Abstract
The gammaretrovirus, xenotropic murine leukemia virus-related virus (XMRV), replicates to high titers in some human cell lines and is able to infect non-human primates. To determine whether APOBEC3 (A3) proteins restrict XMRV infections in a non-human primate model, we sequenced proviral DNA from peripheral blood mononuclear cells of XMRV-infected rhesus macaques. Hypermutation characteristic of A3DE, A3F and A3G activities was observed in the XMRV proviral sequences in vivo. Furthermore, expression of rhesus A3DE, A3F, or A3G in human cells inhibited XMRV infection and caused hypermutation of XMRV DNA. These studies show that some rhesus A3 isoforms are highly effective against XMRV in the blood of a non-human primate model of infection and in cultured human cells.
Keywords: XMRV, APOBEC3, hypermutation, retrovirus, host restriction, innate immunity
Introduction
Viral infections of mammalian cells are subject to inhibition by a diverse group of host restriction factors that target different stages in viral replication cycles(Douville and Hiscott, 2010). Members of the APOBEC3 (A3) family of host restriction factors deaminate deoxycytidines in the minus DNA strand of some retroviruses leading to G to A hypermutation of the coding strand with potent suppression of viral replication(Malim and Emerman, 2008). The xenotropic murine leukemia virus-related virus (XMRV) is a gammaretrovirus originally identified by a virus DNA microarray approach in a study of human prostate cancer tissues obtained following radical prostatectomies (Urisman et al., 2006). XMRV lacks an inhibitor of A3 proteins, such as the HIV-1 Vif protein, and is therefore highly susceptible to inhibition by human (hu) A3G(Bogerd et al., 2011; Groom et al., 2010; Paprotka et al., 2010; Stieler and Fischer, 2010). Prior studies on huA3B, huA3F and murine (mu) A3 produced mixed results, possibly reflecting different experimental conditions such as levels of A3 proteins that were expressed or cell types studied(Bogerd et al., 2011; Groom et al., 2010; Paprotka et al., 2010; Stieler and Fischer, 2010).
Evidence of XMRV infection has been reported in patients with prostate cancer (PCa)(Arnold et al., 2010; Danielson, Ayala, and Kimata, 2010; Schlaberg et al., 2009; Urisman et al., 2006), chronic fatigue syndrome-myalgic encephalomyelitis (CFS-ME)(Lombardi et al., 2009), immunosuppressed with respiratory tract infections(Fischer et al., 2010) and normal healthy controls(Lombardi et al., 2009). However, many studies have failed to detect XMRV in humans raising the issue of laboratory contamination as a source of the infections [reviewed in (Stoye et al., 2010)]. Furthermore, the recombinant origin of XMRV in a cell line derived from human prostate tumor cells implanted in mice has been described(Paprotka et al., 2011), Here we sought to determine the ability of A3 proteins to mutate XMRV DNA in a non-human primate model of infection. In humans, A3G and A3F are widely expressed in hematopoietic cell populations, including T cells, B cells, and myeloid cells and the mRNA level of A3G is higher than that of A3F mRNA(Koning et al., 2009; Refsland et al., 2010). Accordingly, XMRV DNA would be expected to exhibit evidence of A3 editing in its sequence if recovered from these cell populations. We show a high prevalence of G to A mutations in XMRV proviral sequences isolated from PBMC of infected rhesus macaques indicative of A3-mediated mutagenesis. Furthermore, most of the G to A mutations occurred in the context of GA and GC dinucleotides. In contrast, HIV-1 mutations typically occur within GG and GA dinucleotides in human PBMC (Janini et al., 2001; Kieffer et al., 2005). In addition, we have determined that rhesus A3DE, A3G and A3F are active against XMRV in cultured cells. Our results demonstrate that primate A3 isoforms are effective host restriction factors against XMRV in the blood cells of a non-human primate model of infection.
