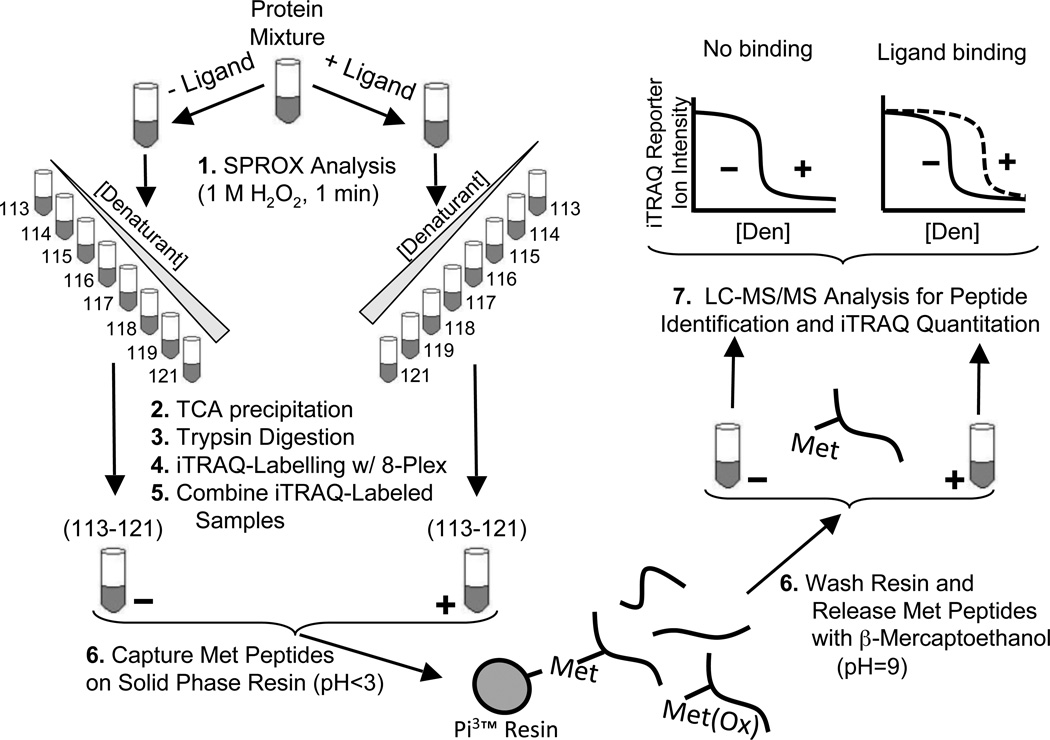
The general strategy used in this work is outlined in Figure 1. In this strategy, a complex biological mixture of proteins (e.g., a cell lysate sample) is subjected to two SPROX analyses,16 one in the presence of ligand and one in the absence of ligand. In each SPROX analysis, the protein sample is reacted with hydrogen peroxide in the presence of increasing concentrations of a chemical denaturant. The H2O2 concentration and reaction time are tuned to selectively oxidize exposed methionine residues in each protein sample. After the oxidation reaction is quenched, the protein in each denaturant-containing buffer is precipitated with TCA and submitted to a quantitative proteomic analysis using isobaric mass tags. As part of the proteomic analysis, the protein samples from each denaturant-containing SPROX buffer are digested with trypsin, and the resulting peptide samples are labeled with an appropriate set of isobaric mass tags (e.g., iTRAQ 8-plex) before the peptide mixtures generated in each SPROX analysis are combined as shown in Figure 1.
Figure 1.
Schematic representation of the protocol used in this work. A protein mixture is incubated in the presence and absence of ligand before diluting into a series of denaturant-containing buffers. Following SPROX analysis, trypsin digestion, and iTRAQ labeling with the eight different iTRAQ reagents bearing the 113 – 121 reporter ions, the samples across the range of denaturants are combined (i.e., the −ligand samples labeled with the eight iTRAQ reagents bearing the 113–121 reporter ions are combined and the +ligand samples labeled with the eight iTRAQ reagents bearing the 113–121 reporter ions are combined). Ultimately, the − and + ligand samples are each subjected to a Met peptide selection strategy. The samples enriched for Met peptides are analyzed by LC-MS/MS, and iTRAQ reporter ions are used to generate chemical denaturation data.
The unoxidized methionine-containing peptides in the combined samples are then isolated using a commercially available resin functionalized with bromoacetyl groups to covalently bind the thioether group of unoxidized methionine residues at low pH. Non-methionine-containing peptides and oxidized methionine-containing peptides are washed away. Ultimately, the methionine-containing peptides are released under reducing conditions at high pH and analyzed by LC-MS/MS. The workflow in Figure 1 is similar to that used in our earlier work,14 except for the inclusion of the methionine-containing peptide enhancement step.
Selection Efficiency and Protein Coverage
The experimental strategy outlined in Figure 1 was used to investigate the binding of two ligands, NAD+ and resveratrol, to the proteins in a yeast cell lysate. During the course of these studies, steps 5 and 6 in Figure 1 (i.e., the methionine enhancement) were performed on a total of 6 different peptide mixtures. These peptide mixtures included the two peptide mixtures generated in the SPROX analyses performed on the yeast cell lysate sample in the presence and in the absence of NAD+ and the four peptide mixtures generated in the duplicate SPROX analyses performed on the yeast cell lysate sample in the presence and absence of resveratrol.
The 6 peptide mixtures generated in this work were each analyzed by LC-MS/MS both before and after they were subjected to the methionine enhancement (i.e., steps 5 and 6 in Figure 1). The frequency of unoxidized methionine-containing peptides identified at the 99% confidence level in the LC-MS/MS analyses of these 6 peptide mixtures averaged 72% in the 6 methionine enhancements that were performed. This was a nearly 5-fold improvement in the average frequency of 15% that was observed for the methionine-containing peptides found in our LC-MS/MS analyses of the same 6 samples prior to enhancement. The methionine-containing peptide selection efficiency observed in our experiments was comparable to the ~70% efficiency previously observed in other experiments using the same selection strategy.24 The presence of non-methionine-containing peptides in the enhanced samples was presumably due to non-specific binding of the peptides to the solid-phase resin, as the covalent binding to the bromoacetyl group is highly selective for unoxidized methionine-containing peptides at pH 2–3.25 The relatively small number of oxidized methionine-containing peptides present after the enhancement was likely oxidized during the methionine enhancement and/or due to nonspecific binding to the resin.
