Abstract
Selecting suitable feature types is crucial to obtain good overall brain–computer interface performance. Popular feature types include logarithmic band power (logBP), autoregressive (AR) parameters, time-domain parameters, and wavelet-based methods. In this study, we focused on different variants of AR models and compare performance with logBP features. In particular, we analyzed univariate, vector, and bilinear AR models. We used four-class motor imagery data from nine healthy users over two sessions. We used the first session to optimize parameters such as model order and frequency bands. We then evaluated optimized feature extraction methods on the unseen second session. We found that band power yields significantly higher classification accuracies than AR methods. However, we did not update the bias of the classifiers for the second session in our analysis procedure. When updating the bias at the beginning of a new session, we found no significant differences between all methods anymore. Furthermore, our results indicate that subject-specific optimization is not better than globally optimized parameters. The comparison within the AR methods showed that the vector model is significantly better than both univariate and bilinear variants. Finally, adding the prediction error variance to the feature space significantly improved classification results.
Keywords: Brain–computer interface, Autoregressive model, Logarithmic band power, Feature extraction, Motor imagery
Introduction
A brain–computer interface (BCI) is a device that measures signals from the brain and translates them into control commands for an application such as a wheelchair, an orthosis, or a spelling device [43]. By definition, a BCI does not use signals from muscles or peripheral nerves. Furthermore, a BCI operates in real-time, presents feedback, and requires goal-directed behavior from the user [27].
Most non-invasive BCIs record the electroencephalogram (EEG) from the surface of the scalp [19]. In general, there are several components which process the raw EEG signals before an actual output of the system is available. Typically, signals are first preprocessed with temporal or spatial filters. Examples of preprocessing techniques include bandpass filters, bipolar filters, or more advanced approaches such as common spatial patterns (CSP) [4]. The next stage extracts suitable features from the preprocessed signals, that is, relevant (discriminative) signal characteristics are isolated. Popular features for BCIs include logarithmic band power (logBP) [25, 26], autoregressive (AR) parameters [35], time-domain parameters [42], and wavelet-based methods [11]. Finally, a classification or regression algorithm translates the features into an output signal for a specific application. Examples of widely used classifiers in BCI research are linear discriminant analysis (LDA), support vector machines, neural networks, and nearest neighbor classifiers [18, 19, 40]. Optionally, and depending on the application, the output of the classification stage can be post-processed, for example by averaging over time or by introducing additional constraints such as a dwell time and refractory period [37].
Selecting suitable features is crucial to obtain good overall BCI performance [7, 8]. In this study, we focus on BCIs based on event-related desynchronization [28] and explore extensions of the simple AR model and compare the resulting features with logBP features. More specifically, we compare the performance of a standard univariate AR (UAR) model, a vector AR (VAR) model, and a bilinear AR (BAR) model on BCI data. We also study the influence of adding the error variance as a feature for all three AR model types. Similar to logBP, AR parameters can be used to estimate the power spectral density [20], but they can also serve directly as features for BCIs [35]. Many groups have used AR parameters as features for BCIs in either way; some groups used short segments of time and fitted an AR model to this data segment [9, 30], whereas others adapted the model coefficients continuously [35, 39] (for example with a Kalman filter approach).
Most studies used UAR models, which means that each EEG channel is described with a separate AR model. This means that information about the relationships between signals is completely neglected. In contrast, a VAR model describes all channels at once and therefore includes information about the correlation between individual signals. Only a few studies have described VAR parameters applied to BCI data, but they reported promising results [2, 24]. Furthermore, the additional information inherent in VAR models can be used to compute explicit coupling measures such as the partial directed coherence and the directed transfer function [34].
Another extension of the AR model is the BAR model. In contrast to the classical linear AR model, a BAR model can describe certain non-linear signal properties [29] such as non-Gaussian signals. Many real-world time series exhibit such behavior, for example the arc-shaped sensorimotor mu rhythm [10] in the case of EEG signals. Consequently, a bilinear model (which is a special case of general non-linear models) should be better suited to model such data.
The objective of this study is to assess the influence of different feature types based on AR models on the performance of a BCI (for example as measured by the classification accuracy). More specifically, we compared standard UAR models with VAR and BAR models, and variants including the prediction error variance as an additional feature. We also used logBP features as state-of-the-art features for comparison. We hypothesized that both VAR and BAR models could yield higher BCI performance than UAR parameters, because they contain more information on the underlying signals and/or describe the signals more accurately. Moreover, adding the error variance as a feature could add discriminative information and thus increase BCI performance.
Methods
Data
We used data set 2a from the BCI Competition IV 1, which comprises data from nine users over two sessions each (recorded on separate days). The data was recorded with prior consent of all participants, and the study conformed to guidelines established by the local ethics commission. In each trial, participants performed one out of four different motor imagery tasks: movement imagination of left hand, right hand, both feet, and tongue. In total, each of the two sessions consists of 288 trials (72 trials per class) in random order.
Subjects were sitting in front of a computer monitor. At the beginning of a trial, a cross appeared on the black screen. In addition, subjects heard a tone indicating trial onset. After 2 s, subjects viewed an arrow that pointed either to the left, right, top or bottom of the screen. They performed the corresponding motor imagery task until the cross disappeared after 6 s. A short break between 1.5 and 2.5 s followed before the next trial.
The data set consists of 22 EEG signals recorded monopolarly (referenced to the left mastoid and grounded to the right mastoid). Signals were sampled at 250 Hz and bandpass-filtered between 0.5 and 100 Hz. An additional 50 Hz notch filter removed line noise. In this study, we used only three bipolar channels, calculated by subtracting channels anterior to C3, Cz, and C4 from sites posterior to these locations (the inter-electrode distance was 3.5 cm).
Features
We compared three different AR variants, namely (1) a UAR model, (2) a VAR model, and (3) a BAR model. In all three cases, we used the corresponding AR coefficients as features. In addition, we enhanced each AR method by adding the prediction error variance to the feature space. In summary, we analyzed six different AR-based feature types, described in more detail in the following paragraphs.
UAR model
A 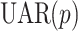 model is defined as
model is defined as
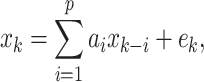 |
1 |
where x k is the value of the time series x at time point k. The current value of x k can be predicted by the weighted sum of p past values x k-i plus an additional error term e k. The weights a i are called the AR parameters. In a typical BCI, x k corresponds to the amplitude of an EEG channel at time k.