Results
XMRV proviral sequences isolated from infected rhesus macaques contain extensive G→A mutations typical of A3 activity
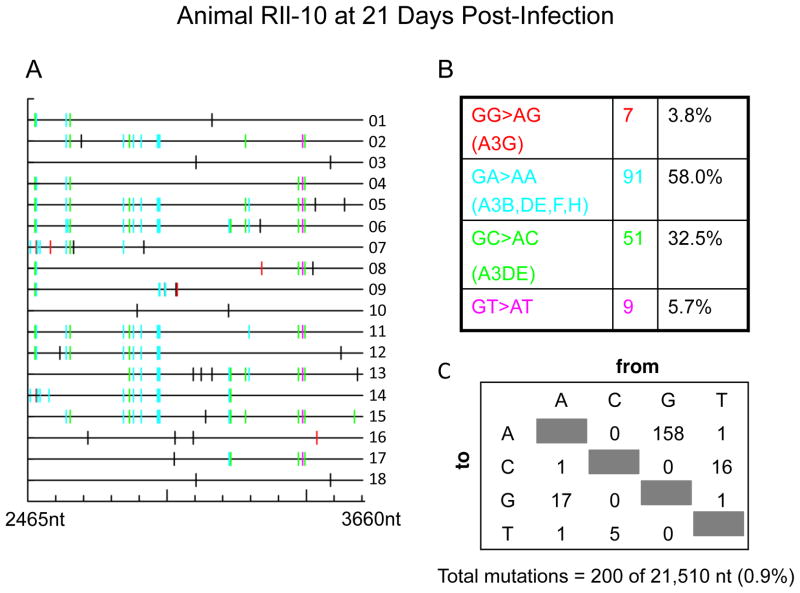
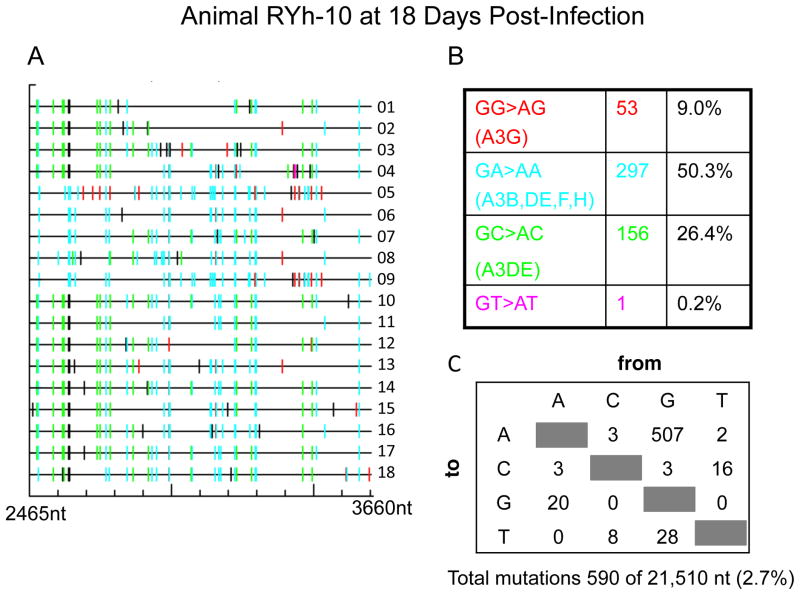
To determine the effect of rhesus A3 on XMRV infections in vivo, PBMC DNA samples from XMRV-infected animals were examined. Three animals were infected with a high dose of virus by the intravenous route (Onlamoon et al., 2011). Virus replication was transient and the two monkeys (RII-10 and RYh-10) that had detectable viremia, as determined by RT-PCR, were used for these studies. Eighteen individual cDNA clones per animal of a 1.2 kb segment from the XMRV pol gene were obtained from PBMC DNA by nested PCR. Single-round PCR was unsuccessful due to low levels of virus replication (Materials and Methods). Most, but not all, of the cDNA clones showed extensive G to A mutations when compared to the pol sequence of XMRV strain VP62 (EF185282), the virus used as the inoculum. In animal RIl-10 there were 158 G to A mutations out of 21,510 nt sequenced in total (Fig. 1), while in animal RYh-10 there were 507 G to A mutations out of 21,510 nt sequenced (Fig. 2). These results are strong evidence of A3 deaminase activity in vivo in PBMC. Surprisingly, in both animals at least half of the A3 editing was in the context of GA→AA typical of A3F activities. The next most frequent context for hypermutation was GC→AC, typical of A3DE, resulting in 32.5% and 26.4% of the mutations in animals RIl-10 and RYh-10, respectively. A3G, which results in GG→AG hypermutation, and the primary mediator of XMRV mutagenesis in human PBMCs culture, caused only 3.8% and 9.0% of the mutations in animals RIl-10 and RYh-10, respectively (Figs. 1 & 2). Examining the sequencing results revealed a combination of different types of dinucleotide context for G→A mutations in individual cDNA clones. The overall rate of mutagenesis was 0.9% for animal RIl-10 and 2.7% for animal RYh-10, respectively. These findings are in contrast to XMRV infected human CEM and H9 T cell lines in which 76.5% and 93% were GG→AG mutations typical of A3G activity(Paprotka et al., 2010). In Mus pahari, XMRV infections caused G→A mutations in gag gene in spleen as a result of muA3 activity, although in an unknown dinucleotide context (Sakuma et al., 2011).
