Abstract
Mutation of the BRCA1 tumor suppressor gene predisposes women to hereditary breast and ovarian cancers. BRCA1 forms a heterodimer with BARD1. The BRCA1/BARD1 heterodimer has ubiquitin ligase activity, considered to play crucial roles in tumor suppression and DNA damage response. Nevertheless, relevant BRCA1 substrates are poorly defined. We have developed a new approach to systematically identify the substrates of ubiquitin ligases by identifying proteins that display enhanced incorporation of His-tagged ubiquitin upon ligase co-expression; using this method, we identified several candidate substrates for BRCA1. These include scaffold attachment factor B2 (SAFB2), Tel2, as well as BARD1. BRCA1 was found to enhance SAFB protein expression and induce Tel2 nuclear translocation. Identification of the ubiquitination substrates has been a major obstacle to understanding the functions of ubiquitin ligases. The quantitative proteomics approach we devised for the identification of BRCA1 substrates will facilitate the identification of ubiquitin ligase-substrate pairs.
Keywords: BRCA1, BARD1, ubiquitination, substrate, quantitative proteomics
INTRODUCTION
Covalent modification of cellular proteins by ubiquitin targets them for degradation or otherwise alters their function. Addition of lysine-48-linked poly-ubiquitin chains targets proteins for degradation by the 26S proteasome whereas addition of other types of poly-ubiquitin chains such as lysine-6-linked chains can exert functions other than protein degradation. The ubiquitin system plays important roles in a wide variety of biological phenomena including tumorigenesis (for reviews see 1,2). A number of oncogene and anti-oncogene products were found to be targets of ubiquitination, leading to the idea that malfunction of protein ubiquitination could modulate the tumorigenic process. The products of several oncogenes and anti-oncogenes display ubiquitin ligase activities, and it can be expected that identification of their substrates would greatly advance our understanding of the mechanism of tumorigenesis.
Women with hereditary BRCA1 mutations have very high risk of breast and ovarian cancers (65% and 40%, respectively). In addition, 30-40% of sporadic breast cancers display reduced or absent BRCA1 expression. The BRCA1 protein has been implicated in a diverse array of biological processes such as the DNA damage response, cell cycle checkpoint, transcription, chromatin remodeling, X chromosome inactivation, and centrosome duplication (for reviews see 3-5). While these processes are essential to all cell types, BRCA1 mutation results in tumors of very restricted tissues. The basis for this tissue-specific tumor suppressor activity is not well understood.
BRCA1 forms a stable heterodimer with BARD1, and this complex displays ubiquitin ligase activity. Many tumor-associated missense mutations target the RING finger domain of BRCA1 which is essential for the ubiquitin ligase activity, suggesting that this activity plays an important role in tumor suppression. In addition, these RING finger domain mutants are also defective for restoring DNA damage checkpoint function in BRCA1-deficient cells, suggesting that the ubiquitin ligase activity of BRCA1 is also critical for the DNA damage response 6. Data mainly from in vitro reactions suggested that BRCA1/BARD1 can ubiquitinate several different proteins, including BRCA1/BARD1 7,8, histones 6,7,9, FANCD2 10, p53 11, RNA polymerase II 12, nucleophosmin 13, and CtIP 14. However, it is not yet established whether these proteins are valid targets in vivo or what kind of BRCA1 functions are mediated by ubiquitination of these proteins.
Interestingly, unlike most known ubiquitin ligases that catalyze the formation of lysine-48-linked poly-ubiquitin chains, BRCA1/BARD1 catalyzes the formation of lysine-6-linked chains, which do not result in protein degradation 8,15. In addition, it was shown that proteins with lysine-6-linked poly-ubiquitin accumulate at sites of DNA damage in a BRCA1-dependent manner, suggesting an important role of this unconventional poly-ubiquitin chains in DNA damage response 16.
A key to the understanding of BRCA1 function is to pinpoint its substrates and to elucidate the role of their ubiquitination. However, the identification of the substrates of a ubiquitin ligase is generally a difficult task because of the lack of an established method. Using quantitative proteomics, we devised an approach to systematically identify the substrates of a ubiquitin ligase through detection of proteins displaying enhanced incorporation of His-tagged ubiquitin upon ligase expression. We applied this method to BRCA1/BARD1 and identified several candidate substrates, including scaffold attachment factor B2 (SAFB2) and Tel2 as well as auto-ubiquitination of BARD1. BRCA1 ubiquitinated SAFB and increased its protein expression whereas Tel2 nuclear translocation was induced by BRCA1. The method reported here is applicable to other ubiquitin and ubiquitin-like protein ligases.
EXPERIMENTAL PROCEDURES
Cell culture
Mouse embryonic fibroblasts from BRCA1-exon11-null and wild-type mice were obtained from Dr. Chu-Xia Deng at the NIDDK/NIH 17. Mouse embryonic fibroblasts and U2OS cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. 293T cells were cultured in DMEM supplemented with 10% calf serum. 786-O cells were cultured in RPMI1640 medium supplemented with 10% fetal calf serum. MCF-7 cells were cultured in minimum essential medium supplemented with 10% fetal calf serum and non-essential amino acids. Calcium phosphate co-precipitation was used for plasmid DNA transfection. Doxorubicin was purchased from Sigma-Aldrich. Human BRCA1 siRNA (P-002111-01-05) and control siRNA (D-001210-02) were purchased from Dharmacon and were transfected using Lipofectamine 2000 reagent (Invitrogen). The target sequences for shRNAs are as follows: human SAFB (targeting both SAFB1 and SAFB2), GCGGAGGAAGAGGACCTATTT; luciferase, GCACTCTGATTGACAAATACGATTT. BRCA1 and empty vector adenoviruses were generated using AdEasy XL adenoviral vector system (Stratagene).