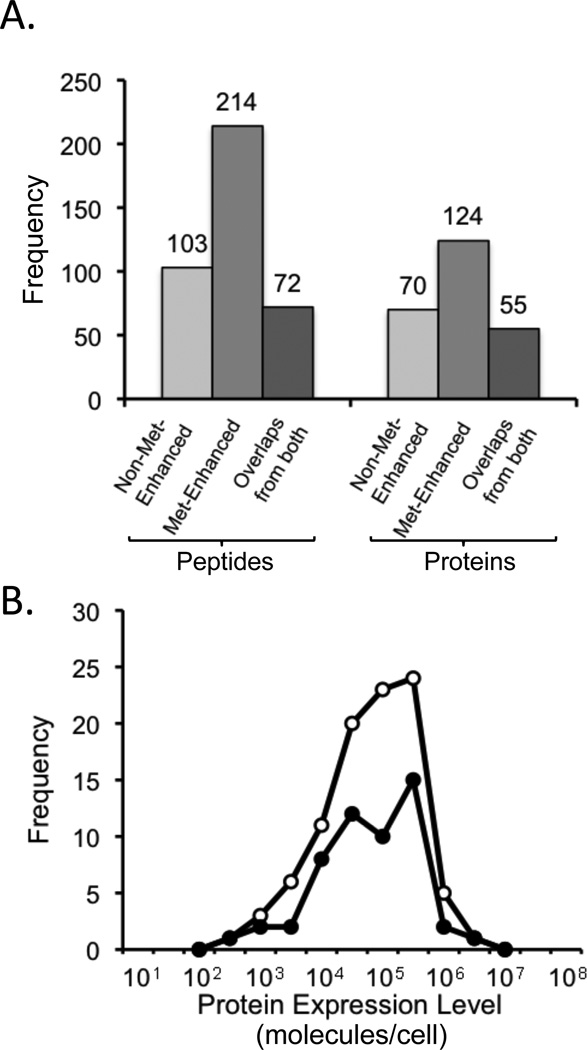
The methionine-enhanced and non-enhanced peptide mixtures generated in the NAD+ binding experiment were subjected to the same number of LC-MS/MS analyses using the same data-dependant acquisition method. Thus, the methionine-containing peptide identifications obtained in our analyses of the methionine-enhanced and nonenhanced peptide mixtures generated in the NAD+ binding experiment could be used to determine the peptide and protein coverage enhancement that was achieved in the SPROX experiment using the methionine-enhancement protocol described here (Figure 2). Our results indicated that 1.5- and 2-fold more protein and peptides, respectively, were observed in the SPROX analyses using the methionine enrichment protocol with a one-dimensional LC-MS/MS analysis than using the one-dimensional LC-MS/MS analysis alone. It is also noteworthy that while the overall peptide and protein coverage was clearly increased in the SPROX experiment using the methionine enhancement protocol, neither lower nor higher abundant proteins were selectively enhanced (see Figure 2).
Figure 2.
Evaluation of the Met-enhancement strategy utilized in this work. (A) Number of methionine-containing peptides and corresponding proteins detected in the NAD+ ligand binding experiment using SPROX with and without the Met-enhancement protocol. The peptide identifications used in this figure were made using a false discovery rate less than 1%. (B) Distribution of the protein expression levels of the proteins identified using SPROX with (open circles) and without (filled circles) the Met-enhancement protocol. The protein expression levels were taken from reference 35.
NAD+ Binding Analysis
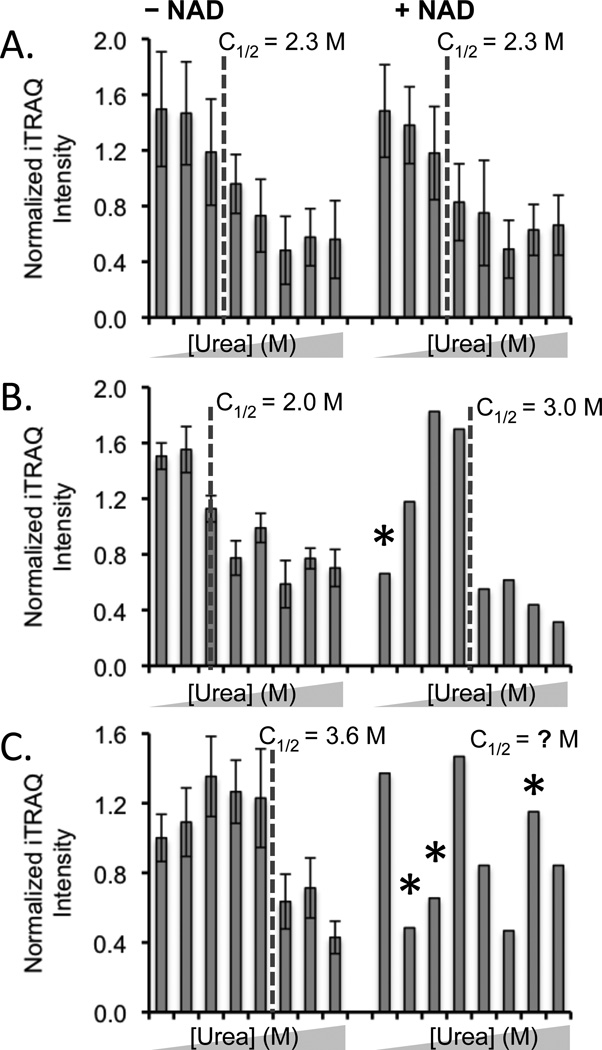
SPROX data (i.e., chemical denaturation data in the presence and absence of ligand) were obtained for 232 different methionine-containing peptides from 122 different proteins that were identified in the LC-MS/MS analyses performed in the NAD+ binding experiment. A total of 297 of the 464 (64%) chemical denaturation data sets generated in the NAD+ binding experiment had clearly defined pre- and post-transition baselines and easily assigned transition midpoints (see Figure 3A and B). The other 167 chemical denaturation data sets generated in the NAD+ binding experiment did not have the expected denaturation data set structure (i.e., a pre- and/or post-transition baseline could not be established according to the rules used in our visual inspection) (see Figure 3C). These poor quality data sets were randomly distributed both in the with- and without-ligand samples and in the samples that were and were not subjected to the methionine-containing peptide isolation. This ultimately meant that only about 40% of the 232 peptides were effectively in the NAD+ binding assay.