Figure 1. Hypermutation of XMRV DNA in rhesus macaque PBMC characteristic of A3 activity in animal RIl-10.
(A) GG to AC mutations were labeled in red, GA to AA in cyan, GC to AC in green, GT to AT in magenta and all other mutations in black. (B&C) Total numbers and the sequence context of mutations are shown.
Figure 2. Hypermutation of XMRV DNA in rhesus macaque PBMC characteristic of A3 activity in animal RYh-10.
(A) GG to AC mutations were labeled in red, GA to AA in cyan, GC to AC in green, GT to AT in magenta and all other mutations in black. (B&C) Total numbers and the sequence context of mutations are shown.
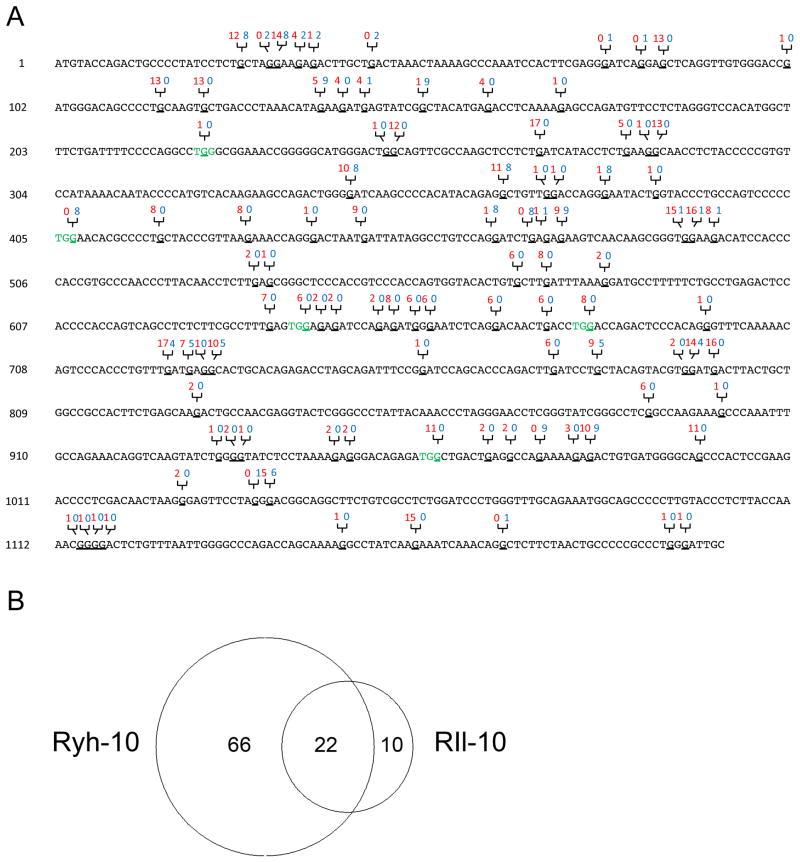
A comparison of XMRV sequences of all 36 clones from both RIl-10 and RYh-10 showed a total of 97 individual G to A mutation sites throughout the 1.2 kb region that were sequenced (Fig. 3A). A Venn diagram showed that mutations at 22 sites were shared in the XMRV sequences from both animals, possibly representing “hotspots” for A3 editing in XMRV genome (Fig. 3B). Hypermutation of XMRV in DU145 cells, the cell line used to produce the viral stock, occurs at a low frequency (Paprotka et al., 2010). However, each macaque also showed characteristic mutation profiles. Sixty-six editing sites were unique to animal RYh-10 while ten editing sites were only identified in animal RIl-10. Importantly, these results provide evidence that the G to A mutations were the result of in vivo mutagenesis and rule out the possibility that these mutations could have existed in the virus stock used to infect the animals.