VAR model
A 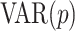 model is an extension of the UAR case described above, because it simultaneously describes several time series. Thus, it is defined as
model is an extension of the UAR case described above, because it simultaneously describes several time series. Thus, it is defined as
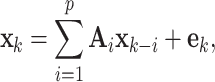 |
2 |
where 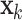 is a vector of time series at time k. The p AR parameters from the UAR model generalize to p matrices
is a vector of time series at time k. The p AR parameters from the UAR model generalize to p matrices 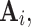 and the error term
and the error term 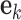 becomes a vector. In contrast to a UAR model, a VAR model explicitly models the correlation between the different time series. Applied to EEG data, VAR models can describe the relationships between different EEG channels, which might contain discriminable information for BCIs [5].
becomes a vector. In contrast to a UAR model, a VAR model explicitly models the correlation between the different time series. Applied to EEG data, VAR models can describe the relationships between different EEG channels, which might contain discriminable information for BCIs [5].
BAR model
In contrast to UAR and VAR models (which are linear time series models), non-linear models can describe non-linear characteristics such as large bursts or extremely rapid and large fluctuations [29]. A 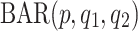 model is an extension of a linear
model is an extension of a linear 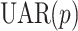 model and a special case of general non-linear models with finite parameters. It is defined as
model and a special case of general non-linear models with finite parameters. It is defined as
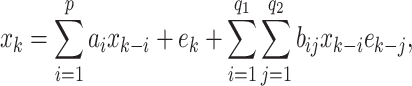 |
3 |
where the first part is a 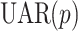 model and the last part describes the bilinear contribution with the
model and the last part describes the bilinear contribution with the 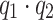 bilinear coefficients b
ij.
bilinear coefficients b
ij.
BAR models might be more suitable to describe EEG signals, because EEG signals may contain non-linear features such as the arc-shaped mu rhythm [10]. Such characteristics cannot be captured by linear time series models [29].
Parameter estimation
We estimated AR parameters adaptively for all AR-based methods (UAR, VAR, and BAR) using a Kalman filter [14]. A Kalman filter operates in the state space, which is defined by the following two equations:
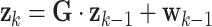 |
4 |
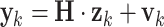 |
5 |
Here, 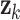 is the state at time
is the state at time 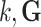 is the state transition matrix, and
is the state transition matrix, and 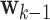 is the process noise with
is the process noise with 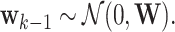 Furthermore,
Furthermore, 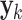 is the measurement vector,
is the measurement vector, 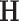 is the measurement sensitivity matrix, and
is the measurement sensitivity matrix, and 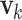 is the measurement noise with
is the measurement noise with 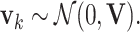 For univariate models UAR and BAR,
For univariate models UAR and BAR, 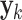 and
and 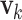 reduce to scalars y
k and v
k (with
reduce to scalars y
k and v
k (with 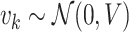 ), respectively.
), respectively.
We used these equations to estimate AR parameters by assigning 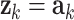 (where
(where 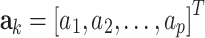 is a vector containing all AR coefficients),
is a vector containing all AR coefficients), 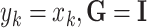 (the identity matrix), and
(the identity matrix), and 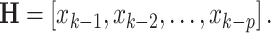 These assignments hold for the UAR model only, but they can be easily generalized for the VAR case by using matrix equivalents of the corresponding variables, and for the BAR model by extending
These assignments hold for the UAR model only, but they can be easily generalized for the VAR case by using matrix equivalents of the corresponding variables, and for the BAR model by extending 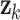 and
and 
We adopted an estimation approach based on results presented in [36] and as recommended and implemented in the BioSig2 toolbox [33] function
tvaar.m
. We implemented this function in C and added a MATLAB3 interface, which speeded up computation time significantly.
In the first step, we tried to find suitable initial values for parameters such as the AR coefficients, the process noise covariance, and the measurement noise covariance. We updated all parameters in this first run over the complete first data session. Once we found initial values with this procedure, we estimated AR parameters in a second run over the session using another update mode, which essentially keeps the process noise and measurement noise covariances constant at the previously found values. In the final evaluation step on the unseen second session, we only used mode the latter mode, but initialized parameters with values found in the optimization step using the first session (see Sects. 2.3, 2.4 for more details).
Features based on AR models
The prediction error 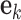 at time k can be estimated by subtracting the prediction
at time k can be estimated by subtracting the prediction 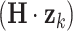 from the measurement
from the measurement 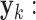
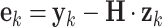 |
6 |
We used the logarithm of the estimated covariance of the prediction error 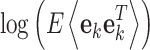 to augment the feature space of UAR, VAR, and BAR models, thus yielding three additional AR feature types termed xUAR, xVAR, and xBAR. Note that we adapted the covariance estimation in each step directly with UC.
to augment the feature space of UAR, VAR, and BAR models, thus yielding three additional AR feature types termed xUAR, xVAR, and xBAR. Note that we adapted the covariance estimation in each step directly with UC.
In summary, we compared the following six AR-based feature extraction methods: (1) UAR, (2) xUAR, (3) VAR, (4) xVAR, (5) BAR, and (6) xBAR.
LogBP
We compared our AR features with results obtained from logBP, which is commonly used in many BCI systems [19]. The calculation procedure is as follows:
Bandpass-filter raw EEG signal in a specific frequency band (we used a fifth order Butterworth filter)
Square samples
Smooth over a one second time window (we used a moving average filter)
Compute the logarithm
Parameter optimization
We conducted two independent parameter optimization procedures. In the first analysis (individual optimization), we optimized parameters for each subject individually. In the second analysis (global optimization), we searched for parameters that were optimal for all subjects in the data set. Importantly, we used only data from the first session in both procedures; we never used data from the second session during parameter optimization.
Individual optimization
For each AR method, we optimized model order(s) and update coefficient UC (a parameter which determines the update speed in each iteration of the Kalman filter algorithm) for each subject individually. We used a grid search to find the optimal parameter combination. Table 1 lists the search spaces for the different methods. In summary, we searched in 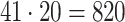 (UAR, xUAR),
(UAR, xUAR), 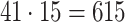 (VAR, xVAR), and
(VAR, xVAR), and 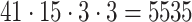 (BAR, xBAR) parameter combinations, respectively.