Protein sample preparation, ICAT (isotope-coded affinity tag) reagent labeling, and mass spectrometry
293T cells (ten 15 cm plates each) were transfected with His-tagged ubiquitin K48R mutant together with BRCA1 and BARD1 or with empty vector. Forty-eight hours after transfection, the cells were lysed in a buffer containing 20 mM Tris, pH 8.0/6 M guanidine HCl/0.5 M NaCl/5 mM imidazole. Proteins conjugated with His-ubiquitin K48R were isolated by nickel-NTA agarose (Qiagen) under denaturing conditions followed by elution with a buffer containing 20 mM Tris, pH 8.0/0.5 M NaCl/1 M imidazole. The samples were concentrated using a Microcon YM-3 column (Millipore) and eluted in 50 mM Tris, pH 8.5. These procedures yielded 355 g and 160 g proteins for BRCA1/BARD1 and empty vector sample, respectively, which were used for labeling with the cleavable ICAT reagent (purchased from Applied Biosystems; BRCA1/BARD1 sample: isotopically light ICAT; empty vector sample: isotopically heavy ICAT). The two labeled samples were combined, digested with trypsin (Promega), and fractionated by off-line cation-exchange chromatography using ICAT Cartridge-Cation Exchange (Applied Biosystems). ICAT reagent-labeled peptides were purified using an avidin affinity column (ICAT Cartridge-Avidin, Applied Biosystems) to capture the biotin tag present in the reagent. The biotin tag was cleaved from the ICAT-labeled peptides by treatment with trifluoroacetic acid.
Released peptides were analyzed by capillary high performance liquid chromatography-tandem mass spectrometry (HPLC-ESI-MS/MS), using a Thermo Fisher LTQ linear ion trap mass spectrometer fitted with a New Objective PicoView 550 nanospray interface. On-line HPLC separation of the digests was accomplished with an Eksigent NanoLC micro HPLC: column, PicoFrit™ (New Objective; 75 m i.d.) packed to 10 cm with C18 adsorbent (Vydac; 218MSB5, 5 m, 300 Å); mobile phase A, 0.5% acetic acid (HAc)/0.005% TFA; mobile phase B, 90% acetonitrile/0.5% HAc/0.005% TFA; gradient 2 to 42% B in 1 hr; flow rate, 0.4 l/min. MS conditions were: ESI voltage, 2.9 kV; isolation window for MS/MS, 3; relative collision energy, 35%; scan strategy, survey scan followed by acquisition of data dependent collision-induced dissociation (CID) spectra of the seven most intense ions in the survey scan above a set threshold.
Mass spectrometry data analysis
Raw data were converted to mzXML format using ReAdW and were searched against the human the IPI human protein database (v 3.24; 66,923 protein entries) using SEQUEST Cluster 3.1 SR1. Variable modifications considered in the searches included methionine oxidation and addition of light (+227) and heavy (+236) ICAT tags to cysteine. The peptide mass tolerance was set as 3.0 Da. The SEQUEST search result was analyzed by the Trans-Proteomic Pipeline [for a review see 18] version 3.0. Peptide/protein identifications were validated by Peptide/ProteinProphet 19,20. A ProteinProphet score of 0.7 was used as a cutoff, which corresponded to false identification rate of 2.1 %. Protein abundance ratios were calculated using ASAPRatio 21. Sample-dependent normalization was not employed because of the significant difference in the quantity of protein between the BRCA1/BARD1 and empty vector samples.
Immunoprecipitation and immunoblotting
Immunoprecipitation was performed as described 22. The indicated amounts of cell lysates were separated by SDS-PAGE and analyzed by immunoblotting as described 23. The following antibodies were used: mouse monoclonal anti-HA (16B12. Covance); mouse monoclonal anti-SAFB (6F7, Millipore); rabbit polyclonal anti-BRCA1 (C-20, Santa Cruz Biotechnology); rabbit polyclonal anti-BARD1 (59N, gift from Dr. Richard Baer, Columbia University); mouse monoclonal anti-FLAG (M2, Sigma-Aldrich); mouse monoclonal anti-tubulin (DM1A, Sigma-Aldrich): and rabbit polyclonal anti- Myb-binding protein p160 (40-1200, Zymed/Invitrogen).
RESULTS
Quantitative proteomic identification of ubiquitination substrates for BRCA1/BARD1
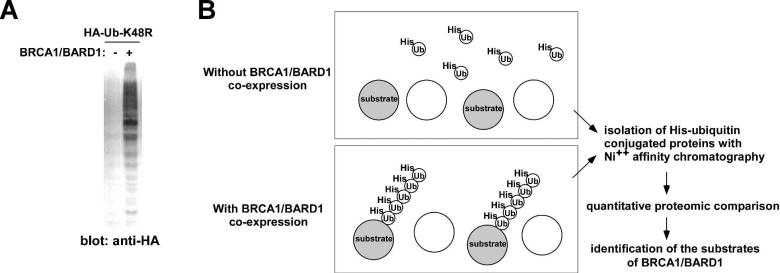
BRCA1/BARD1 catalyzes the formation of lysine-6-linked, but not lysine-48-linked polyubiquitin chains. As shown in Figure 1A, expression of BRCA1/BARD1 dramatically enhanced the conjugation of K48R mutant ubiquitin to cellular proteins. To identify the proteins ubiquitinated by BRCA1/BARD1, we compared the His-tagged ubiquitin-K48R-conjugated proteins in the presence or absence of co-expressed BRCA1/BARD1. We expressed Hisubiquitin-K48R in human embryonic kidney 293T cells, with and without co-expressed BRCA1/BARD1, purified the His-ubiquitin-K48R conjugated proteins using Ni++ affinity column under denaturing conditions (6M Guanidine-HCl), and compared the His-ubiquitin-K48R-conjugated proteins by using quantitative proteomics (Figure 1B). It was anticipated that the substrates of BRCA1/BARD1 would be more efficiently ubiquitinated in the presence of co-expressed BRCA1/BARD1. Using this approach, we detected enhanced ubiquitination of a number of proteins by BRCA1/BARD1 as well as auto-ubiquitination of BARD1 (Table 1).
Figure 1. Quantitative proteomic strategy for the identification of BRCA1/BARD1 ubiquitination substrates.