Figure 3.
Typical SPROX data obtained in the NAD+ binding experiment. (A) SPROX data obtained on the TVPFVPISGWNGDNMIEATTNAPWYK peptide from the Tef2p protein did not display NAD+ binding. (B) SPROX data obtained on the GDIESISDKTMYK peptide from NAD+-dependent glutamate dehydrogenase, a protein that did display NAD+ binding. (C) SPROX data obtained on the STAISLYSEMSDEDVKEIK peptide from Eft2p, that has an uninterpretable data set in the presence of NAD. The urea concentrations in each data set were (from left to right) 0, 1.0, 2.0, 2.7, 3.3, 4.0, 4.9, and 6.0 M. Error bars representing ± one standard deviation are shown in cases were the data from multiple product ion spectra were averaged. Error bars are not shown in the (+) NAD data in (B) and (C) because the data were obtained from a single product ion scan. An asterisk represents an outlying data point (see Experimental).
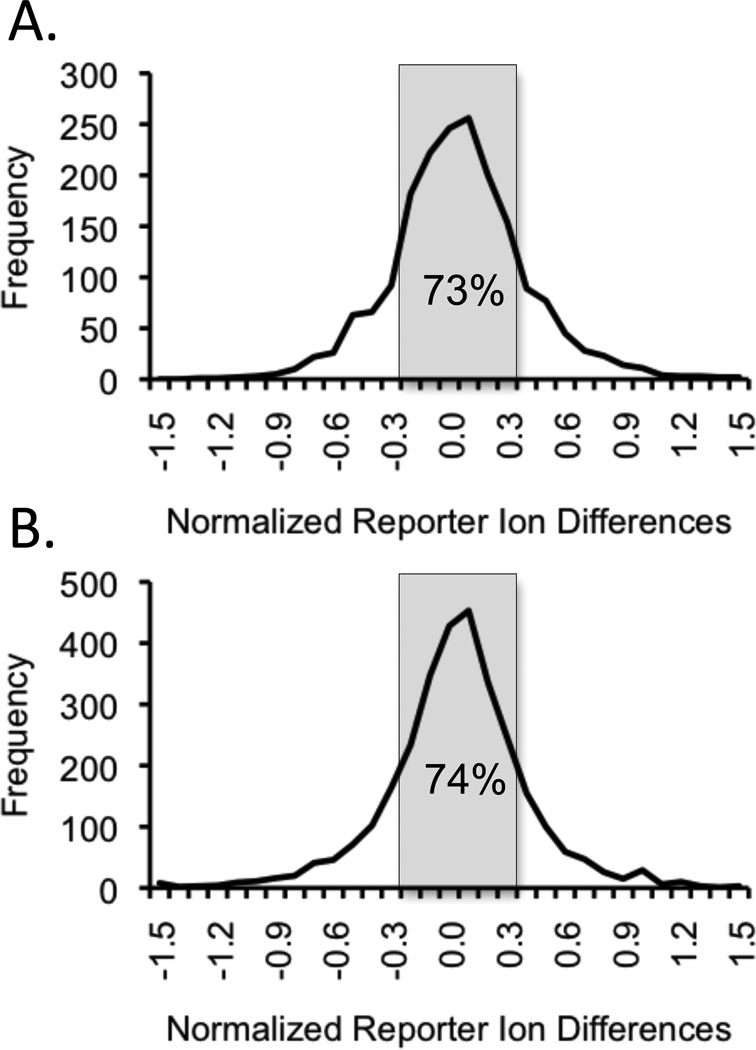
The chemical denaturation data generated for peptides from proteins with no NAD+ binding interactions were expected to have the same transition midpoint in the SPROX analyses performed in the presence and in the absence of NAD+. However, peptides from proteins with NAD+ interactions (either direct or indirect) were expected to have transition midpoints that were shifted to either a higher or lower denaturant concentration in the presence of ligand. Peptides with shifted transition midpoints were identified by examining the differences between the normalized iTRAQ reporter ion intensities observed for a given methionine-containing peptide in the presence and in the absence of ligand. Normalized iTRAQ reporter ion differences greater than 0.3 or less than −0.3 were deemed significant based on the observed distribution of all the reporter ion differences (Figure 4A), which revealed that approximately 73% of the differences from all the reporter ions from all the peptides were within 0.3 of the average difference of approximately 0. We also reasoned that significant transition midpoint shifts (≥ 1 M urea) would require a significant iTRAQ reporter ion intensity difference at two (or more) consecutive denaturant concentrations.
Figure 4.
Distributions of normalized iTRAQ reporter ion intensity difference observed between the SPROX data sets generated in the presence and in the absence of ligand. The distribution of differences obtained at all of the denaturant concentrations for the 232 and 410 peptides identified in the NAD+ and resveratrol binding experiments are shown in (A) and (B), respectively.