Figure 3. Locations and numbers of A3-mediated G to A mutations in a 1.2 kb region of XMRV pol sequence.
(A) Nucleotide locations of G to A mutations in the 1.2 kb XMRV pol proviral sequence from both rhesus macaques. The number of clones from RIl-10 (red) and RYh-10 (blue) that contains the specific mutation are labeled above the sequence. Codons mutated by A3 to stop codons are shown in green. Mutated G nucleotides are underlined. (B) Shared and unique numbers of G to A mutations from both rhesus macaques depicted in a Venn diagram generated online at the website http://jura.wi.mit.edu/bioc/tools/venn.php (Whitehead Institute for Biomedical Research, Cambridge, MA).
A possible mechanism responsible for the A3-mediated viral restriction is the accumulation of deleterious mutations in the viral genome, including nonsense mutations. Nucleotides 126–1203 of the 1.2 kb region overlap with the 5′-terminal part of the open reading frame (ORF) of XMRV reverse transcriptase (RT) gene. We were able to identify a total of 5 newly generated stop codons due to G to A transition at different locations in the pol open reading frame. These nonsense mutations, mediated by different isoforms of rhA3 proteins, occurred at different frequencies in the two macaques (Table 1). Nonsense mutation at position 407 was exclusively identified in RYh-10 and thus resulted in production of shortened and non-functional XMRV RT in 8 out of 18 clones. The other 4 nonsense mutations (at positions 223, 641, 680 and 962) were only identified in RIl-10, each at different frequencies. Seven of the clones from RIl-10 contained more than one nonsense mutation. Altogether, only 3 of 18 clones from RIl-10 have the potential to encode full length RT. The other clones encode RT polypeptides truncated at different locations. These data suggest that rhA3 proteins would drastically reduce the production of infectious progeny virus by disrupting genes of key enzymes essential for viral replication.
Table 1.
Stop codons generated by A3 mediated mutations in XMRV pol.
| Nucleotide numbers (relative positions in the 1.2 kb pol segment) | Initial Sequence | Sequence after infection | Presumptive A3 isoform responsible for mutation | Number of clones |
|---|---|---|---|---|
| 223–226 | TGGG | TAGG | A3G | RIl-10: 1/18 RYh-10: 0/18 |
| 407–410 | TGGA | TGAA | A3F | RIl-10: 0/18 RYh-10: 8/18 |
| 641–644 | TGGA | TGAA | A3F | RIl-10: 6/18 RYh-10: 0/18 |
| 680–683 | TGGA | TGAA | A3F | RIl-10: 8/18 RYh-10: 0/18 |
| 962–965 | TGGC | TGAC | A3DE | RIl-10: 11/18 RYh-10: 0/18 |
XMRV is susceptible to rhA3 isoforms in cultured cells
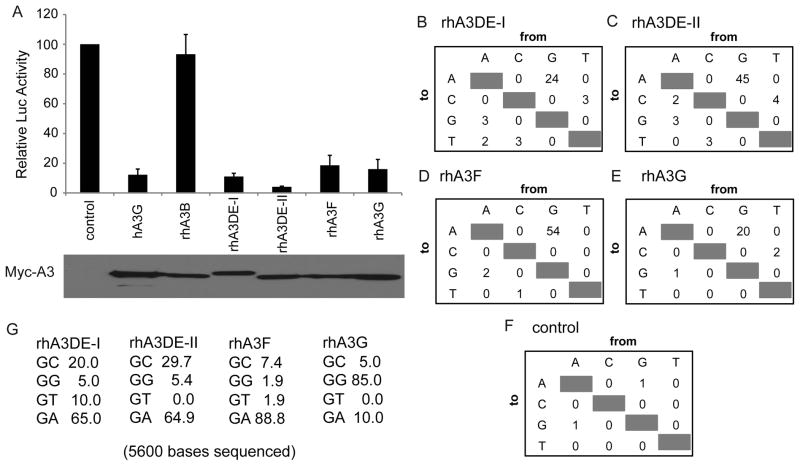
To address whether different rhA3 isoforms are indeed able to effectively inhibit XMRV infectivity, we performed a single-cycle infectivity assay in 293T cells as described previously(Bogerd et al., 2011). In brief, 293T cells were co-transfected with the MLV-based luciferase indicator construct pFB-Luc, together with plasmids encoding the XMRV Env protein and XMRV Gag-Pol and also different members of the human (hA3G) or rhesus (rhA3B, rhA3DE-I, rhA3DE-II, rhA3G and rhA3F)(Virgen and Hatziioannou, 2007) A3 family. The empty parental vector served as a control. When XMRV virions released from transfected producer cells were used to infect naïve 293T target cells, we observed very efficient inhibition of XMRV infectivity by not only rhA3G but also by both isoforms of rhA3DE, rhA3DE-I and rhA3DE-II, as well as rhA3F (Fig. 4A). Interestingly, rhA3B, differed from human A3B in being an ineffective inhibitor of XMRV infection (Bogerd et al., 2011; Groom et al., 2010; Paprotka et al., 2010; Stieler and Fischer, 2010).