(A) The BRCA1/BARD1 heterodimer enhances the conjugation of HA-ubiquitin-K48R to cellular proteins. 293T cells were transfected with HA-ubiquitin-K48R with or without BRCA1/BARD1. Forty-eight hours after transfection, the incorporation of HA-ubiquitin-K48R to cellular proteins was examined by anti-HA immunoblotting using 30 μg whole cell lysate.
(B) Outline of the proteomic identification of the BRCA1/BARD1 ubiquitination substrates. Co-expression of BRCA1/BARD1 results in enhanced incorporation of His-ubiquitin-K48R to substrate proteins. His-ubiquitin-K48R conjugated proteins can be purified by nickel affinity chromatography under stringent denaturing conditions. BRCA1/BARD1 substrates can be identified as proteins displaying enhanced conjugation with His-ubiquitin-K48R upon BRCA1/BARD1 co-expression.
Table 1.
Proteins displaying increased ubiquitination upon BRCA1/BARD1 co-expression
| Relative abundance +BRCA1/BARD1 : -BRCA1/BARD1a | |
|---|---|
| BARD1 | 19.0 |
| SAFB2 | 14.4 |
| Splicing factor 3a subunit 2 | 3.2 |
| Tel2 | 3.2 |
| Hox-c4 | 2.9 |
| Stromal cell-derived factor 2-like protein 1 | 2.7 |
| PAI1 RNA- binding protein 1 | 2.5 |
| FMRP interacting protein | 2.4 |
| DKFZP434K1421 protein | 2.3 |
| Eukaryotic initiation factor 5a | 2.3 |
| Ataxin-2 | 2.2 |
| Histidine triad nucleotide-binding protein 1 | 2.2 |
| NADH-ubiquinone oxidoreductase | 2.1 |
| Chd7 | 2.0 |
Relative abundance of each protein in the His-ubiquitin-K48R-conjugated protein fraction with and without BRCA1/BARD1 co-expression as determined by ICAT
Among the ubiquitination substrates identified, SAFB2 was particularly interesting. SAFB2 is 75 % homologous to SAFB1, and these proteins have been implicated in a variety of cellular processes such as chromatin organization, transcriptional regulation, RNA splicing, and stress response 24. Both SAFB1 and SAFB2 have been shown to bind to and inhibit the transcriptional activity of estrogen receptor α (ERα). 25-29. Importantly, the SAFB1 and SAFB2 genes map to chromosome 19p13 and display a high rate of loss of heterozygosity (22~47%) in sporadic breast cancers 30.
BRCA1 ubiquitinates SAFB and increases its expression
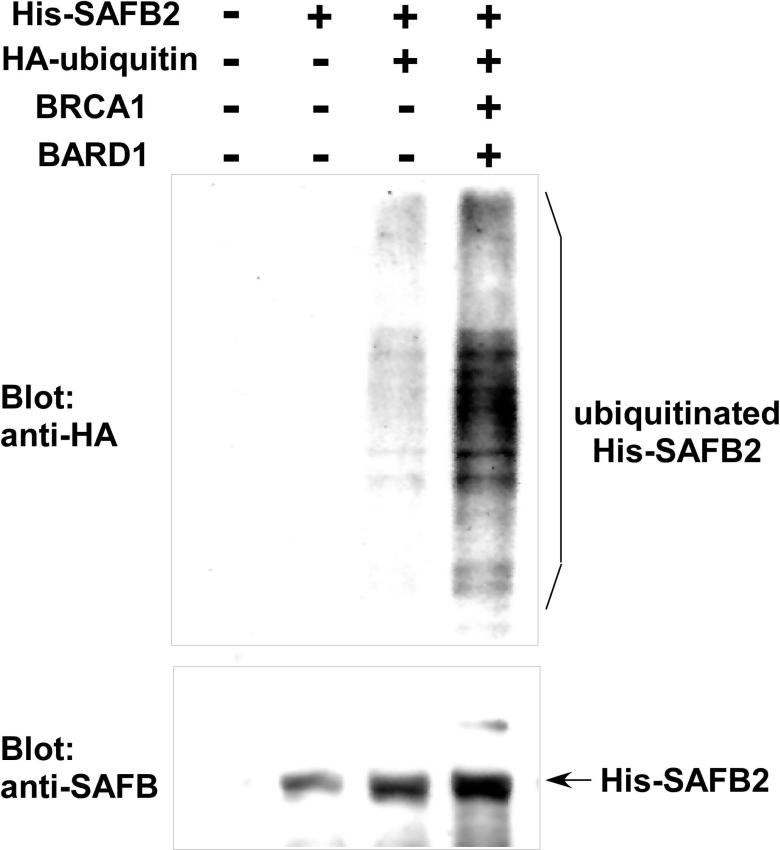
We confirmed that co-expression of BRCA1, BARD1, and HA-ubiquitin induces ubiquitination of His-tagged SAFB2 in vivo (Figure 2). 293T cells were transfected with the indicated expression constructs and His-SAFB2 was purified under denaturing conditions. Purified His-SAFB2 was analyzed for ubiquitination by anti-HA immunoblotting. As shown in Figure 2 top, incorporation of HA-ubiquitin to His-SAFB2 was significantly enhanced by co-expression of BRCA1 and BARD1. Interestingly, ubiquitination of SAFB2 appears to result in increased SAFB2 protein levels (Figure 2, bottom), which may be due to the formation of lysine-6-linked poly-ubiquitin chains that can compete with the formation of lysine-48-linked polyubiquitin chains. Consistent with this notion, we found that adenoviral expression of BRCA1 in 786-O renal carcinoma cells results in a significant increase in SAFB protein expression (Figure 3A). (Note that the anti-SAFB antibody recognizes both SAFB1 and SAFB2, which co-migrate).
Figure 2. SAFB2 ubiquitination by BRCA1/BARD1.
293 T cells were transfected with His-SAFB2, HA-ubiquitin, BRCA1, and BARD1 as indicated. Forty-eight hours after transfection, His-SAFB2 was isolated using nickel affinity chromatography under denaturing conditions. Ubiquitination of His-SAFB2 was analyzed by anti-HA immunoblotting (top), and His-SAFB2 was detected by anti-SAFB immunoblotting (bottom).