The difference analysis described above identified 66 of the 232 peptides with normalized iTRAQ reporter ion intensity value differences of > 0.3 (or < −0.3) in at least two consecutive denaturant concentrations. Visual inspection of the chemical denaturation data generated in the presence and absence of ligand for these 66 peptides revealed 8 peptides from 8 different proteins (see Table 1) with transition midpoints that were shifted ≥1.0 M urea in the presence of NAD+ (Figure 3B) using the “rules” defined above (see Experimental). A visual inspection of the chemical denaturation data generated for all 232 peptides revealed 4 additional peptides with transition midpoint shifts of ≥1.0 M urea in the presence of NAD+ (see Table 1), again using the “rules” defined above (see Experimental). In total 12 peptides from 11 different proteins were identified as NAD+ binding “hits.” All 12 of the hit peptides were identified in the LC-MS/MS analyses with greater than a 95% confidence level.
Table 1.
Peptide and Protein Hits Identified in NAD+ Binding SPROX Analysis.
| Sequence | Protein | Midpoint shift (M urea) |
|---|---|---|
| SVELGVEDIVLGMAHR* | α-ketoglutarate dehydrogenase | 1.0 |
| MPELIPVLSETMWDTK** | translation elongation factor 3A | −1.5 |
| IALSRPNVEVVALNDPFITNDYAAYM(ox)FK** | glyceraldehyde 3-phosphate dehydrogenase | 1.6 |
| GGNIPMIPGWVMDFPTGK* | hexokinase 2 | −1.2 |
| LLIVDDNGNLVSMLSR** | inosine monophosphate dehydrogenase | −1.5 |
| GFTPEEPDGLNNALKDTDMVLIPAGVPR* | mitochondrial malate dehydrogenase | −2.1 |
| GDIESISDKTMYK** | NAD+-dependent glutamate dehydrogenase | 1.0 |
| LIDDMVAQMIK** | NADP+-specific isocitrate dehydrogenase | 1.9 |
| NIPMMPAGEPVLEAIFEVDANGILK** | Ssb1 (member of HSP70 family) | 1.0 |
| TFSPQEISAMVLTK* | Ssb1 (member of HSP70 family) | −1.3 |
| ELQDIANPIMSK** | Ssa1 (member of HSP70 family) | ≥2.4 |
| ANGTTVLVGMPAGAK** | alcohol dehydrogenase | ≥2.4 |
Identified by visual inspection of the chemical denaturation data.
Identified in difference analysis and by visual inspection of the chemical denaturation data.
All 12 hit peptides were ultimately selected as hits according to the transition midpoint shift of the chemical denaturation data collected in the absence and presence of NAD+. The transition midpoint shifts were all determined by visual inspection of the chemical denaturation data using the same set of “rules.” Use of the difference analysis prior to the visual inspection was advantageous because it reduced the number of data sets that needed to be visually inspected. This may become important for the analysis of larger proteomic data sets. However, visual inspection of all the data does appear important for producing the most comprehensive hit list. It is also possible that use of the difference analysis prior to the visual inspection may slightly reduce the false discovery rate (see below).
It is not surprising that most of the protein hits (7 of the 11) were dehydrogenases, as dehydrogenases generally have NAD+ binding domains. Indeed 5 of the 7 dehydrogenase protein hits (including glyceraldehyde 3-phosphate dehydrogenase GAPDH, glutamate dehydrogenase, inosine monophosphate dehydrogenase, α-ketoglutarate dehydrogenase, and malate dehydrogenase) have known binding interactions with NAD+.26 One of the other dehydrogenase hits (isocitrate dehydrogenase) is known to be a NADP+-specific enzyme, and it has been shown to bind NAD+ in other organisms.27–29 To our knowledge, alcohol dehydrogenase is the only yeast dehydrogenase protein hit identified here with a previously measured NAD+ binding affinity.30 The Kd value of 6 µM that can be determined from the SPROX transition midpoint shift observed for alcohol dehydrogenase in this work (see Fig. S2) is in reasonable agreement (i.e., within 10-fold) with the previously determined NAD+ binding affinity of this protein (i.e., a KM value of 54 µM).30 Reported NAD+ binding affinities (e.g., Kd or KM values) for other dehydrogenases from other organisms range from 4–470 µM.30–32
The methionine-containing peptides from inosine monophosphate dehydrogenase and malate dehydrogenase had midpoints that shifted to lower denaturant concentration with added NAD+. These results suggest the domains from which these peptides originated were destabilized upon the addition of NAD+. Both of these proteins are multi-domain proteins that are known to undergo large conformational changes upon NAD+ binding.33, 34 The transition midpoint shifts observed for the peptides detected from these two proteins are likely a result of such conformational changes. Similar conformational changes may have also occurred in the other dehydrogenase hits in Table 1. However, it appears the methionine-containing peptides identified from these other dehydrogenase hits were only those peptides derived from regions of these proteins’ structure that were stabilized upon NAD+ binding.
Two of the hit proteins detected, SSA1 and SSB1, are not dehydrogenases, but have been shown to interact with NAD+, albeit indirectly.35 These proteins are members of the Hsp70 family, which are known to interact with Hsp80. It has been shown that the Hsp70-Hsp80 interaction is strengthened in the presence of NAD+, which is known to interact with Hsp80.35 A methionine-containing peptide from Hsp80 was not identified in our LC-MS/MS analyses, therefore this protein was not included in our assay. However, it was likely present and interacting with SSA1 and SSB1 in the protein mixture. This ability to detect such indirect protein-ligand binding events is an important advantage of the SPROX-based approach.15
Based on information collected from the UniProt protein database,26 a total of 5 additional NAD+-binding enzymes (Hst4, Ald4, Ald6, Yhb1, and Mdh3) appeared in the list of 122 proteins that were successfully identified in the proteomics readout. However, the peptides from these proteins had at least one chemical denaturation data set that was uninterpretable (i.e., a transition midpoint in at least one data set could not be assigned because a pre- and/or post-transition baseline could not be determined). Therefore, these proteins were not effectively in our NAD+ binding assay. The presence of poor quality data sets appears to be due to random error in the experiment (see above). Therefore, it is likely that binding data for these proteins could be obtained if of additional data (i.e., additional product ion spectra) were acquired on the methionine-containing peptides from these proteins.