Figure 4. The rhesus A3DE, A3G and A3F proteins inhibit XMRV infectivity and edit XMRV reverse transcripts.
(A) 293T cells were transfected with MLV-based indicator plasmid expressing the firefly luciferase pFB-Luc, the XMRV Gag-Pol expression plasmid pCAG/XGP, the XMRV Env expressing plasmid pCAG/XE and the indicated human and rhesus APOBEC3 expression vectors. The empty parental vector was used as a control. Induced luciferase levels in infect naïve 293T cells were assayed ~44 h later (upper panel). Data are given are relative to cells transfected with the empty control plasmid, with standard deviations indicated. The lower panel presents a Western Blot of the 293T virus producer cells using an antibody specific for the myc-tag present on each of the encoded A3 proteins. (B–F) Total numbers and (G) the sequence context of these mutations were determined and are shown for rhA3DE-I, rhA3DE-II, rhA3F, rhA3G, and empty control vector control transfected cells. GG→AA double mutations were not used in the determination of dinucleotide recognition sequences. These numbers were determined by sequencing 14 independent cDNA clones obtained from each infected cell culture, covering an identical 400-bp segment of the vector-encoded firefly luciferase gene.
To prove that rhA3DE-I, rhA3DE-II, rhA3F and rhA3G are indeed able to effectively edit newly formed proviruses during acute XMRV infection in cell culture, we generated XMRV virions containing these rhA3 isoforms with the same co-transfection method in 293T cells and then used the resultant XMRV virions to infect naïve 293T cells. Fourteen independent 400-bp segments of the firefly luciferase gene present in the newly formed proviruses were then recovered from genomic DNA by PCR and subjected to sequencing analysis. As expected, most (88.8%) of the mutations caused by rhA3F were G to A transitions in the GA context (Fig. 4D&G). Similarly, most (65%) of the mutations caused by rhA3DE-I and rhA3DE-II were in the GA context. In addition, however, 20% and 29.7% of the mutations caused by rhA3DE-I and rhA3DE-II, respectively, were in the GC context. In contrast, rhA3G caused 85% of the G to A mutations in the context of GG, as expected (Fig. 4E&G). In the control cells infected by XMRV virions produced in the absence of ectopic A3 proteins, almost no mutations were detected (Fig. 3D). It is worth noting that the human isoforms of A3G and A3B (but not A3F) were active against XMRV(Bogerd et al., 2011), while rhesus A3F and A3G (but not rhA3B) inhibited XMRV (Fig. 4). Our findings support the notion that A3 members have been under strong positive selective pressure during evolution resulting in different antiviral activities in different mammalian species(Zennou and Bieniasz, 2006).
Discussion
Previously we reported that XMRV could be consistently detected in CD4+ and CD8+ T cells and in NK cells from acutely IV infected rhesus macaques(Onlamoon et al., 2011). Results presented here indicate G to A substitutions in XMRV sequences characteristic of rhA3 editing in PBMC of rhesus macaques, which are consistent with previous studies showing that XMRV is highly susceptible to inhibition by certain huA3 isoforms. However, it is also possible that the virus replicated first in other rhesus cell types and then infected PBMC. Our results show that rhA3 can cause extensive mutations throughout XMRV proviral sequences as well as introducing nonsense mutations into the viral RT. The restriction effect of rhA3 might have contributed to the transient viremia that disappeared within one month from initial infection. It is also worth noting that although XMRV sequences from our analysis seem to be predominantly mutated by rhA3F, the two rhesus macaques in our study displayed distinct mutation patterns in terms of the nucleotide positions of G to A transition as well as frequency. This suggested that similar to humans, the expression levels of A3 proteins also vary between individual rhesus macaques(Koning et al., 2009).