Figure 3. BRCA1/BARD1 increases SAFB protein expression.
(A) Adenoviral expression of BRCA1 increases SAFB protein levels. 786-O renal carcinoma cells were infected with BRCA1 or empty vector adenovirus. Forty-eight hours after infection, the expression levels of BRCA1, SAFB, and tubulin were examined by immunoblotting using 30 μg whole cell lysate. Note that anti-SAFB antibody recognizes both SAFB1 and SAFB2, which co-migrate.
(B) BRCA1 knock-down results in reduced SAFB protein expression. 786-O cells were transfected with BRCA1 or control siRNA and 48 hours after transfection, the expression levels of BRCA1, SAFB, and tubulin were examined by immunoblotting using 30 μg whole cell lysate.
(C) BRCA1-exon 11-null mouse embryonic fibroblasts display reduced SAFB protein expression.
The expression levels of BRCA1, SAFB, and tubulin in BRCA1-exon 11-null or wild-type mouse embryonic fibroblasts were examined by immunoblotting using 30 μg whole cell lysate.
The role of BRCA1 in increasing SAFB protein levels was further examined by downregulating BRCA1 expression. As shown in Figure 3B, siRNA-mediated knock-down of BRCA1 in 786-O cells resulted in a concomitant decrease in SAFB protein expression, suggesting that BRCA1 normally functions in increasing SAFB protein expression. We also analyzed SAFB protein expression in BRCA1-exon 11-null mouse embryonic fibroblasts. As shown in Figure 3C, SAFB protein expression was significantly reduced in BRCA1-exon11-null mouse embryonic fibroblasts. Although exon 11 is the largest BRCA1 exon and encodes approximately 60% of the protein, BRCA1-exon11-null mouse cells can still express a defective BRCA1 fragment with an N-terminal RING finger ubiquitin ligase domain, which may explain the residual expression of SAFB.
These results suggest that the BRCA1/BARD1 heterodimer ubiquitinates SAFB and increases SAFB protein levels. By regulating the expression of SAFB, which is a co-repressor of ERα, BRCA1/BARD1 may control estrogen-dependent transcription.
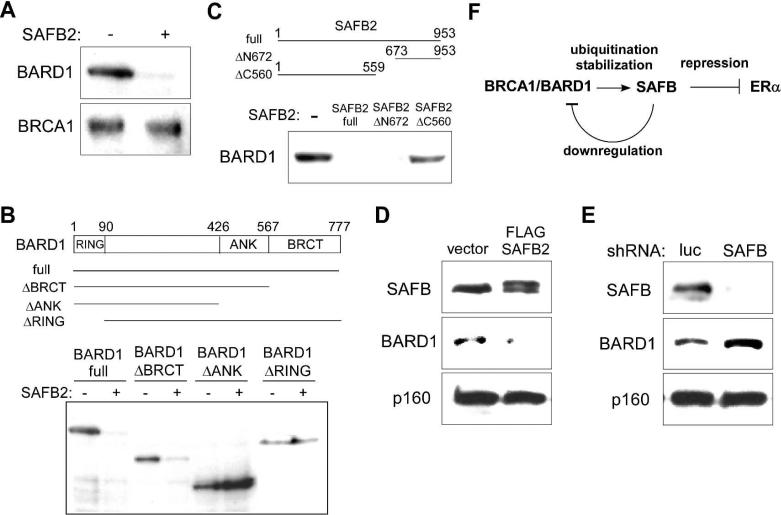
SAFB reduces BARD1 protein expression
During the course of our analysis of molecular interactions between BRCA1/BARD1 and SAFB, we found that co-expression of SAFB2 in 293T cells resulted in dramatic reduction of the levels of BARD1, but not BRCA1 (Figure 4A). To map the domains in BARD1 and SAFB2 that are required for the SAFB2-mediated reduction of BARD1, we used a series of deletion mutants of BARD1 and SAFB2. The expression of a BARD1 deletion mutant that lacks the C-terminal BRCT domain was reduced by SAFB2, but when the deletion included the ankyrin repeat domain of BARD1, the levels of BARD1 were not reduced in the presence of SAFB2 (Figure 4B). The RING finger domain of BARD1 was also required for reduced protein expression by SAFB2 (Figure 4B). Regarding the SAFB2 domains, N-terminal deletion to residue 672 had no effect on BARD1 reduction, but deletion from the C-terminus to residue 560 abolished BARD1 reduction, indicating that SAFB2 C-terminus is required for BARD1 reduction (Figure 4C). Furthermore, when FLAG-SAFB2 was exogenously expressed in MCF-7 breast cancer cells using a lentiviral vector, the endogenous BARD1 protein expression was significantly reduced (Figure 4D).
Figure 4. SAFB reduces BARD1 protein expression.
(A) SAFB2 co-expression reduces BARD1 protein expression in 293T cells. 293T cells were transfected with BARD1 and BRCA1 with or without SAFB2. Forty-eight hours after transfection, the expression levels of BARD1 and BRCA1 expression were examined by immunoblotting using 30 μg whole cell lysate.
(B) Ankyrin repeat domain and RING finger domain of BARD1 are required for BARD1 reduction by SAFB2. 293T cells were transfected with full-length BARD1 or a BARD1 deletion mutant lacking the BRCT domain, BRCT and ankyrin repeat domains, or RING finger domain (all N-terminally FLAG-tagged) in the presence or absence of co-transfected SAFB2. The expression of full-length BARD1 and deletion mutants was examined by anti-FLAG immunoblotting using 30 μg whole cell lysate.
(C) SAFB2 C-terminus is required for BARD1 reduction.
293T cells were transfected with BARD1 alone or together with full-length SAFB2 (encoding amino acids 1-953) or SAFB2 mutant lacking the N-terminal 672 amino acids (encoding amino acids 673-953) or the C-terminal 394 amino acids (encoding amino acids 1-559). BARD1 expression was examined by immunoblotting using 30 μg whole cell lysate.