Resveratrol Binding Assay
Resveratrol is a stilbenoid found in red grapes that has been associated with various health benefits;36, 37 however, the protein targets of resveratrol have remained elusive. As part of this work the experimental strategy described above was used to identify yeast protein targets of resveratrol. In total, the resveratrol binding experiment was performed in duplicate, and proteomic data (i.e., peptide identifications and iTRAQ reporter ion intensities) obtained from the two experiments were combined together to generate a single SPROX data set.
The SPROX data set generated in the resveratrol binding experiment included chemical denaturation data (both in the presence and absence of ligand) for 410 different methionine-containing peptides from 243 different proteins. A total of 645 out of 820 (79%) chemical denaturation data sets generated had clearly defined pre- and post-transition baselines and easily assigned transition midpoints. The other 175 chemical denaturation data sets generated in the resveratrol binding experiment were uninterpretable because they did not have the expected structure of chemical denaturation data (i.e., a pre- and/or post-transition baseline could not be established). The fraction of uninterpretable chemical denaturation data sets in the resveratrol binding experiment, 21%, was smaller than that in the NAD+ binding experiment (36%). This was most likely because of the larger number of LC-MS/MS analyses that were performed in the resveratrol binding experiment. This suggests that some source of random error is most likely responsible for the uninterpretable chemical data sets obtained in our analyses. This random error effectively reduced from 410 to ~250 the number of peptides identified in our proteomics readout that were effectively assayed in our resveratrol binding assay.
A difference analysis, similar to that described above in the NAD+ binding experiment, identified 82 of the 410 peptides with significant normalized iTRAQ reporter ion intensity value differences (Figure 4B) in at least two consecutive denaturant concentrations. Visual inspection of the chemical denaturation data generated in the presence and absence of ligand for these 82 peptides revealed 5 peptides from 5 different proteins (see Table 2 and Supplemental Figure S3) with transition midpoints that were shifted > 0.5 M GdmCl in the presence of resveratrol using the “rules” defined above (see Experimental). A visual inspection of the chemical denaturation data generated for all 410 peptides revealed only two additional peptides with transition midpoint shifts of > 0.5 M GdmCl in the presence of resveratrol (see Table 2 and Supplemental Figure S3), again using the “rules” defined above (see Experimental). Ultimately, 7 peptides from 7 different proteins were identified as resveratrol binding “hits.” The 7 peptide hits were all identified in our LC-MS/MS analyses with greater than a 97% confidence level.
Table 2.
Peptide and Protein Hits Identified in Resveratrol Binding SPROX Analysis.
| Sequence | Protein | Midpoint shift (M GdmCl) |
|---|---|---|
| QQFDTIMNYIDIGK** | cytosolic aldehyde dehydrogenase | 0.9 |
| QIVNIPSFMVR* | 40S ribosomal protein S9-A | 1.1 |
| MGVPYAIVK* | 60S ribosomal protein L8-A | 0.7 |
| MPELIPVLSETMWDTK** | translation elongation factor 3A | 0.7 |
| AHNGDLVNAIMSLSK** | nascent polypeptide-associated complex subunit alpha | 1.0 |
| EQAIIDMAK** | NADPH-dependent aldo-keto reductase | 1.1 |
| IAKLPWTEEEMIQKKSVVEDDAANNSLVK** | 2-dehydropantoate 2-reductase | 0.8 |
Identified by visual inspection of the chemical denaturation data.
Identified in difference analysis and by visual inspection of the chemical denaturation data.
One of the 7 protein hits (cytosolic aldehyde dehydrogenase) identified in the SPROX experiment was shown in a previous study38 to have a direct interaction with resveratrol. The results of catalytic activity studies performed in this earlier work, suggested that resveratrol binds to the aldehyde substrate-binding pocket of aldehyde dehydrogenase.38 It was shown in this earlier work that resveratrol increased the activity of aldehyde dehydrogenase at high concentrations of substrate, but inhibited it at low concentrations of substrate. The magnitude of the midpoint shift in the iTRAQ data can be used to estimate a Kd value of 10 µM for the aldehyde dehydrogenase-resveratrol complex.
Four of the 7 protein hits identified here (including elongation factor 3A (EF-3A), ribosomal proteins S9-A and L8-A, and the nascent polypeptide-associated complex subunit alpha (NACα)) are associated with the protein translation machinery. Previous reports have suggested that resveratrol acts on the protein translation machinery.12, 39 Thus, it is not surprising that the ribosomal proteins and elongation factors were identified as hits in the resveratrol binding experiment. Additionally, many ribosomal proteins were previously identified as targets of resveratrol-mediated effects using a protease digestion assay.12 However, we note that the two ribosomal proteins identified in the SPROX experiment were not previously identified in this earlier experiment. The remaining two protein hits from this study were NADPH-dependent aldo-keto reductase and 2-dehydropantoate 2-reductase. It is unclear what significance, if any, these two potential protein targets of resveratrol possess.
The eukaryotic initiation factor 4A (eIF4A) was previously identified as a potential protein target of resveratrol using a protease digestion assay.12 Four Met peptides from eIF4A were identified and assayed in the SPROX experiment described here (see Fig. S4). However, no significant changes in transition midpoints were observed for these peptides. The results of a SPROX experiment performed on purified eIF4A (see Figure S5) also showed no evidence of a direct binding interaction between eIF4A and resveratrol. However, it should be noted that the experiments on the purified eIF4A involved an eIF4A construct that contained a C-terminal 19-kDa purification tag. It is possible that this purification tag could have precluded the potential binding interaction. It is also possible that eIF4A binds resveratrol with a lower binding affinity that could be detected in our SPROX experiments. If the eIF4A-resveratrol complex had a Kd value of 50 µM or larger, it would go undetected in the SPROX experiment performed here, as such a weak binding affinity would not be expected to produce a measurable transition midpoint shift (i.e., > 0.5 M GdmCl) in the chemical denaturation data generated here. A higher free ligand concentration than that used here (i.e., 400 µM) would be required to see a measurable shift for complexes with Kd values > 50 µM.