(BAR, xBAR) parameter combinations, respectively.
Table 1.
Search spaces for the AR-based feature extraction methods
| Methods | log(UC) | p | q 1 | q 2 |
|---|---|---|---|---|
| UAR, xUAR |
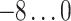
|
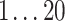
|
– | – |
| VAR, xVAR |
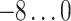
|
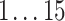
|
– | – |
| BAR, xBAR |
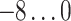
|
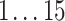
|
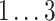
|
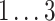
|
We varied linear and bilinear model orders p, q
1, and q
2 in steps of 1, and the logarithmic update coefficient 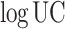 in steps of 0.2
in steps of 0.2
For each parameter combination and method, we performed the following steps:
Find best segment for classifier setup using a running classifier [22] (we divided a trial into 1 s segments with 0.5 s overlap and used all samples within a segment for the running classifier procedure; see Sect. 2.4 for more details on the classifier)
Leave-8-trials-out cross-validation (train a classifier on best segment found in the previous step, test on whole trial)
Use 0.9 quantile of classification accuracy p 0 as performance measure
Finally, we selected the parameter combination associated with the highest performance measure.
In contrast to the grid search optimization for AR methods, we used a method based on neurophysiological principles instead of classification results to optimize logBP features; we refer to this method as band power difference maps [3], which is similar to the approach described in [4]. The procedure is as follows (applied to each EEG channel separately):
Compute time-frequency maps of signal power for each motor imagery task and the three remaining tasks combined (using only data from within trials)
Calculate difference maps by subtracting the map of one task from the map of the three remaining tasks combined
Iteratively find and remove connected patches in maps (corresponding to largest differences)
Combine adjacent or overlapping bands.
We calculated time–frequency maps with high time and frequency resolution (we varied time from 0–8 s in steps of 0.04 s and frequency from 5 to 40 Hz with 1 Hz bands in steps of 0.1 Hz). We also calculated confidence intervals for each time–frequency point by first applying a Box-Cox transformation and then computing confidence intervals from the normal distribution.
In summary, we calculated eight time–frequency maps for the following motor imagery tasks and combination of tasks: 1, 2, 3, 4, 234, 134, 124, and 123 (the numbers 1, 2, 3, and 4 correspond to left hand, right hand, feet, and tongue motor imagery, respectively; the numbers 234, 134, 124, and 123 are combinations of these tasks). Next, we calculated four difference maps, namely 1–234, 2–134, 3–124, and 4–123. Within each difference map, we iteratively searched for connected significant patches (inspired by a four-way flood fill algorithm), starting with the pixel with the largest difference. If the area of such a patch was over a predefined threshold of 1 s Hz, we used its upper and lower frequency borders to define a band for the logBP feature extraction method. We then removed this patch from the map and repeated the search procedure, searching again for the pixel with the largest difference. We continued this procedure until the algorithm had removed all patches from the map. Finally, we combined all frequency bands found in the four difference maps and combined adjacent or overlapping frequency bands.
Global optimization
In addition to the individual optimization, we also tried to find parameters that are optimal for all subjects. For each AR method, we averaged the performance measures (calculated for all parameter combinations) over all nine subjects. From these averaged results, we selected the combination of linear model order(s) and update coefficient with the highest performance measure.
For logBP, we simply selected standard frequency bands 8–12 and 16–24 Hz (containing alpha and beta bands) for all channels.
Evaluation
We evaluated all feature extraction methods in two different ways. First, we calculated the cross-validated (XV) classification accuracy p 0 on the second session. Second, we estimated the session transfer (ST) by calculating classifier weights on the first session and computing the classification accuracy p 0 on the second session. We carried out this evaluation for both individually and globally optimized features (see Sect. 2.2.4).
Cross-validation (XV)
With the optimized parameter values found in the optimization step (using data from the first session only), we calculated the cross-validated classification accuracy p 0 on the second session. Therefore, we used a similar classification procedure as described in Sect. 2.2.4. First, we extracted features from the second session. Next, we determined the best segment for classifier setup using a running classifier [22]. As before, we divided each trial into 1 s segments with 0.5 s overlap. We used a combination of LDA classifiers in a one-versus-rest scheme; this classifier assigned one out of four classes to the class with the highest discriminant value. We performed a leave-8-trials-out cross-validation, which means that we used segments of 280 trials to train and eight trials to test a classifier. We repeated this procedure until all segments had been used as a test set once. Finally, we averaged over all folds, and we calculated the 0.9 quantile of the cross-validated classification accuracy. That is, instead of reporting the maximum of the classification accuracy within a trial, we chose the 0.9 quantile as a more robust measure of performance, because it effectively removes outliers.
Session transfer
The ST estimates the performance of a real-world BCI system more realistically, but it requires a sufficiently high number of unseen test data trials. In this analysis, we determined optimal parameters and classifier weights from the first session. After that we extracted features from the second session and applied the classifier from the previous step. We used the same one-versus-rest classifier scheme as in the cross-validation analysis.
Statistical analysis
We used repeated measures analysis of variance (ANOVA) to statistically analyze the classification results. First, we checked the sphericity assumption with Mauchly’s spericity test. Then, we performed the ANOVA and corrected degrees of freedom if necessary. If we found significant effects, we used Newman–Keuls post-hoc tests to determine significant differences.
Basically, we performed ANOVAs for XV and ST results separately. First, we wanted to assess differences over all seven feature extraction methods (factor “method”; 7 levels; UAR, xUAR, VAR, xVAR, BAR, xBAR, and logBP) and optimization strategies (factor “optimization”; 2 levels; individual and global). Second, we were also interested in differences between the three AR-based methods only (factor “method”; 3 levels; U, V, and B), the influence of the prediction error variance feature (factor “x”; 2 levels; yes or no), and the optimization strategies (factor “optimization”; 2 levels; individual or global).
We repeated these analyses with both XV and ST results combined into a factor “ST/XV” (2 levels; ST and XV). In summary, we performed six repeated measures ANOVAs.
Results
Parameter optimization
Tables 2 and 3 show the results of the optimization procedure for both the individual and global optimization, respectively. On average, univariate methods (UAR, BAR, xUAR, and xBAR) require a higher model order p as opposed to vector models (VAR and xVAR). The optimized values of the update coefficient UC are similar for all methods, except in the case of BAR for subjects A01 and A02, where the UC is significantly lower (see Fig. 1). This might be due to our optimization procedure, where we selected the parameter combination with the highest fitness function. However, only a slightly lower classification accuracy is associated with a log(UC) around −2.5, a value found for all other subjects.