(D) Exogenous SAFB2 expression results in reduced BARD1 in MCF-7 cells.
MCF-7 breast cancer cells were infected with FLAG-SAFB2 or empty vector lentivirus and the expression of SAFB, BARD1, and Myb-binding protein p160 was examined by immunoblotting using 30 μg whole cell lysate.
(E) SAFB knock-down increases BARD1 expression in MCF-7 cells.
MCF-7 cells were infected with lentivirus expressing short hairpin RNA against SAFB (targeting both SAFB1 and SAFB2) or luciferase and the expression of SAFB, BARD1, and Myb-binding protein p160 was examined by immunoblotting using 30 μg whole cell lysate.
(F) Model for the regulation of SAFB and BARD1 protein levels.
BRCA1/BARD1 ubiquitinates SAFB and increases SAFB protein levels, which may mediate the repression of estrogen receptor α transcriptional activity. SAFB, in turn, downregulates BARD1 expression, which may constitute a feedback mechanism to regulate the SAFB and BARD1 protein levels.
We then asked whether downregulation of SAFB would result in increased BARD1 protein levels. We employed lentiviral expression of short hairpin RNA (shRNA) targeting both SAFB1 and SAFB2 to knock down SAFB expression in MCF-7 cells (using luciferase shRNA as control). As shown in Figure 4E, knock-down of SAFB resulted in an increase of endogenous BARD1, indicating that endogenous SAFB can downregulate the BARD1 protein levels. Since the ubiquitination of SAFB by BRCA1/BARD1 appears to result in increased SAFB protein expression (Figure 2 and 3), our results suggest that there may a feedback loop regulating the SAFB and BARD1 protein levels (Figure 4F).
Tel2 ubiquitination and nuclear translocation by BRCA1
In our quantitative proteomic analysis of BRCA1/BARD1 substrates, we also found a 3.2-fold enhancement of ubiquitination of Tel2 by BRCA1/BARD1 (Table 1). Tel2 is a human homolog of a regulator of telomere length, TEL2, in budding yeast 31 and a DNA damage/S-phase checkpoint protein, CLK-2/RAD5, in C. elegans 32-34. Tel2 interacts with all six phosphatidylinositol 3-kinase-related protein kinases (PIKKs) and maintains their protein expression levels 35. A recent structural and functional study demonstrated that Tel2 forms a complex with Tit1 and Tit2 and participates in Hsp90-dependent maturation of PIKKs 36. In addition, budding yeast TEL2 was shown to be required for localization of TEL1/ATM (a PIKK and a central regulator of DNA damage response) to a DNA break 37. Identification of Tel2 as BRCA1/BARD1 ubiquitination substrate is of particular interest because both BRCA1/BARD1 and PIKKs such as ATM and ATR play important roles in DNA damage signaling.
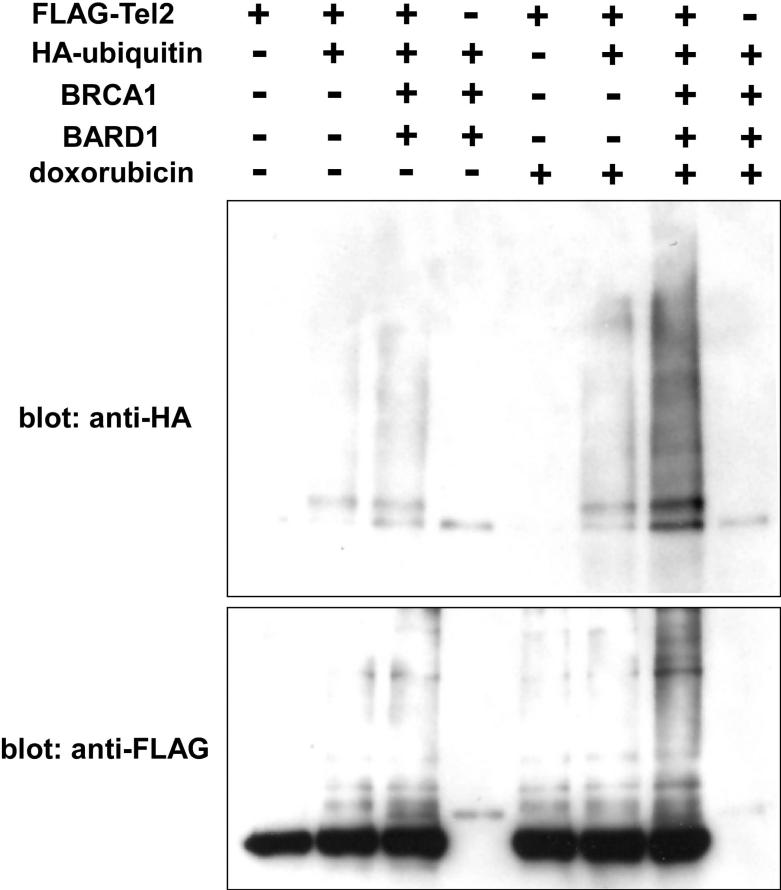
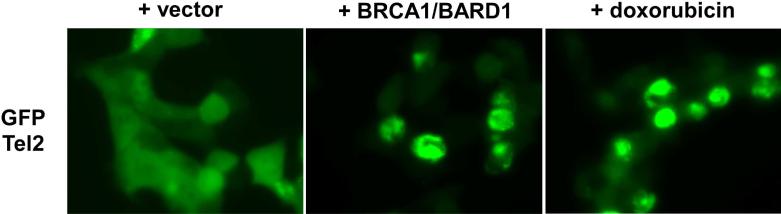
We constructed an N-terminally FLAG-tagged human Tel2 expression vector and analyzed the ubiquitination of Tel2 by BRCA1/BARD1 using co-expression in 293T cells. The FLAG-Tel2 was immunoprecipitated under denaturing conditions and the conjugation of HA-ubiquitin to FLAG-Tel2 was analyzed by anti-HA immunoblotting. As shown in Figure 5, BRCA1/BARD1 induced ubiquitination of FLAG-Tel2; moreover, the level of ubiquitination was dramatically enhanced upon treatment with a DNA damaging chemical, doxorubicin. Although the levels of Tel2 were not affected by the presence of BRCA1, we observed nuclear accumulation of GFP-Tel2 upon BRCA1/BARD1 co-expression or doxorubicin treatment (Figure 6). These results suggest that BRCA1 ubiquitinates Tel2 and modulates Tel2 subcellular location.