Our results suggest that A3 proteins will be potent inhibitors of XMRV replication in PBMC of humans. Indeed, a recent study showed little or no replication of XMRV in activated PBMC from humans in which mutations at GG dinucleotides characteristic of A3G activity occurred(Chaipan et al., 2011). It is unlikely that a virus that it is potently inhibited by A3 could circulate in the human population. One remote possibility is that XMRV might escape hypermutation by infecting cell types that are deficient in A3 proteins, such as prostate cancer cells(Paprotka et al., 2010). Prostate provides a potential environment for XMRV replication because its androgen rich environment could drive viral transcription through an androgen response element in the LTR U3 region(Dong and Silverman, 2010; Rodriguez and Goff, 2010) and prostate cancer cells are often deficient in A3G (Paprotka et al., 2010). Moreover, during acute infection, the most prominent XMRV replication observed in rhesus macaques was in the prostate (Onlamoon et al., 2011; Sharma, 2011). Interestingly, viremia was reactivated in monkey RIl-10–16 days after immunization with a cocktail of recombinant XMRV proteins and incomplete Freund’s Adjuvant, suggesting potential reservoir sites protected from the antiviral effect of APOBEC proteins (Onlamoon et al., 2011).
Only less than 10% of the G to A mutations from our sequence analysis occurred in the GG dinucleotides context, mediated by rhA3G. However, rhA3G exhibited potent (comparable to that of rhA3F) inhibitory effect on XMRV replication in a single cycle infectivity assay in 293T cells and was also capable of causing its signature mutations on XMRV proviral DNA in the same assay. One possible explanation is that rhA3G might possess in vivo inhibitory mechanisms distinct from simple editing. It has already been shown hA3G is able to inhibit HIV-1 cDNA accumulation in the absence of hypermutation possibly by binding directly to viral RNA and impeding the translocation of reverse transcriptase along its template(Bishop et al., 2008). Whether rhA3G shares similar properties with its human ortholog in restricting XMRV replication will be an interesting topic for future investigation.
In summary, in PBMC from XMRV infected rhesus macaques we identified extensive G to A mutations in the context of GA and to a lesser extent, at GC and GG dinucleotides. In addition, rhA3DE, rhA3F and rhA3G strongly inhibited XMRV replication in single cycle infectivity assay in 293T cells. These data support a role for rhA3 proteins in controlling XMRV spread in experimentally infected rhesus macaques and suggest that huA3 proteins would have a similar effect of restricting XMRV infection in humans.
Materials and Methods
XMRV inoculation of rhesus macaques
Methods of rhesus macaque inoculation with XMRV were described previously(Onlamoon et al., 2011). In brief, two adult healthy rhesus macaques (India origin) were housed at Yerkes National Primate Research Center at Emory University and maintained in accordance with the instructions of the Committee on the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council, and the U.S. Public Health Service (PHS) Guidelines, Guide for the Care and Use of Laboratory Animals as described previously(Onlamoon et al., 2011). XMRV was grown in DU145 prostate cancer cells. The monkeys were inoculated with 3.6×106 TCID50 by the IV route. Blood was collected by venipuncture at days 21 (male RIl-10) and 18 (female RYh-10) post-infection. Peripheral blood mononuclear cells (PBMCs) were purified through ficoll density centrifugation.
PCR Amplification of XMRV proviral DNA and sequence analysis
DNA from total PBMC was extracted using QIAamp Blood DNA Mini Kit following manufacturer’s protocol (Qiagen). Nested PCR was used to amplify a 1.2 kb segment of the XMRV pol gene (2454–3656 nt) from genomic DNA of PBMC with the following primers: 5′-ACCACGGATCGCAAAGTACA-3′ (first round, forward); 5′-TTGCGTTAGGACGCCTTTGG-3′ (first round, reverse); 5′-TCACCCACTCTTTCCTCCATGTACC-3′ (second round, forward) and 5′-GAGTTCAAAGGGCTTAGTCAAATCTGG-3′ (second round, reverse). Reactions in a total volume of 50 μl contained 100 to 250 ng genomic DNA, 2 μl of 25 mM MgCl2, 25 μl of HotStart-IT FideliTaq Master Mix (USB Corporation, Cleveland, OH) and 0.75 μl of each of 20 μM forward and reverses primers. PCR conditions were 94°C for 4 min initial denaturation followed by 45 cycles consisting of 94°C for 30 s, 57°C for 30 s and 72°C for 1.5 min with a final extension of 5 min at 72°C. Five μl of first round PCR products were used as template in the second round PCR with the same reaction conditions. Nested PCR without template were performed along with testing samples as a negative control to exclude possible contamination. PCR products were purified and cloned in pCR 2.1 TOP vector (Invitrogen, CA). Positive clones were selected by colony PCR using the same second round oligos. Eighteen positive clones were selected from each monkey and sequenced followed by analysis using Hypermut program(Rose and Korber, 2000).