Table 2.
Results of parameter optimization for AR-based methods UAR, VAR, and BAR without the prediction error variance feature
| |
UAR | VAR | BAR | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p 0 | p | log(UC) | p 0 | p | log(UC) | p 0 | p | q | log(UC) | |
| A01 | 0.582 | 13 | −2.8 | 0.612 | 4 | −2.6 | 0.601 | 8 | 2, 2 | −0.8 |
| A02 | 0.446 | 6 | −3.0 | 0.461 | 6 | −2.8 | 0.461 | 14 | 1, 1 | −0.6 |
| A03 | 0.573 | 12 | −2.6 | 0.625 | 2 | −2.8 | 0.578 | 12 | 1, 3 | −2.6 |
| A04 | 0.418 | 10 | −2.2 | 0.395 | 4 | −2.2 | 0.421 | 12 | 2, 2 | −2.6 |
| A05 | 0.406 | 4 | −2.6 | 0.410 | 2 | −2.4 | 0.418 | 5 | 1, 2 | −2.2 |
| A06 | 0.429 | 15 | −2.2 | 0.434 | 12 | −2.2 | 0.457 | 15 | 1, 1 | −2.6 |
| A07 | 0.544 | 14 | −2.6 | 0.533 | 13 | −2.4 | 0.559 | 14 | 1, 3 | −2.6 |
| A08 | 0.635 | 15 | −2.4 | 0.673 | 4 | −2.4 | 0.639 | 5 | 1, 2 | −2.4 |
| A09 | 0.614 | 3 | −2.2 | 0.640 | 3 | −2.0 | 0.623 | 7 | 1, 2 | −2.2 |
| Global | 0.494 | 13 | −2.6 | 0.507 | 4 | −2.6 | 0.499 | 13 | 1, 1 | −2.4 |
All nine subjects (A01, A02,  ) are shown. Columns show the 0.9 quantile of the classification accuracy p
0, linear model order p, bilinear model order q, and update coefficient logUC. The last row shows the results of the global optimization
) are shown. Columns show the 0.9 quantile of the classification accuracy p
0, linear model order p, bilinear model order q, and update coefficient logUC. The last row shows the results of the global optimization
Table 3.
Results of parameter optimization for AR-based methods xUAR, xVAR, and xBAR (including the prediction error variance feature)
| |
xUAR | xVAR | xBAR | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p 0 | p | log(UC) | p 0 | p | log(UC) | p 0 | p | q | log(UC) | |
| A01 | 0.619 | 12 | −2.6 | 0.626 | 4 | −2.6 | 0.619 | 13 | 1, 1 | −2.6 |
| A02 | 0.509 | 8 | −2.8 | 0.506 | 4 | −3.0 | 0.509 | 8 | 1, 1 | −2.8 |
| A03 | 0.654 | 5 | −2.6 | 0.651 | 2 | −2.8 | 0.651 | 5 | 1, 1 | −2.6 |
| A04 | 0.410 | 18 | −2.0 | 0.400 | 3 | −2.0 | 0.425 | 15 | 2, 2 | −2.2 |
| A05 | 0.418 | 2 | −2.8 | 0.410 | 6 | −2.4 | 0.414 | 5 | 1, 2 | −2.2 |
| A06 | 0.436 | 2 | −2.2 | 0.434 | 13 | −2.2 | 0.457 | 15 | 1, 2 | −2.6 |
| A07 | 0.556 | 14 | −2.6 | 0.541 | 13 | −2.4 | 0.563 | 15 | 2, 3 | −2.0 |
| A08 | 0.654 | 16 | −2.4 | 0.677 | 4 | −2.6 | 0.639 | 4 | 1, 1 | −2.6 |
| A09 | 0.629 | 3 | −2.2 | 0.653 | 3 | −2.0 | 0.640 | 7 | 1, 2 | −2.2 |
| Global | 0.511 | 13 | −2.6 | 0.518 | 4 | −2.4 | 0.513 | 12 | 1, 1 | −2.4 |
The notation is the same as in Table 2
Fig. 1.
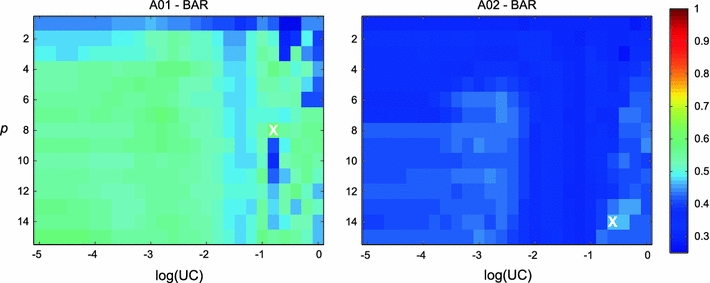
Optimization results for subjects A01 (left) and A02 (right) for BAR with the best bilinear model order q. Maps show the 0.9 quantile of the classification accuracy for all parameter combinations of log(UC) (x-axis) and model order p (y-axis). The white cross marks the location of the maximum
Finally, note that we used the achieved classification accuracies only within our optimization procedure. We report it here only for the sake of completeness, and stress that we did not use these accuracies for evaluation of the methods. The evaluation results are described in the next section.
Evaluation
Using the optimal parameter combinations found in the optimization step, we evaluated the methods on the second session. Table 4 shows the results for the ST analysis, whereas Table 5 shows the cross-validated (XV) results. As expected, classification accuracies are generally higher in the cross-validated case than in the ST analysis. In both cases, there is no obvious difference in the means for the individual and global optimization. The following paragraphs describe the outcomes of the statistical analyses.
Table 4.