Platform Efficiency
The overall efficiency of the described platform is ultimately dependent on how reproducibly transition midpoints can be determined in the chemical denaturation data sets. As part of this work the transition midpoints of the methionine-containing peptides generated in the yeast lysate sample analyzed here in the absence of resveratrol were compared to the transition midpoints of the same methionine-containing peptides generated in a yeast lysate sample analyzed in the absence of cyclosporine A as part of our earlier study.14 The yeast cell lysate samples used in both studies were essentially identical with exception of the specific overexpression of cyclophilin and calcinuerin in the yeast cell lysate sample analyzed in the cyclosporin A binding study. The GdmCl concentrations used to construct the chemical denaturation data in both experiments were nearly identical in the 0.5 – 2.0 M GdmCl range, which is where the large majority of transition midpoints fall. Both experiments also involved the use of similar isobaric mass tagging strategies, even though they employed isobaric mass tags from two different commercial vendors.
A set of 93 methionine-containing peptides with overlapping methionine residues were identified from the 410 and 886 methionine-containing data sets generated as part of the resveratrol binding study in this work and our earlier cyclosporine A study,14 respectively. The data analysis methods described in this work were used to assign transition midpoints for the 93 overlapping methionine peptides using the chemical denaturation data obtained in each study. Only one of the transition midpoints determined for the 93 methionine-containing peptides differed in the two experiments by more than 1 M, and only 4 out of the 93 peptides had transition midpoint differences greater than 0.5 M GdmCl between the two experiments (Table S3). This provides a false discovery rate estimate of ~4% for peptide hits with transition midpoint shifts between 0.5 and 1 M GdmCl (or between ~1 and ~2 M urea, which is a weaker denaturant),22 and suggests that the random error of the experiment is likely to produce ~4 peptide hits with such midpoint shifts for every 100 peptides that appear in the assay with interpretable data in both the plus and minus ligand data sets. The above analysis of the 93 overlapping methionine peptides also suggests that the identification of protein targets with larger midpoint shifts (i.e., > 1 M GdmCl or >2 M urea) is subject to a lower false discovery rate (~1% based on the above analysis in which only one of the 93 overlapping peptides had a transition midpoint difference > 1 M GdmCl).
The NAD+ binding study in this work included approximately 100 methionine-containing peptides with interpretable chemical denaturation data both in the presence and in the absence of ligand. Therefore, a 4% false discovery rate is expected to generate approximately 4 peptides (i.e., false positives) with small transition midpoint shifts (i.e., shifts of 1 to 2 M urea) in the NAD+ binding study. We note that 2 of the 12 peptide hits in our NAD+ binding study were from proteins with previously unknown binding interactions with these target ligands, and both proteins displayed transition midpoints shifts in the 1 and 2 M urea. Given the above false discovery rates expected for proteins with such small transition midpoint shifts (~4%), it is possible that these previously unknown protein targets of NAD+ identified in our experiment may be false positives. If these previously unknown protein targets are assumed to be false positives, a false positive rate of 17% (i.e., 2 erroneous peptide hits out 12 total peptide hits) can be calculated along with a false discovery rate of about ~2% (i.e., 2 erroneous peptide hits out of 100 total peptides assayed). This false discovery rate is close to the 4% that was estimated above.
It is interesting to note that one of the previously unknown protein targets of NAD+, hexokinase, was not identified using the difference analysis prior to the visual inspection. Thus, if the difference analysis and visual inspection strategy is exclusively used to analyze the data then the false discovery and false positive rates are slightly improved from 2 to 1% and from 17 to 13 %, respectively. However, such a strategy appears to have an adverse effect on the false negative rate, as the exclusive use of such a strategy to analyze the NAD+ binding data results in one expected hit, α-ketoglutarate dehydrogenase, appearing as a false negative.
Conclusions
A solid phase-based chemo-selection selection strategy for the enrichment of unoxidized methionine-containing peptides in shotgun proteomics-based SPROX experiments was developed in the context of a protein-ligand binding study designed to identify the proteins in a yeast cell lysate that bind a well-known enzyme co-factor, NAD+. The NAD+ binding study enabled an evaluation of the increased peptide and protein coverage that the chemo-selection strategy affords. Using the selection strategy in the proteomics platform described here, a 1.5- and 2-fold increase in protein and peptide coverage, respectively, was realized. The NAD+ binding experiment also enabled an evaluation of the protocol’s efficiency. If the peptide hits from the two previously unknown NAD+ binding proteins, hexokinase and translation elongation factor 3A, are considered to be false positives, the false positive rate in the NAD+ binding study would be ~17%.
The proteins assayed in this work for NAD+ binding represent a relatively small subset of the proteins in the yeast proteome, and there are clearly more NAD+ binding proteins outside of this subset. Likewise, there may be more protein targets of resveratrol that are outside of the proteins that appeared in the resveratrol binding assay in this work. A limitation of the SPROX-based protein-ligand binding assay used in this work is that for a protein to be included in the assay it must be identified with a methionine-containing peptide in the shotgun proteomics experiment. The assay’s false negative rate would clearly be very high if all the NAD+ binding proteins in the yeast proteome were considered in the calculation. However, we note all 9 of the known NAD+ binding proteins that appeared in our NAD+ binding assay and yielded interpretable chemical denaturation data were successfully detected as hits.