Assay for the effect of A3 proteins on XMRV infectivity
The assay to measure the effect of different human (hu) and rhesus (rh) A3 proteins on XMRV infectivity were performed as previously described(Bogerd et al., 2011). HuA3G, huA3F, rhA3DE-I, rhA3DE-II, rhA3G and rhA3F(Virgen and Hatziioannou, 2007) were cloned into pCR1-3-based expression plasmids that expresses full-length A3 protein linked to an amino-terminal Myc tag. XMRV virions containing empty vector and each A3 protein were generated by co-transfection of 293T cells of different A3 expression plasmids (hA3G, rhA3B, rhA3DE-I, rhA3DE-II, rhA3G and rhA3F) with XMRV Env (pCAG/XE), XMRV gag-pol (pCAG/XGP) expression clone and MLV based luciferase containing vector pFB-Luc (Stratagene). Conditioned culture medium were harvested in 48 h, filtered and used to infect naïve 293T cells. Luciferase activity was measured about 44 h post-infection. Expression of A3 proteins was monitored by Western blot analysis of XMRV producer cell lysates using a rabbit anti-Myc polyclonal antiserum (Abcam), followed by visualization using enhanced chemiluminescence.
Analysis of XMRV editing by rhesus A3 proteins in cell culture
To analyze editing of XMRV transcripts in by rhesus macaque A3 proteins in culture, total genomic DNA was harvested 48 h after infection of 293T cells with XMRV virions containing rhA3DE-I, rhA3DE-II, rhA3G or rhA3F. A total of 14 independent 400 bp segments of the firefly luciferase gene were then recovered from genomic DNA by PCR and subjected to sequence analysis as described above.
Acknowledgments
These studies were supported by Mal and Lea Bank (to R. H. S.), the Charlotte Geyer Foundation(to R. H. S.), Abbott Laboratories (to R. H. S. and F. V.), the Milton and Tamar Maltz Family Foundation (to E. A. K.), NCRR support to the Yerkes NPRC DRR000165 (to F. V.) and by NIH grant AI065301 to B.R.C. The authors thank Theodora Hatziioannou for the gift of rhesus macaque A3B, A3DE-I, A3DE-II, A3F and A3G expression plasmids. The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pcDNA3.1 Human APOBEC3G-Myc-6Xhis from Dr. David Kabat.
Footnotes
Conflict of Interest Statement
R.H.S., J.D.G., and E.A.K. are inventors on patents licensed to Abbott Laboratories.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnold RS, Makarova NV, Osunkoya AO, Suppiah S, Scott TA, Johnson NA, Bhosle SM, Liotta D, Hunter E, Marshall FF, Ly H, Molinaro RJ, Blackwell JL, Petros JA. XMRV infection in patients with prostate cancer: novel serologic assay and correlation with PCR and FISH. Urology. 2010;75(4):755–61. doi: 10.1016/j.urology.2010.01.038. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4(12):e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Zhang F, Bieniasz PD, Cullen BR. Human APOBEC3 proteins can inhibit xenotropic murine leukemia virus-related virus infectivity. Virology. 2011;410(1):234–9. doi: 10.1016/j.virol.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaipan C, Dilley KA, Paprotka T, Delviks-Frankenberry KA, Venkatachari NJ, Hu WS, Pathak VK. Severe Restriction of Xenotropic Murine Leukemia Virus-Related Virus Replication and Spread in Cultured Human Peripheral Blood Mononuclear Cells. J Virol. 2011 doi: 10.1128/JVI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson BP, Ayala GE, Kimata JT. Detection of xenotropic murine leukemia virus-related virus in normal and tumor tissue of patients from the southern United States with prostate cancer is dependent on specific polymerase chain reaction conditions. J Infect Dis. 2010;202(10):1470–7. doi: 10.1086/656146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Silverman RH. Androgen stimulates transcription and replication of xenotropic murine leukemia virus-related virus. J Virol. 2010;84(3):1648–51. doi: 10.1128/JVI.01763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville RN, Hiscott J. The interface between the innate interferon response and expression of host retroviral restriction factors. Cytokine. 2010;52(1–2):108–15. doi: 10.1016/j.cyto.