ST evaluation results (0.9 quantile of the classification accuracy) for each feature extraction method and optimization strategy on the second session
| |
Individual | Global | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UAR | xUAR | VAR | xVAR | BAR | xBAR | LogBP | UAR | xUAR | VAR | xVAR | BAR | xBAR | LogBP | |
| A01 | 0.471 | 0.571 | 0.521 | 0.550 | 0.275 | 0.600 | 0.650 | 0.554 | 0.611 | 0.521 | 0.575 | 0.511 | 0.593 | 0.596 |
| A02 | 0.340 | 0.376 | 0.351 | 0.372 | 0.294 | 0.351 | 0.340 | 0.312 | 0.379 | 0.390 | 0.411 | 0.326 | 0.390 | 0.351 |
| A03 | 0.357 | 0.555 | 0.452 | 0.529 | 0.379 | 0.548 | 0.645 | 0.360 | 0.467 | 0.563 | 0.599 | 0.419 | 0.511 | 0.601 |
| A04 | 0.273 | 0.282 | 0.291 | 0.379 | 0.282 | 0.273 | 0.410 | 0.260 | 0.282 | 0.317 | 0.292 | 0.269 | 0.300 | 0.441 |
| A05 | 0.258 | 0.298 | 0.273 | 0.258 | 0.273 | 0.265 | 0.287 | 0.291 | 0.291 | 0.258 | 0.276 | 0.273 | 0.284 | 0.305 |
| A06 | 0.374 | 0.308 | 0.350 | 0.336 | 0.369 | 0.369 | 0.369 | 0.360 | 0.355 | 0.294 | 0.318 | 0.369 | 0.355 | 0.369 |
| A07 | 0.239 | 0.239 | 0.239 | 0.326 | 0.239 | 0.239 | 0.395 | 0.239 | 0.290 | 0.239 | 0.250 | 0.239 | 0.264 | 0.471 |
| A08 | 0.467 | 0.407 | 0.581 | 0.567 | 0.563 | 0.533 | 0.641 | 0.481 | 0.481 | 0.548 | 0.585 | 0.552 | 0.504 | 0.641 |
| A09 | 0.498 | 0.498 | 0.487 | 0.597 | 0.327 | 0.498 | 0.608 | 0.259 | 0.304 | 0.380 | 0.517 | 0.270 | 0.430 | 0.601 |
| Mean | 0.364 | 0.393 | 0.394 | 0.435 | 0.333 | 0.408 | 0.483 | 0.346 | 0.384 | 0.390 | 0.425 | 0.359 | 0.403 | 0.486 |
| SD | 0.10 | 0.12 | 0.12 | 0.13 | 0.10 | 0.14 | 0.15 | 0.11 | 0.11 | 0.13 | 0.15 | 0.11 | 0.11 | 0.13 |
The last two rows show the mean and standard deviation (SD)
Table 5.
Cross-validated evaluation results (0.9 quantile of the classification accuracy) for each feature extraction method and optimization strategy on the second session
| |
Individual | Global | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UAR | xUAR | VAR | xVAR | BAR | xBAR | LogBP | UAR | xUAR | VAR | xVAR | BAR | xBAR | LogBP | |
| A01 | 0.621 | 0.664 | 0.611 | 0.629 | 0.318 | 0.657 | 0.650 | 0.639 | 0.664 | 0.611 | 0.646 | 0.618 | 0.664 | 0.614 |
| A02 | 0.382 | 0.420 | 0.427 | 0.444 | 0.299 | 0.392 | 0.375 | 0.406 | 0.410 | 0.417 | 0.392 | 0.385 | 0.406 | 0.377 |
| A03 | 0.502 | 0.603 | 0.610 | 0.658 | 0.518 | 0.603 | 0.680 | 0.496 | 0.599 | 0.627 | 0.647 | 0.518 | 0.610 | 0.603 |
| A04 | 0.425 | 0.463 | 0.408 | 0.421 | 0.379 | 0.454 | 0.458 | 0.430 | 0.451 | 0.391 | 0.405 | 0.421 | 0.434 | 0.524 |
| A05 | 0.411 | 0.377 | 0.406 | 0.400 | 0.364 | 0.363 | 0.293 | 0.375 | 0.367 | 0.404 | 0.407 | 0.393 | 0.389 | 0.329 |
| A06 | 0.332 | 0.355 | 0.309 | 0.356 | 0.326 | 0.333 | 0.424 | 0.343 | 0.350 | 0.358 | 0.378 | 0.356 | 0.343 | 0.424 |
| A07 | 0.489 | 0.504 | 0.482 | 0.518 | 0.496 | 0.411 | 0.418 | 0.486 | 0.507 | 0.532 | 0.543 | 0.493 | 0.500 | 0.489 |
| A08 | 0.620 | 0.612 | 0.642 | 0.645 | 0.599 | 0.609 | 0.647 | 0.600 | 0.598 | 0.645 | 0.632 | 0.602 | 0.602 | 0.643 |
| A09 | 0.599 | 0.613 | 0.619 | 0.646 | 0.608 | 0.615 | 0.615 | 0.581 | 0.577 | 0.672 | 0.683 | 0.581 | 0.570 | 0.624 |
| Mean | 0.487 | 0.512 | 0.502 | 0.524 | 0.434 | 0.493 | 0.507 | 0.484 | 0.503 | 0.517 | 0.526 | 0.485 | 0.502 | 0.514 |
| SD | 0.11 | 0.11 | 0.12 | 0.12 | 0.12 | 0.13 | 0.14 | 0.10 | 0.11 | 0.13 | 0.13 | 0.10 | 0.11 | 0.12 |
The last two rows show the mean and standard deviation (SD)
Overall comparison
A two-way repeated measures ANOVA for the ST case (factors “method” and “optimization”) found a significant main effect of “method” (F 6,48 = 8.104, Greenhouse-Geisser-adjusted P < 0.01). A Newman–Keuls post-hoc test found that logBP is significantly better than all six AR-based methods (mean classification accuracies of 0.355, 0.389, 0.392, 0.430, 0.346, 0.406, and 0.485 for UAR, xUAR, VAR, xVAR, BAR, xBAR, and logBP, respectively). Furthermore, xVAR is significantly better than both UAR and BAR. The factor “optimization” was not significant (F 1,8 = 0.030, P = 0.87).
In the XV case, an ANOVA with the same factors as in the ST analysis also found a significant main effect of “method” (F 6,48 = 3.247, P < 0.01). A Newman–Keuls post-hoc test revealed that BAR (mean accuracy of 0.460) is significantly worse than xUAR, VAR, xVAR, and logBP (mean accuracies of 0.507, 0.509, 0.525, and 0.510, respectively). Again, the factor “optimization” was not significant (F 1,8 = 2.901, P = 0.13).