Figure 5. Tel2 ubiquitination by BRCA1/BARD1.
293 T cells were transfected with FLAG-Tel2, HA-ubiquitin, BRCA1, and BARD1 as indicated. Where indicated, the cells were also treated with 1 μM doxorubicin for 12 hours before harvesting the cells. Forty-eight hours after transfection, FLAG-Tel2 was purified by anti-FLAG immunoprecipitation under denaturing conditions. Ubiquitination of FLAG-Tel2 was analyzed by anti-HA immunoblotting (top) and FLAG-Tel2 was detected by anti-FLAG immunoblotting (bottom).
Figure 6. BRCA1/BARD1 induces Tel2 nuclear accumulation.
U2OS cells were co-transfected with GFP-Tel2 and empty vector or BRCA1 and BARD1. Where indicated, GFP-Tel2 transfected cells were also treated with 1 μM doxorubicin for 12 hours. The subcellular location of GFP-Tel2 was examined by immunofluorescence microscopy.
DISCUSSION
In this report, we describe a new strategy to identify the substrates of ubiquitin ligases based on detection of enhanced His-ubiquitin conjugation after ubiquitin ligase co-expression. Using this approach, we identified several candidate substrates for BRCA1/BARD1. We confirmed the ubiquitination of two substrates, SAFB2 and Tel2, and demonstrated that BRCA1 increases SAFB protein expression whereas nuclear translocation of Tel2 is induced by BRCA1.
BRCA1 plays a key role in the cellular response to DNA damage, which is conserved in different cell types, but germline inactivation of BRCA1 predominantly results in cancers of breast and ovary. The basis for this tissue-specific tumor suppressor activity of BRCA1 has not, as yet been elucidated. One model to explain this activity involves a role for BRCA1 in the regulation of estrogen signaling. BRCA1 was shown to strongly inhibit the transcriptional activity of ERα 38,39, but the mechanism of transcriptional inhibition by BRCA1 was not clear.
SAFB2 and its close homolog SAFB1 are transcriptional co-repressors that bind to ERα and inhibit its transcriptional activity. The SAFB1 and SAFB2 loci map to chromosome 19p13 and display a high rate of loss of heterozygosity (22 – 47%) in sporadic breast cancers. Taken together, these observations suggest that SAFB might be a missing link between BRCA1 and estrogen receptor activity and that ubiquitination of SAFB by BRCA1/BARD1 might contribute to the tissue-specific tumor suppressor activity of BRCA1. In future studies, more detailed analysis of the deletion and mutation of the SAFB locus in breast and ovary cancers as well as their relation to ERα expression status and BRCA1 mRNA/protein levels may provide important insight into the role of the BRCA1-SAFB-ERα pathway in hormone-dependent tumorigenesis.
In our studies, we also found evidence suggesting that SAFB reduces the BARD1 protein expression. SAFB expression reduced the protein levels of both transfected and endogenous BARD1, while RNAi suppression of SAFB resulted in increased endogenous BARD1 protein levels (Figure 4). Besides its function as a heterodimerization partner for BRCA1, BARD1 was proposed to have BRCA1-independent functions 40. BARD1 was shown to bind and stabilize p53 and overexpression of BARD1 resulted in p53-dependent apoptosis 41. BRCA1 is not only dispensable for apoptosis induction by BARD1, but also counteracts BARD1-induced apoptosis 41. Knock-down of BRCA1 expression by siRNA or the disruption of BRCA1/BARD1 heterodimer formation by peptide competition resulted in cytoplasmic translocation of BARD1 and subsequent apoptosis 42,43. An excess of BARD1 over BRCA1 and cytoplasmic localization of BARD1 have been linked to induction of apoptosis by BARD1 41. If the relative abundance of BRCA1 and BARD1 determines cell survival, then the levels of these two proteins need to be coordinately regulated. Downregulation of BARD1 by SAFB may explain how the expression of BRCA1 and BARD1 are coordinately regulated.
Tel2 has been shown to bind and stabilize PIKKs 35, which include ATM, ATR, DNA-PKcs, mammalian target of rapamycin (mTOR), suppressor with morphological effect on genitalia 1 (SMG1), and transformation/transcription domain-associated protein (TRRAP). A recent study also uncovered that Tel2 in conjunction with Tit1 and Tit2 acts as co-chaperone for Hsp90-dependent maturation of PIKK complexes 36. PIKKs such as ATM and ATR play central roles in the cellular response to DNA damage. There is also evidence that budding yeast Tel1/ATM requires Tel2 for localization to the site of a DNA break 37. Thus, Tel2 appears to be an important regulator of DNA damage signaling. BRCA1/BARD1 also play a pivotal role in DNA damage response and BRCA1 was shown to act downstream of ATR/ATM 44.
Our proteomic screen identified Tel2 as a BRCA1/BARD1 substrate. Further analysis demonstrated that BRCA1 is able to induce Tel2 ubiquitination and cause nuclear translocation of Tel2. By quantitative immunoblotting, it was estimated that HeLa cells and HTC75 fibrosarcoma cells contain 11,000 molecules of Tel2, 14,000 – 22,000 molecules of ATM, and 80,000 – 200,000 molecules of mTOR per cell 36. This suggests that the quantity of Tel2 is limiting compared to PIKKs and that subcellular distribution of Tel2 may impact the activity of different PIKKs located in different subcellular compartments. BRCA1-induced nuclear accumulation of Tel2 may be a mechanism to allocate limiting Tel2 to nuclear PIKKs such as ATM and ATR upon DNA damage.