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Fischer N, Schulz C, Stieler K, Hohn O, Lange C, Drosten C, Aepfelbacher M. Xenotropic murine leukemia virus-related gammaretrovirus in respiratory tract. Emerg Infect Dis. 2010;16(6):1000–2. doi: 10.3201/eid1606.100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom HC, Yap MW, Galao RP, Neil SJ, Bishop KN. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc Natl Acad Sci U S A. 2010;107(11):5166–71. doi: 10.1073/pnas.0913650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janini M, Rogers M, Birx DR, McCutchan FE. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J Virol. 2001;75(17):7973–86. doi: 10.1128/JVI.75.17.7973-7986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol. 2005;79(3):1975–80. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83(18):9474–85. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, Dean M, Silverman RH, Mikovits JA. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326(5952):585–9. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3(6):388–98. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Onlamoon N, Das Gupta J, Sharma P, Rogers K, Suppiah S, Rhea J, Molinaro RJ, Gaughan C, Dong B, Klein EA, Qiu X, Devare S, Schochetman G, Hackett J, Jr, Silverman RH, Villinger F. Infection, Viral Dissemination, and Antibody Responses of Rhesus Macaques Exposed to the Human Gammaretrovirus XMRV. J Virol. 2011;85(9):4547–57. doi: 10.1128/JVI.02411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paprotka T, Delviks-Frankenberry KA, Cingoz O, Martinez A, Kung HJ, Tepper CG, Hu WS, Fivash MJ, Jr, Coffin JM, Pathak VK. Recombinant Origin of the Retrovirus XMRV. Science. 2011 doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paprotka T, Venkatachari NJ, Chaipan C, Burdick R, Delviks-Frankenberry KA, Hu WS, Pathak VK. Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs. J Virol. 2010;84(11):5719–29. doi: 10.1128/JVI.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38(13):4274–84. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JJ, Goff SP. Xenotropic murine leukemia virus-related virus establishes an efficient spreading infection and exhibits enhanced transcriptional activity in prostate carcinoma cells. J Virol. 2010;84(5):2556–62. doi: 10.1128/JVI.01969-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G --> A hypermutation. Bioinformatics. 2000;16(4):400–1. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- Sakuma T, Tonne JM, Squillace KA, Ohmine S, Thatava T, Peng KW, Barry MA, Ikeda Y. Early events in retrovirus XMRV infection of the wild-derived mouse Mus pahari. J Virol. 2011;85(3):1205–13. doi: 10.1128/JVI.00886-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaberg R, Choe DJ, Brown KR, Thaker HM, Singh IR. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci U S A. 2009;106(38):16351–6. doi: 10.1073/pnas.0906922106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sharma P, Rogers KA, Suppiah S, Molinaro RJ, Onlamoon N, Hackett J, Schochetman G, Klein EA, Silverman RH, Villinger F. Sexual transmission of XMRV: a potential infection route. Advances in Virology. 2011 doi: 10.1155/2011/965689. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieler K, Fischer N. Apobec 3G efficiently reduces infectivity of the human exogenous gammaretrovirus XMRV. PLoS ONE. 2010;5(7):e11738. doi: 10.1371/journal.pone.0011738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye JP, Silverman RH, Boucher CA, Le Grice SF. The Xenotropic Murine Leukemia Virus-Related Retrovirus Debate Continues at First International Workshop. Retrovirology. 2010;7(1):113. doi: 10.1186/1742-4690-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, Silverman RH, Derisi JL. Identification of a Novel Gammaretrovirus in Prostate Tumors of Patients Homozygous for R462Q RNASEL Variant. PLoS Pathog. 2006;2(3):e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Virgen CA, Hatziioannou T. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J Virol. 2007;81(24):13932–7. doi: 10.1128/JVI.01760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Bieniasz PD. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349(1):31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]