We also conducted a repeated measures ANOVAs as described above for the combined evaluation results (that is, we combined ST and XV results and introduced a new factor “ST/XV”). This analysis yielded significant main effects “ST/XV” (F 1,8 = 22.797, P < 0.01) and “method” (F 6,48 = 6.700, P < 0.01), as well as a significant interaction between “ST/XV” and “method” (F 6,48 = 5.746, Greenhouse-Geisser-adjusted P < 0.01). Post-hoc tests showed that XV results (mean accuracy 0.499) are significantly higher than ST results (0.400). Furthermore, logBP yielded significantly higher results than UAR, VAR, BAR, and xBAR. BAR was significantly worse than xUAR, VAR, xVAR, and logBP. Finally, xVAR was significantly better than UAR. The mean accuracies for UAR, xUAR, VAR, xVAR, BAR, xBAR, and logBP were 0.420, 0.448, 0.451, 0.477, 0.403, 0.452, and 0.497, respectively.
Comparison of AR-based methods
We also analyzed the six AR-based methods in more detail and performed three-way repeated measures ANOVAs (factors “method”, “x”, and “optimization”). In the ST case, we found significant main effects of “method” (F 2,16 = 3.939, P < 0.05) and “x” (F 1,8 = 6.324, P < 0.05). Post-hoc tests revealed that vector methods (mean accuracy of 0.411) are significantly better than bilinear methods (mean accuracy of 0.376). Furthermore, methods including the prediction error variance are significantly better (mean accuracy 0.408) than their counterparts without this additional feature (mean accuracy 0.364). In the XV case, we found a significant main effect of “method” (F 2,16 = 6.753, P < 0.01). Post-hoc tests showed that vector models (mean accuracy of 0.517) are significantly better than bilinear models (mean accuracy of 0.479).
Finally, we analyzed the six AR methods for the combined ST and XV results (by introducing the factor “ST/XV”). We found significant main effects of “ST/XV” (F 1,8 = 20.604, P < 0.01), “method” (F 2,16 = 5.597, P < 0.05), and “x” (F 1,8 = 6.778, P < 0.05). Post-hoc tests showed that cross-validated results (mean accuracy 0.497) were significantly higher than ST results (mean accuracy 0.386). Furthermore, vector models (mean accuracy of 0.464) were significantly better than both univariate and bilinear models (mean accuracies of 0.434 and 0.427, respectively). Finally, results were significantly higher for methods using the prediction error variance feature (mean accuracy of 0.459) compared to methods that did not use this feature (mean accuracy of 0.425).
Discussion
In summary, logBP features yielded the highest classification results in this study. In the ST analysis, where features and classifiers are determined on the first session and then applied to the second (completely unseen) session, logBP was significantly better than all AR-based methods. When assessing this result in more detail, we found out that it might be due to our optimization and evaluation procedure, which resembles a practical BCI setup. In such a setup, users control the BCI in different sessions on different days, and only data from previous sessions can be used to tune parameters. However, this only works if the features are stable over sessions, that is, the bias of the classifiers does not change significantly. In fact, it turned out that all AR methods led to a much higher bias in the second session compared to logBP features, where the bias was about as small as in the first session. A statistical analysis comparing all feature extraction methods after adapting the bias in the second session resulted in no significant differences in the ST analysis. Therefore, adapting the bias of the classifier [15] or using adaptive classifiers [12, 38, 41] to improve ST is necessary for AR features.
Due to the high dimensionality of the feature space in our globally optimized features (see Tables 2, 3), and because similarly high classification accuracies could be obtained for lower model orders in the optimization step, we assessed the performance of univariate models with a lower model order of p = 5 for all subjects. It turned out that classification accuracies improved slightly, but statistical analyses showed that the overall results did not change. That is, all results described above are also valid for univariate models with lower model orders. Therefore, we can safely rule out overfitting effects that might have explained the inferior performance of (univariate) AR models, especially in the ST analysis. Other studies such as [20] have also found similarly high or higher model orders (although they did not use AR coefficients directly for classification, but calculated the power spectrum).
Furthermore, we have shown that optimizing parameters for individual subjects does not result in better classification rates. Indeed, there was no significant difference between globally and individually optimized parameters. This implies that using logBP with default bands (8–12 and 16–24 Hz) works as well as with subject-specific bands. Note that we used bipolar channels in this study, which is very common in BCI research [1, 6, 16, 17, 23, 32, 31, 40]. Had we used subject-specific spatial filters such as CSP, subject-specific bands might have yielded better results than default bands [4].
The comparison of all analyzed AR methods showed that vector models yielded higher classification results than both univariate and bilinear models. On the one hand, this is not surprising, because vector models consider more information, namely the relationships between individual signals. On the other hand, the potentially more accurate signal description with bilinear models could not be translated into improved classification results. This could be due to two reasons: first, the EEG might not contain signal characteristics that cannot be described by linear models; or second, although bilinear signal properties might improve the model fit, they do not contribute discriminative information for BCIs.
Clearly, all AR methods benefited from the inclusion of the prediction error variance as an additional feature. This feature makes initialization of parameters even more important, because the prediction error variance is updated directly with the update coefficient UC. Without initialization to suitable values, it would take a long time until this feature was in its operating range. This underscores the importance of estimating good initial values, for example with a first run over the optimization data set as implemented in our study.
In conclusion, logBP is superior to AR-based methods, at least with the procedure and implementation used in this study. However, as described above, the performance of AR features can be improved when adapting the bias of the classifiers in new sessions [21, 41]. We also found that low model orders generalized better, and the high model orders determined in our optimization step on the first session resulted in significantly lower classification accuracies on the unseen second session. Moreover, for the settings used in this study (which is very common in BCI experiments), it is not necessary to optimize features for each user individually globally optimized parameters for all users yield equally high classification rates. In particular, we recommend using low model orders (such as a model order of 5) for univariate models to ensure generalization of the features. Finally, vector models should be preferred over univariate models, and the prediction error variance improved classification performance of all AR models. Future study should apply these findings to online BCIs, where users receive feedback based on their brain patterns, for example to control a prosthesis [13]. Although we are confident that our results will generalize to online sessions with feedback, we are currently working on an online study to verify our findings. Another follow-up study could explore the combination of AR and logBP features to assess whether they contain complimentary information on the data.