BRCA1/BARD1 generates non-conventional lysine-6-linked poly-ubiquitin chains. Therefore, we employed a His-tagged ubiquitin K48R mutant for proteomic identification of BRCA1/BARD1 substrates. Compared with wild-type ubiquitin, there is less incorporation of the K48R mutant into cellular proteins in the absence of co-expressed BRCA1/BARD1. This allowed more sensitive detection of BRCA1/BARD1-dependent ubiquitination events. However, we also observed that incorporation of wild-type ubiquitin into cellular proteins was enhanced by co-expression of E3 ubiquitin ligases that generate conventional lysine-48-linked poly-ubiquitin chains. This suggests that His-tagged wild-type ubiquitin can be used for the identification of the substrates of these E3 ligases. In addition, we also found that several SUMO E3 ligases can significantly enhance the incorporation of SUMO to cellular proteins upon co-expression, suggesting that the SUMO E3 ligase substrates can be identified by His-SUMO quantitative proteomics approach similar to the one presented in this article.
It is estimated that there are several hundred E3 ubiquitin ligases encoded in the human genome, but, in most cases, their ubiquitination substrates have not been identified. Ubiquitin ligases and their substrates often do not interact tightly, which has hampered the identification of ligase-substrate pairs. We recently reported a protein expression profiling approach to identify candidate ubiquitination substrates by their increased abundance in cells without functional ubiquitin ligase compared with cells that contain the ubiquitin ligase 45. Using this approach, we were able to identify known and novel substrates for the VHL ubiquitin ligase 45. However, a major limitation of the protein expression profiling approach is that mass spectrometry-based quantitative proteomics cannot currently detect every protein in mammalian whole cell proteome. The analysis of sub-proteomes such as subcellular fractions could increase the proteome coverage by protein expression profiling approach. The His-ubiquitin quantitative proteomics approach presented in this article and its further refinement will complement the protein expression profiling approach, and together they will increase our knowledge about ubiquitin ligase-substrate pairs.
Supplementary Material
Supporting Information
The supplementary table lists the proteins displaying a two-fold or higher enrichment in Hisubiquitin-K48R-conjugated protein fraction upon BRCA1/BARD1 co-expression.
Table S1. List of proteins displaying increased ubiquitination upon BRCA1/BARD1 co-expression The supplementary figure shows the tandem mass spectra of peptides derived from the proteins that are listed in Table 1.
Figure S1. Tandem mass spectra of peptides derived from the proteins in Table 1.
These materials are available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank Dr. Richard Baer for generous gifts of BRCA1 and BARD1 plasmids as well as anti-BARD1 antibody and Mr. Barron Blackman for assistance with proteomics informatics. This work was supported in part by NIH grants CA125020 (to Y.S.) and CA054174 (Cancer Therapy and Research Center at UTHSCSA - Mass Spectrometry Shared Resource).
REFERENCES
- 1.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23(21):4776–89. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A, Schwartz AL. The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochim Biophys Acta. 2004;1695(1-3):3–17. doi: 10.1016/j.bbamcr.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Parvin JD. Overview of history and progress in BRCA1 research: the first BRCA1 decade. Cancer Biol Ther. 2004;3(6):505–8. doi: 10.4161/cbt.3.6.839. [DOI] [PubMed] [Google Scholar]
- 4.Rosen EM, Fan S, Pestell RG, Goldberg ID. BRCA1 in hormone-responsive cancers. Trends Endocrinol Metab. 2003;14(8):378–85. doi: 10.1016/j.tem.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95(11):866–71. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci U S A. 2001;98(9):5134–9. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen A, Kleiman FE, Manley JL, Ouchi T, Pan ZQ. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J Biol Chem. 2002;277(24):22085–92. doi: 10.1074/jbc.M201252200. [DOI] [PubMed] [Google Scholar]
- 8.Wu-Baer F, Lagrazon K, Yuan W, Baer R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem. 2003;278(37):34743–6. doi: 10.1074/jbc.C300249200. [DOI] [PubMed] [Google Scholar]
- 9.Mallery DL, Vandenberg CJ, Hiom K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. Embo J. 2002;21(24):6755–62. doi: 10.1093/emboj/cdf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandenberg CJ, Gergely F, Ong CY, Pace P, Mallery DL, Hiom K, Patel KJ. BRCA1-independent ubiquitination of FANCD2. Mol Cell. 2003;12(1):247–54. doi: 10.1016/s1097-2765(03)00281-8. [DOI] [PubMed] [Google Scholar]
- 11.Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, Shiekhattar R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12(5):1087–99. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 12.Starita LM, Horwitz AA, Keogh MC, Ishioka C, Parvin JD, Chiba N. BRCA1/BARD1 ubiquitinate phosphorylated RNA polymerase II. J Biol Chem. 2005;280(26):24498–505. doi: 10.1074/jbc.M414020200. [DOI] [PubMed] [Google Scholar]
- 13.Sato K, Hayami R, Wu W, Nishikawa T, Nishikawa H, Okuda Y, Ogata H, Fukuda M, Ohta T. Nucleophosmin/B23 is a candidate substrate for the BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279(30):30919–22. doi: 10.1074/jbc.C400169200. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20(13):1721–6. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikawa H, Ooka S, Sato K, Arima K, Okamoto J, Klevit RE, Fukuda M, Ohta T. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279(6):3916–24. doi: 10.1074/jbc.M308540200. [DOI] [PubMed] [Google Scholar]