Application of the shotgun proteomics-based SPROX protocol to the detection and quantitation of resveratrol binding to proteins in the yeast proteome identified 7 potential protein targets of resveratrol. These targets included one previously known interaction of resveratrol with cytosolic aldehyde dehydrogenase.38 Considering the protocol’s false discovery rate of 2–4% that was evaluated as part of this work, it is likely that at least several of the previously unknown resveratrol protein targets identified here are false positives (especially those with relatively small transition midpoint shifts < 1.0 M GdmCl). Further studies are needed to determine if the 6 newly identified protein targets are indeed direct and/or indirect targets of resveratrol or are false positives. However, it is interesting to note that four of the newly identified protein targets of resveratrol (EF-3A, NACα, and the 2 ribosomal proteins) are associated with the protein translation machinery, which has been implicated in resveratrol’s mode-of-action.12, 39
Supplementary Material
Acknowledgment
This work was supported in part by a National Institutes of Health Grant GM084174 (to M.C.F.) and in part by a National Science Foundation Grant CHE-0848462 (to M.C.F.). The National Science Foundation Grant, which was made possible with funds from the American Recovery and Reinvestment Act (ARRA), is jointly funded by the Analytical and Surface Chemistry Program in the Chemistry Division at NSF and by the Biomolecular Systems Cluster in the Division of Molecular and Cellular Biosciences at NSF.
Footnotes
Supporting Information
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org. The supporting information for this manuscript includes: Additional text, Figures S1–S5, and Tables S1–S3.
References
- 1.Fields S, Song OK. A novel genetic system to detect protein protein interactions. Nature. 1989;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 3.Bartel PL, Roecklein JA, SenGupta D, Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 1996;12(1):72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- 4.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang MJ, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403(6770):623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 5.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang LY, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CWV, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 6.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 7.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 2001;98(8):4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li SM, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JDJ, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li QR, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu HY, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, van den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303(5657):540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Licitra EJ, Liu JO. A three-hybrid system for detecting small ligand-protein receptor interactions. Proc. Nat. Acad. Sci. U.S.A. 1996;93(23):12817–12821. doi: 10.1073/pnas.93.23.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong SE, Schenone M, Margolin AA, Li XY, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, Tolliday NJ, Koehler AN, Marcaurelle LA, Golub TR, Gould RJ, Schreiber SL, Carr SA. Identifying the Proteins to Which Small Molecule Probes and Drugs Bind in Cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106(12):4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bach S, Knockaert M, Reinhardt J, Lozach O, Schmitt S, Baratte B, Koken M, Coburn SP, Tang L, Jiang T, Liang DC, Galons H, Dierick JF, Pinna LA, Meggio F, Totzke F, Schachtele C, Lerman AS, Carnero A, Wan YQ, Gray N, Meijer L. Roscovitine targets, protein kinases and pyridoxal kinase. J. Biol. Chem. 2005;280(35):31208–31219. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 12.Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang JN, Wu RP, Gomez F, Loo JA, Wohlschlegel JA, Vondriska TM, Pelletier J, Herschman HR, Clardy J, Clarke CF, Huang J. Target identification using drug affinity responsive target stability (DARTS) Proc. Natl. Acad. Sci. U.S.A. 2009;106(51):21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu PF, Kihara D, Park C. Energetics-based discovery of protein-ligand interactions on a proteomic scale. J. Mol. Biol. 408(1):147–162. doi: 10.1016/j.jmb.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West GM, Tucker CL, Xu T, Park SK, Han XM, Yates JR, Fitzgerald MC. Quantitative proteomics approach for identifying protein-drug interactions in complex mixtures using protein stability measurements. Proc. Natl. Acad. Sci. U.S.A. 2010;107(20):9078–9082. doi: 10.1073/pnas.1000148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeArmond PD, West GM, Huang HT, Fitzgerald MC. Stable Isotope Labeling Strategy for Protein-Ligand Binding Analysis in Multi-Component Protein Mixtures. J. Am. Soc. Mass Spectrom. 2011;22(3):418–430. doi: 10.1007/s13361-010-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West GM, Tang L, Fitzgerald MC. Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass spectrometry-based strategy. Anal. Chem. 2008;80(11):4175–4185. doi: 10.1021/ac702610a. [DOI] [PubMed] [Google Scholar]
- 17.Reid GE, Roberts KD, Simpson RJ, O'Hair RAJ. Selective identification and quantitative analysis of methionine containing peptides by charge derivatization and tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2005;16(7):1131–1150. doi: 10.1016/j.jasms.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 18.Gevaert K, Van Damme J, Goethals M, Thomas GR, Hoorelbeke B, Demol H, Martens L, Puype M, Staes A, Vandekerckhove J. Chromatographic isolation of methionine-containing peptides for gel-free proteome analysis: identification of more than 800 Escherichia coli proteins. Mol. Cell. Proteomics. 2002;1(11):896–903. doi: 10.1074/mcp.m200061-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Grunert T, Pock K, Buchacher A, Allmaier G. Selective solid-phase isolation of methionine-containing peptides and subsequent matrix-assisted laser desorption/ionisation mass spectrometric detection of methionine- and of methionine sulfoxide-containing peptides. Rapid Commun. Mass Spectrom. 2003;17(16):1815–1824. doi: 10.1002/rcm.1110. [DOI] [PubMed] [Google Scholar]