Acknowledgments
This study was supported by the Austrian Science Fund FWF, P20848-N15 (Coupling Measures in BCIs), PASCAL-2 ICT-216886, and MU 987/3-2. This publication only reflects the authors’ views. Funding agencies are not liable for any use that may be made of the information contained herein. We would also like to thank Alois Schlögl for helpful discussions and Vera Kaiser for support with statistical questions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
References
- 1.Allison BZ, Brunner C, Kaiser V, Müller-Putz GR, Neuper C, Pfurtscheller G. Toward a hybrid brain–computer interface based on imagined movement and visual attention. J Neural Eng. 2010;7:026007. doi: 10.1088/1741-2560/7/2/026007. [DOI] [PubMed] [Google Scholar]
- 2.Anderson CW, Stolz EA, Shamsunder S. Multivariate autoregressive models for classification of spontaneous electroencephalographic signals during mental tasks. IEEE Trans Biomed Eng. 1998;45:277–286. doi: 10.1109/10.661153. [DOI] [PubMed] [Google Scholar]
- 3.Billinger M, Kaiser V, Neuper C, Brunner C (2011) Automatic frequency band selection for BCIs with ERDS difference maps. In: Proceedings of the fifth international brain–vomputer interface conference. Graz
- 4.Blankertz B, Tomioka R, Lemm S, Kawanabe M, Müller KR. Optimizing spatial filters for robust EEG single-trial analysis. IEEE Signal Process Mag. 2008;25:41–56. doi: 10.1109/MSP.2008.4408441. [DOI] [Google Scholar]
- 5.Brunner C, Scherer R, Graimann B, Supp G, Pfurtscheller G. Online control of a brain–computer interface using phase synchronization. IEEE Trans Biomed Eng. 2006;53:2501–2506. doi: 10.1109/TBME.2006.881775. [DOI] [PubMed] [Google Scholar]
- 6.Brunner C, Allison BZ, Krusienski DJ, Kaiser V, Müller-Putz GR, Pfurtscheller G, Neuper C. Improved signal processing approaches in an offline simulation of a hybrid brain–computer interface. J Neurosci Methods. 2010;188:165–173. doi: 10.1016/j.jneumeth.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrera AF, Farina D, Dremstrup K. Comparison of feature selection and classification methods for a brain–computer interface driven by non-motor imagery. Med Biol Eng Comput. 2010;48:123–132. doi: 10.1007/s11517-009-0569-2. [DOI] [PubMed] [Google Scholar]
- 8.Dias NS, Kamrunnahar M, Mendes PM, Schiff SJ, Correia JH. Feature selection on movement imagery discrimination and attention detection. Med Biol Eng Comput. 2010;48:331–341. doi: 10.1007/s11517-010-0578-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett D, Peterson DA, Anderson CW, Thaut MH. Comparison of linear, nonlinear, and feature selection methods for EEG signal classification. IEEE Trans Neural Syst Rehabil Eng. 2003;11:141–144. doi: 10.1109/TNSRE.2003.814441. [DOI] [PubMed] [Google Scholar]
- 10.Gastaut H, Dongier M, Courtois G. On the significance of “wicket rhythms” (“rhythmes en arceau”) in psychosomatic medicine. Electroencephalogr Clin Neurophysiol. 1954;6:687. doi: 10.1016/0013-4694(54)90099-1. [DOI] [Google Scholar]
- 11.Graimann B, Huggins JE, Levine SP, Pfurtscheller G. Toward a direct brain interface based on human subdural recordings and wavelet-packet analysis. IEEE Trans Biomed Eng. 2004;51:954–962. doi: 10.1109/TBME.2004.826671. [DOI] [PubMed] [Google Scholar]
- 12.Hasan BAS, Gan JQ. Unsupervised movement onset detection from EEG recorded during self-paced real hand movement. Med Biol Eng Comput. 2010;48:245–253. doi: 10.1007/s11517-009-0550-0. [DOI] [PubMed] [Google Scholar]
- 13.Horki P, Solis-Escalante T, Neuper C, Müller-Putz G. Combined motor imagery and SSVEP based BCI control of a 2 DoF artificial upper limb. Med Biol Eng Comput. 2011;49:567–577. doi: 10.1007/s11517-011-0750-2. [DOI] [PubMed] [Google Scholar]
- 14.Kalman RE. A new approach to linear filtering and prediction problems. Trans ASME J Basic Eng. 1960;82:35–45. [Google Scholar]
- 15.Krauledat M, Tangermann M, Blankertz B, Müller KR. Towards zero training for brain–computer interfacing. PLoS ONE. 2008;3:e2967. doi: 10.1371/journal.pone.0002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leeb R, Lee F, Keinrath C, Scherer R, Bischof H, Pfurtscheller G. Brain–computer communication: motivation, aim and impact of exploring a virtual apartment. IEEE Trans Neural Syst Rehabil Eng. 2007;15:473–482. doi: 10.1109/TNSRE.2007.906956. [DOI] [PubMed] [Google Scholar]
- 17.Leeb R, Settgast V, Fellner DW, Pfurtscheller G. Self-paced exploring of the Austrian National Library through thoughts. Int J Bioelectromagn. 2007;9:237–244. [Google Scholar]
- 18.Lotte F, Congedo M, Lécuyer A, Lamarche F, Arnaldi B. A review of classification algorithms for EEG-based brain–computer interfaces. J Neural Eng. 2007;4:R1–R13. doi: 10.1088/1741-2560/4/2/R01. [DOI] [PubMed] [Google Scholar]
- 19.Mason SG, Bashashati A, Fatourechi M, Navarro KF, Birch GE. A comprehensive survey of brain interface technology designs. Ann Biomed Eng. 2007;35:137–169. doi: 10.1007/s10439-006-9170-0. [DOI] [PubMed] [Google Scholar]
- 20.McFarland DJ, Wolpaw JR. Sensorimotor rhythm-based brain–computer interface (BCI): model order selection for autoregressive spectral analysis. J Neural Eng. 2008;5:155–162. doi: 10.1088/1741-2560/5/2/006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McFarland DJ, Krusienski DJ, Sarnacki WA, Wolpaw JR. Emulation of computer mouse control with a noninvasive brain–computer interface. J Neural Eng. 2008;5:101–110. doi: 10.1088/1741-2560/5/2/001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller-Gerking J, Pfurtscheller G, Flyvbjerg H. Classification of movement-related EEG in a memorized delay task experiment. Clin Neurophysiol. 2000;111:1353–1365. doi: 10.1016/S1388-2457(00)00345-X. [DOI] [PubMed] [Google Scholar]