- 16.Morris JR, Solomon E. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet. 2004;13(8):807–17. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- 17.Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17(2):201–13. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, Martin DB, Nesvizhskii AI, Aebersold R. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 10(6):1150–9. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 20.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 21.Li XJ, Zhang H, Ranish JA, Aebersold R. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal Chem. 2003;75(23):6648–57. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 22.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci U S A. 2003;100(23):13225–30. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiio Y, Donohoe S, Yi EC, Goodlett DR, Aebersold R, Eisenman RN. Quantitative proteomic analysis of Myc oncoprotein function. Embo J. 2002;21(19):5088–96. doi: 10.1093/emboj/cdf525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oesterreich S. Scaffold attachment factors SAFB1 and SAFB2: Innocent bystanders or critical players in breast tumorigenesis? J Cell Biochem. 2003;90(4):653–61. doi: 10.1002/jcb.10685. [DOI] [PubMed] [Google Scholar]
- 25.Oesterreich S, Zhang QP, Lee AV. Inhibition of oestrogen receptor activity by the co-repressor HET/SAF-B is relieved by blockade of histone deacetylase activity. Eur J Cancer. 2000;36(Suppl 4):S43–4. doi: 10.1016/s0959-8049(00)00221-5. [DOI] [PubMed] [Google Scholar]
- 26.Townson SM, Kang K, Lee AV, Oesterreich S. Structure-function analysis of the estrogen receptor alpha corepressor scaffold attachment factor-B1: identification of a potent transcriptional repression domain. J Biol Chem. 2004;279(25):26074–81. doi: 10.1074/jbc.M313726200. [DOI] [PubMed] [Google Scholar]
- 27.Jiang S, Meyer R, Kang K, Kent Osborne C, Wong J, Oesterreich S. Scaffold attachment factor SAFB1 suppresses ER{alpha}-mediated transcription in part via interaction with N-CoR. Mol Endocrinol. 2005 doi: 10.1210/me.2005-0100. [DOI] [PubMed] [Google Scholar]
- 28.Townson SM, Dobrzycka KM, Lee AV, Air M, Deng W, Kang K, Jiang S, Kioka N, Michaelis K, Oesterreich S. SAFB2, a new scaffold attachment factor homolog and estrogen receptor corepressor. J Biol Chem. 2003;278(22):20059–68. doi: 10.1074/jbc.M212988200. [DOI] [PubMed] [Google Scholar]
- 29.Oesterreich S, Zhang Q, Hopp T, Fuqua SA, Michaelis M, Zhao HH, Davie JR, Osborne CK, Lee AV. Tamoxifen-bound estrogen receptor (ER) strongly interacts with the nuclear matrix protein HET/SAF-B, a novel inhibitor of ER-mediated transactivation. Mol Endocrinol. 2000;14(3):369–81. doi: 10.1210/mend.14.3.0432. [DOI] [PubMed] [Google Scholar]
- 30.Oesterreich S, Allredl DC, Mohsin SK, Zhang Q, Wong H, Lee AV, Osborne CK, O'Connell P. High rates of loss of heterozygosity on chromosome 19p13 in human breast cancer. Br J Cancer. 2001;84(4):493–8. doi: 10.1054/bjoc.2000.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Runge KW, Zakian VA. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16(6):3094–105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed S, Alpi A, Hengartner MO, Gartner A. C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr Biol. 2001;11(24):1934–44. doi: 10.1016/s0960-9822(01)00604-2. [DOI] [PubMed] [Google Scholar]
- 33.Dengg M, Garcia-Muse T, Gill SG, Ashcroft N, Boulton SJ, Nilsen H. Abrogation of the CLK-2 checkpoint leads to tolerance to base-excision repair intermediates. EMBO Rep. 2006;7(10):1046–51. doi: 10.1038/sj.embor.7400782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Muse T, Boulton SJ. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. Embo J. 2005;24(24):4345–55. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131(7):1248–59. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 36.Takai H, Xie Y, de Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 2010;24(18):2019–30. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson CM, Korkin D, Smith DL, Makovets S, Seidel JJ, Sali A, Blackburn EH. Tel2 mediates activation and localization of ATM/Tel1 kinase to a double-strand break. Genes Dev. 2008;22(7):854–9. doi: 10.1101/gad.1646208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, Rosen EM. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284(5418):1354–6. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 39.Zheng L, Annab LA, Afshari CA, Lee WH, Boyer TG. BRCA1 mediates ligand-independent transcriptional repression of the estrogen receptor. Proc Natl Acad Sci U S A. 2001;98(17):9587–92. doi: 10.1073/pnas.171174298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irminger-Finger I, Jefford CE. Is there more to BARD1 than BRCA1? Nat Rev Cancer. 2006;6(5):382–91. doi: 10.1038/nrc1878. [DOI] [PubMed] [Google Scholar]
- 41.Irminger-Finger I, Leung WC, Li J, Dubois-Dauphin M, Harb J, Feki A, Jefford CE, Soriano JV, Jaconi M, Montesano R, Krause KH. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol Cell. 2001;8(6):1255–66. doi: 10.1016/s1097-2765(01)00406-3. [DOI] [PubMed] [Google Scholar]
- 42.Jefford CE, Feki A, Harb J, Krause KH, Irminger-Finger I. Nuclear-cytoplasmic translocation of BARD1 is linked to its apoptotic activity. Oncogene. 2004;23(20):3509–20. doi: 10.1038/sj.onc.1207427. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez JA, Schuchner S, Au WW, Fabbro M, Henderson BR. Nuclear-cytoplasmic shuttling of BARD1 contributes to its proapoptotic activity and is regulated by dimerization with BRCA1. Oncogene. 2004;23(10):1809–20. doi: 10.1038/sj.onc.1207302. [DOI] [PubMed] [Google Scholar]
- 44.Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. Embo J. 2006;25(10):2178–88. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai Y, Qiao M, Song M, Weintraub ST, Shiio Y. Quantitative Proteomics Identifies the Myb-Binding Protein p160 as a Novel Target of the von Hippel-Lindau Tumor Suppressor. PLoS One. 2011;6(2):e16975. doi: 10.1371/journal.pone.0016975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
The supplementary table lists the proteins displaying a two-fold or higher enrichment in Hisubiquitin-K48R-conjugated protein fraction upon BRCA1/BARD1 co-expression.
Table S1. List of proteins displaying increased ubiquitination upon BRCA1/BARD1 co-expression The supplementary figure shows the tandem mass spectra of peptides derived from the proteins that are listed in Table 1.
Figure S1. Tandem mass spectra of peptides derived from the proteins in Table 1.
These materials are available free of charge via the Internet at http://pubs.acs.org.