- 20.Weinberger SR, Viner RI, Ho P. Tagless extraction-retentate chromatography: a new global protein digestion strategy for monitoring differential protein expression. Electrophoresis. 2002;23(18):3182–3192. doi: 10.1002/1522-2683(200209)23:18<3182::AID-ELPS3182>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Ma L, Fitzgerald MC. A new H/D exchange- and mass spectrometry-based method for thermodynamic analysis of protein-DNA interactions. Chem. Biol. 2003;10(12):1205–1213. doi: 10.1016/j.chembiol.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 22.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4(10):2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schellman JA. Macromolecular Binding. Biopolymers. 1975;14:999–1018. [Google Scholar]
- 24.Shen M, Guo L, Wallace A, Fitzner J, Eisenman J, Jacobson E, Johnson RS. Isolation and isotope labeling of cysteine- and methionine-containing tryptic peptides: application to the study of cell surface proteolysis. Mol. Cell. Proteomics. 2003;2(5):315–324. doi: 10.1074/mcp.M300028-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Shechter Y, Rubinstein M, Patchornik A. Selective covalent binding of methionyl-containing peptides and proteins to water insoluble polymeric reagent and their regeneration. Biochemistry. 1977;16(7):1424–1430. doi: 10.1021/bi00626a029. [DOI] [PubMed] [Google Scholar]
- 26.Apweiler R, Martin MJ, O'Donovan C, Magrane M, Alam-Faruque Y, Antunes R, Barrell D, Bely B, Bingley M, Binns D, Bower L, Browne P, Chan WM, Dimmer E, Eberhardt R, Fazzini F, Fedotov A, Foulger R, Garavelli J, Castro LG, Huntley R, Jacobsen J, Kleen M, Laiho K, Legge D, Lin QA, Liu WD, Luo J, Orchard S, Patient S, Pichler K, Poggioli D, Pontikos N, Pruess M, Rosanoff S, Sawford T, Sehra H, Turner E, Corbett M, Donnelly M, van Rensburg P, Xenarios I, Bougueleret L, Auchincloss A, Argoud-Puy G, Axelsen K, Bairoch A, Baratin D, Blatter MC, Boeckmann B, Bolleman J, Bollondi L, Boutet E, Quintaje SB, Breuza L, Bridge A, deCastro E, Coudert E, Cusin I, Doche M, Dornevil D, Duvaud S, Estreicher A, Famiglietti L, Feuermann M, Gehant S, Ferro S, Gasteiger E, Gateau A, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Hulo N, James J, Jimenez S, Jungo F, Kappler T, Keller G, Lara V, Lemereier P, Lieberherr D, Martin X, Masson P, Moinat M, Morgat A, Paesano S, Pedruzzi I, Pilbout S, Poux S, Pozzato M, Redaschi N, Rivoire C, Roechert B, Schneider M, Sigrist C, Sonesson K, Staehli S, Stanley E, Stutz A, Sundaram S, Tognolli M, Verbregue L, Veuthey AL, Wu CH, Arighi CN, Arminski L, Barker WC, Chen CM, Chen YX, Dubey P, Huang HZ, Mazumder R, McGarvey P, Natale DA, Natarajan TG, Nchoutmboube J, Roberts NV, Suzek BE, Ugochukwu U, Vinayaka CR, Wang QH, Wang YQ, Yeh LS, Zhang JA, Consortium U. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrlich RS, Colman RF. Selectivity in the binding of NAD(P)+ analogues to NAD− and NADP-dependent pig heart isocitrate dehydrogenases. A nuclear magnetic resonance study. Biochemistry. 1992;31(49):12524–12531. doi: 10.1021/bi00164a032. [DOI] [PubMed] [Google Scholar]
- 28.Ehrlich RS, Colman RF. Histidine in the nucleotide-binding site of NADP-linked isocitrate dehydrogenase from pig heart. Eur. J. Biochem. 1978;89(2):575–587. doi: 10.1111/j.1432-1033.1978.tb12562.x. [DOI] [PubMed] [Google Scholar]
- 29.Hurley JH, Dean AM, Koshland DE, Jr, Stroud RM. Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes. Biochemistry. 1991;30(35):8671–8678. doi: 10.1021/bi00099a026. [DOI] [PubMed] [Google Scholar]
- 30.Suhadolnik RJ, Lennon MB, Uematsu T, Monahan JE, Baur R. Role of adenine ring and adenine ribose of nicotinamide adenine dinucleotide in binding and catalysis with alcohol, lactate, and glyceraldehyde-3-phosphate dehydrogenases. J. Biol. Chem. 1977;252(12):4125–4133. [PubMed] [Google Scholar]
- 31.Plapp BV. Conformational changes and catalysis by alcohol dehydrogenase. Arch. Biochem. Biophys. 2010;493(1):3–12. doi: 10.1016/j.abb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalziel K, Egan RR. The binding of oxidized coenzymes by glutamate dehydrogenase and the effects of glutarate and purine nucleotides. Biochem. J. 1972;126(4):975–984. doi: 10.1042/bj1260975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eklund H, Samama J-P, Jones TA. Crystallographic investigations of nicotinamide adenine dinucleotide binding to horse liver alcohol dehydrogenase. Biochemistry. 1984;23(25):5982–5996. doi: 10.1021/bi00320a014. [DOI] [PubMed] [Google Scholar]
- 34.Ovadi J, Osman IR, Batke J. Local conformational changes induced by successive nicotinamide adenine dinucleotide binding to dissociable tetrameric D-glyceraldehyde-3-phosphate dehydrogenase. Quantitative analysis of a two-step dissociation process. Biochemistry. 1982;21(25):6375–6382. doi: 10.1021/bi00268a009. [DOI] [PubMed] [Google Scholar]
- 35.Ouimet PM, Kapoor M. Analysis of complex formation between Hsp80 and Hsp70, cytosolic molecular chaperones of Neurospora crassa, by enzyme-linked immunosorbent assays (ELISA) Biochem. Cell Biol. 1998;76(1):97–106. [PubMed] [Google Scholar]
- 36.Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275(5297):218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 37.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 38.Kitson TM, Kitson KE, Moore SA. Interaction of sheep liver cytosolic aldehyde dehydrogenase with quercetin, resveratrol and diethylstilbestrol. Chem. Biol. Interact. 2001;130–132(1–3):57–69. doi: 10.1016/s0009-2797(00)00222-2. [DOI] [PubMed] [Google Scholar]
- 39.Cao ZX, Fang J, Xia C, Shi XL, Jiang B-H. Trans-3,4,5 '-trihydroxystibene inhibits hypoxia-inducible factor 1 alpha and vascular endothelial growth factor expression in human ovarian cancer cells. Clin. Cancer Res. 2004;10(15):5253–5263. doi: 10.1158/1078-0432.CCR-03-0588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.