- 23.Müller-Putz GR, Scherer R, Pfurtscheller G, Rupp R. EEG-based neuroprosthesis control: a step towards clinical practice. Neurosci Lett. 2005;382:169–174. doi: 10.1016/j.neulet.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 24.Pei XM, Zheng CX (2004) Feature extraction and classification of brain motor imagery task based on MVAR model. In: Proceedings of the third international conference on machine learning and cybernetics doi:10.1109/ICMLC.2004.1380465
- 25.Pfurtscheller G, Kalcher J, Neuper C, Flotzinger D, Pregenzer M. On-line EEG classification during externally-paced hand movements using a neural network-based classifier. Electroencephalogr Clin Neurophysiol. 1996;99:416–425. doi: 10.1016/S0013-4694(96)95689-8. [DOI] [PubMed] [Google Scholar]
- 26.Pfurtscheller G, Neuper C, Flotzinger D, Pregenzer M. EEG-based discrimination between imagination of right and left hand movement. Electroencephalogr Clin Neurophysiol. 1997;103:642–651. doi: 10.1016/S0013-4694(97)00080-1. [DOI] [PubMed] [Google Scholar]
- 27.Pfurtscheller G, Allison BZ, Brunner C, Bauernfeind G, Solis-Escalante T, Scherer R, Zander TO, Müller-Putz G, Neuper C, Birbaumer N. The hybrid BCI. Front Neurosci. 2010;4:30. doi: 10.3389/fnpro.2010.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfurtscheller G, Brunner C, Leeb R, Scherer R, Müller-Putz GR, Neuper C (2010b) The Graz brain–computer interface. In: Graimann B, Allison BZ, Pfurtscheller G (eds) Brain–computer interfaces: revolutionizing human–computer interaction. Springer, Berlin, pp 79–96
- 29.Priestley MB (1988) Non-linear and non-stationary time series analysis. Academic Press, London
- 30.Sajda P, Gerson A, Müller KR, Blankertz B, Parra L. A data analysis competition to evaluate machine learning algorithms for use in brain–computer interfaces. IEEE Trans Neural Syst Rehabil Eng. 2003;11:184–185. doi: 10.1109/TNSRE.2003.814453. [DOI] [PubMed] [Google Scholar]
- 31.Scherer R, Müller GR, Neuper C, Graimann B, Pfurtscheller G. An asynchronously controlled EEG-based virtual keyboard: improvement of the spelling rate. IEEE Trans Neural Syst Rehabil Eng. 2004;51:979–984. doi: 10.1109/TBME.2004.827062. [DOI] [PubMed] [Google Scholar]
- 32.Scherer R, Lee F, Schlögl A, Leeb R, Bischof H, Pfurtscheller G. Toward self-paced brain–computer communication: navigation through virtual worlds. IEEE Trans Biomed Eng. 2008;55:675–682. doi: 10.1109/TBME.2007.903709. [DOI] [PubMed] [Google Scholar]
- 33.Schlögl A, Brunner C. BioSig: a free and open source software library for BCI research. IEEE Comput Mag. 2008;41:44–50. [Google Scholar]
- 34.Schlögl A, Supp G (2006) Analyzing event-related EEG data with multivariate autoregressive parameters. In: Neuper C, Klimesch W (eds) Event-related dynamics of brain oscillations. Elsevier, Amsterdam, pp 135–147 [DOI] [PubMed]
- 35.Schlögl A, Flotzinger D, Pfurtscheller G. Adaptive autoregressive modeling used for single-trial EEG classification. Biomed Tech. 1997;42:162–167. doi: 10.1515/bmte.1997.42.6.162. [DOI] [PubMed] [Google Scholar]
- 36.Schlögl A, Vidaurre C, Müller KR (2010) Adaptive methods in BCI research—an introductory tutorial. In: Graimann B, Allison BZ, Pfurtscheller G (eds) Brain–computer interfaces: revolutionizing human–computer interaction. Springer, Berlin, pp 331–355
- 37.Townsend G, Graimann B, Pfurtscheller G. Continuous EEG classification during motor imagery—simulation of an asynchronous BCI. IEEE Trans Neural Syst Rehabil Eng. 2004;12:258–265. doi: 10.1109/TNSRE.2004.827220. [DOI] [PubMed] [Google Scholar]
- 38.Tsui CSL, Gan JQ, Roberts SJ. A self-paced brain–computer interface for controlling a robot simulator: an online event labelling paradigm and an extended Kalman filter based algorithm for online training. Med Biol Eng Comput. 2009;47:257–265. doi: 10.1007/s11517-009-0459-7. [DOI] [PubMed] [Google Scholar]
- 39.Vidaurre C, Schlögl A, Cabeza R, Scherer R, Pfurtscheller G. Adaptive on-line classification for EEG-based brain computer interfaces with AAR parameters and band power estimates. Biomed Tech. 2005;50:350–354. doi: 10.1515/BMT.2005.049. [DOI] [PubMed] [Google Scholar]
- 40.Vidaurre C, Scherer R, Cabeza R, Schlögl A, Pfurtscheller G. Study of discriminant analysis applied to motor imagery bipolar data. Med Biol Eng Comput. 2007;45:61–68. doi: 10.1007/s11517-006-0122-5. [DOI] [PubMed] [Google Scholar]
- 41.Vidaurre C, Schlögl A, Cabeza R, Scherer R, Pfurtscheller G. Study of on-line adaptive discriminant analysis for EEG-based brain computer interfaces. IEEE Trans Biomed Eng. 2007;54:550–556. doi: 10.1109/TBME.2006.888836. [DOI] [PubMed] [Google Scholar]
- 42.Vidaurre C, Krämer N, Blankertz B, Schlögl A. Time domain parameters as a feature for EEG-based brain computer interfaces. Neural Netw. 2009;22:1313–1319. doi: 10.1016/j.neunet.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 43.Wolpaw JR, Birbaumer N, McFarland DJ, Pfurtscheller G, Vaughan TM. Brain–computer interfaces for communication and control. Clin Neurophysiol. 2002;113:767–791. doi: 10.1016/S1388-2457(02)00057-3. [DOI] [PubMed] [Google Scholar]


