Abstract
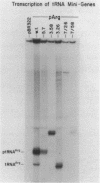
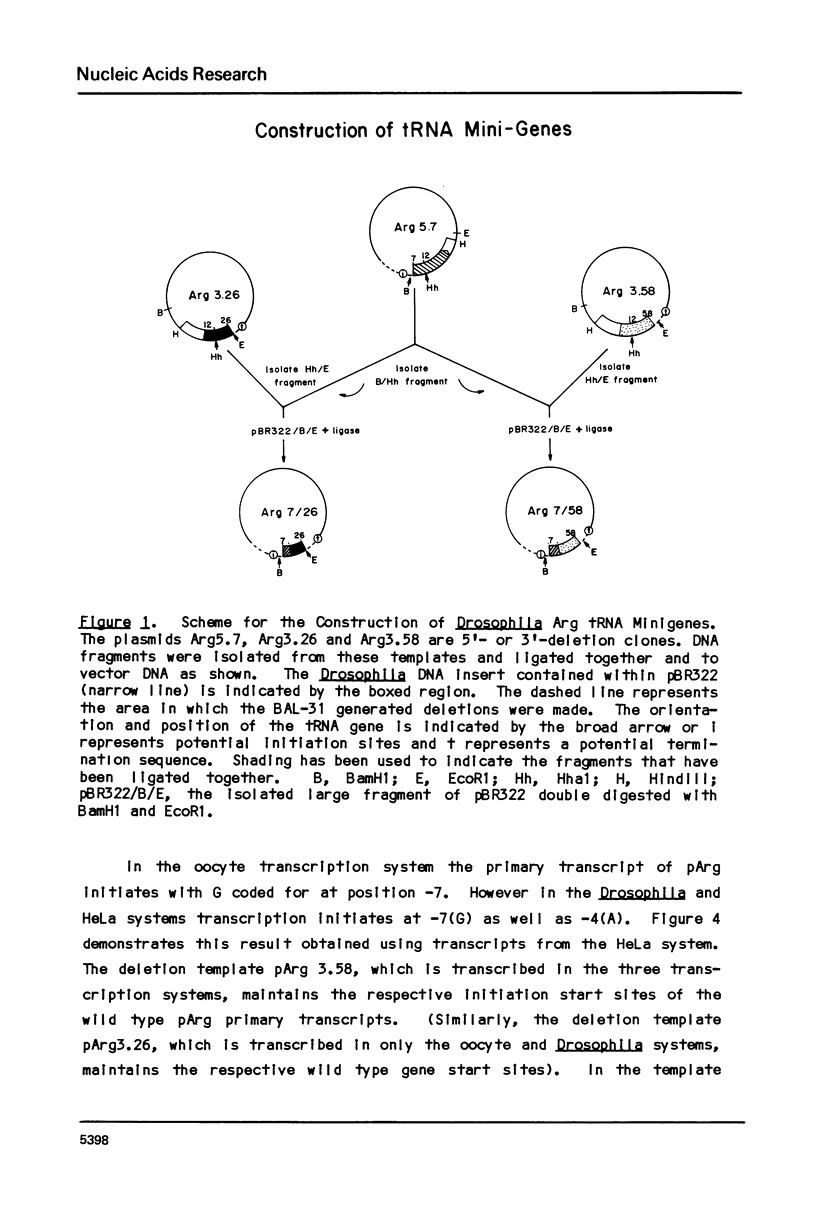
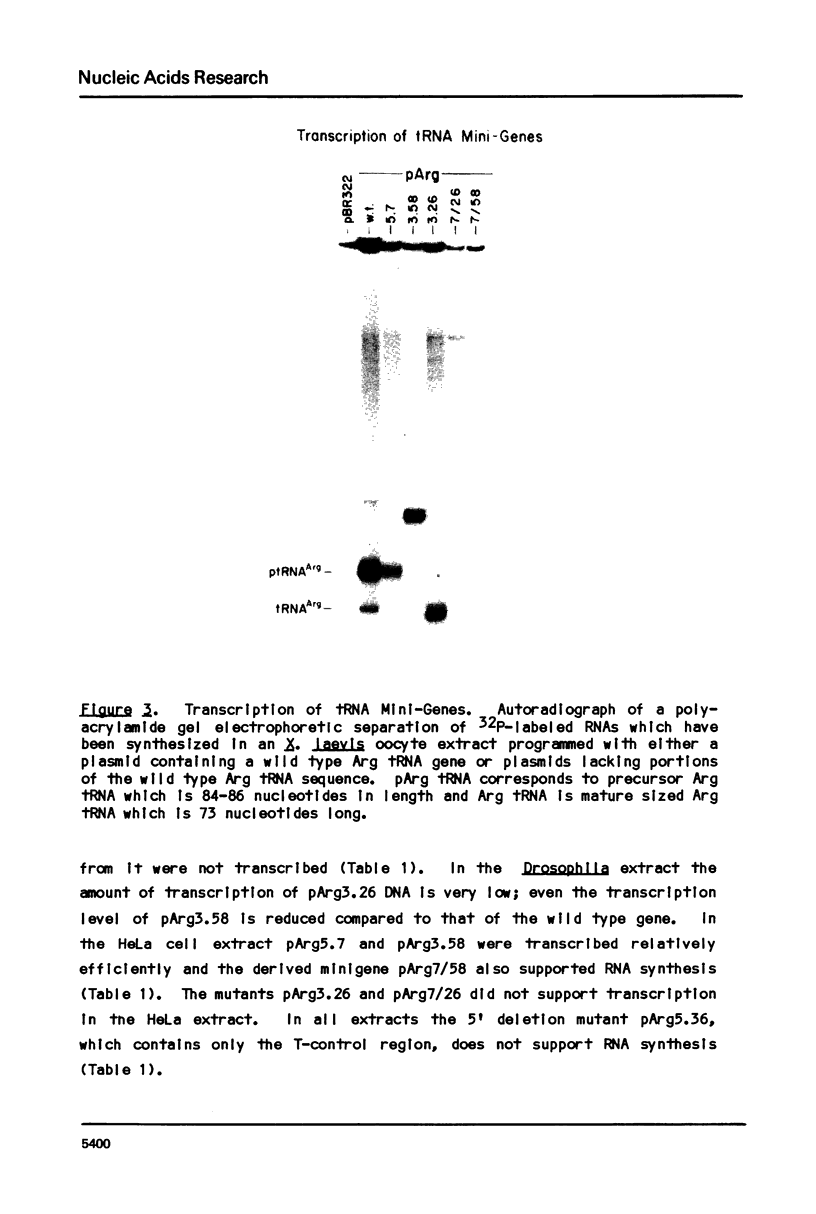
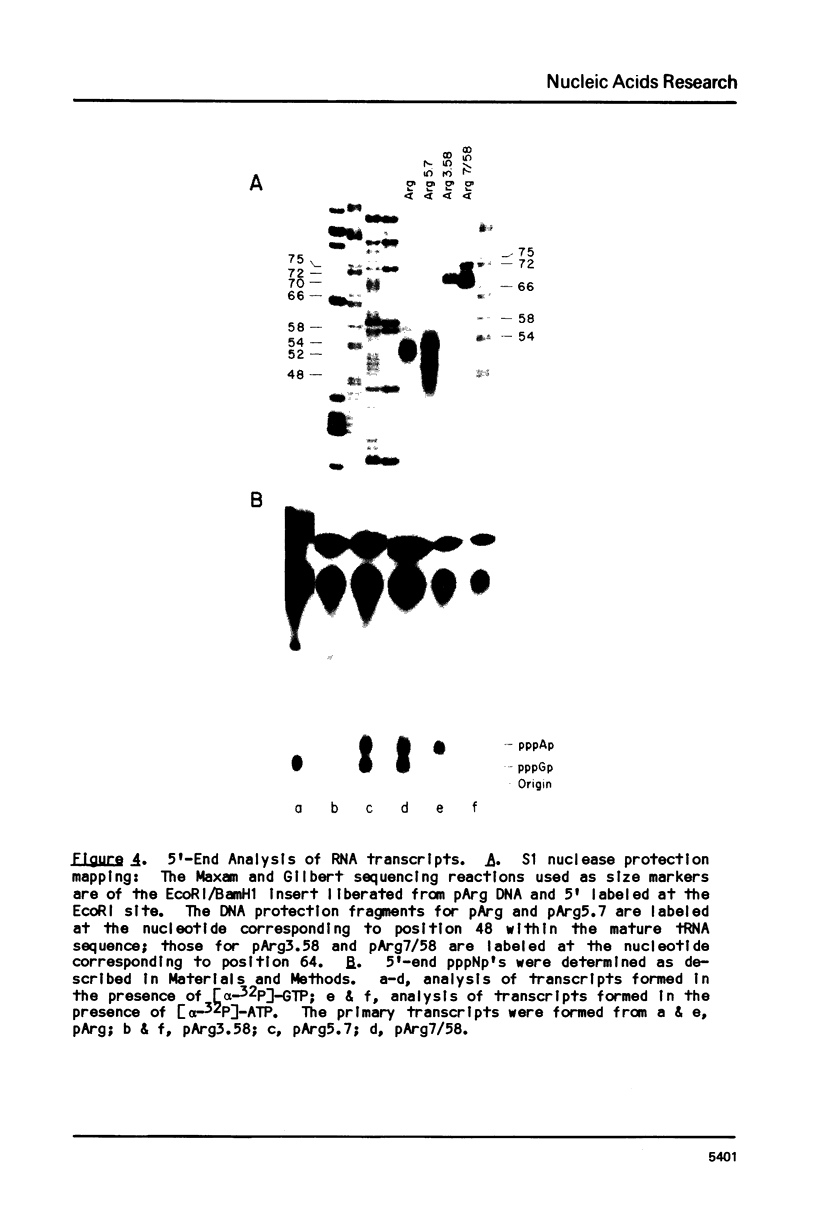
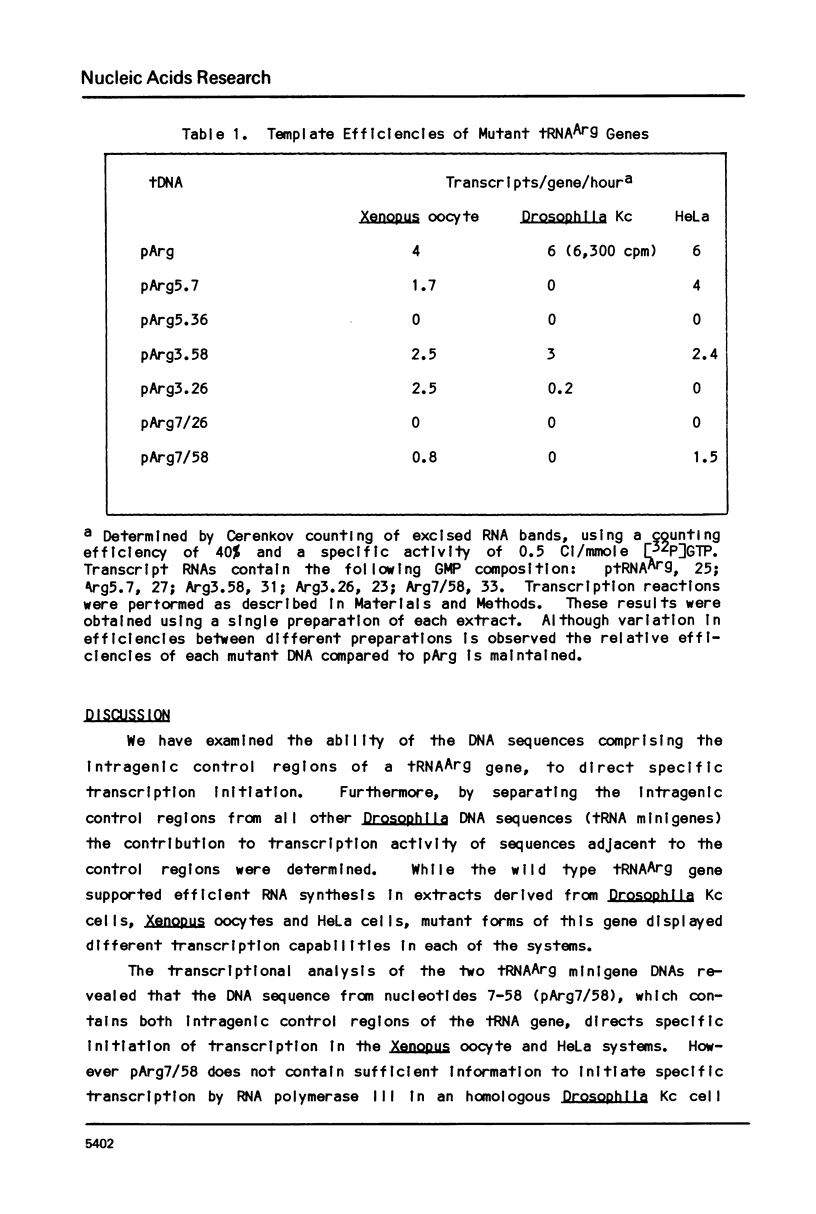
Transcription of eukaryotic tRNA genes is controlled by two intragenic regions, the D-control region (which in the tRNA codes for the D-stem and -loop) and the T-control region (which in the tRNA codes for the T psi C loop). To determine whether these sequences alone are sufficient to promote tRNA gene transcription in vitro, the two control regions of a Drosophila tRNAArg gene were cloned separately from the context of the parental DNA (these constructions are called tRNA minigenes). The tRNA minigene that contains both intragenic control regions supports in vitro RNA synthesis in Xenopus laevis oocyte and HeLa cell transcription systems. The mutant which has deletions to nucleotide 7 within the mature tRNA coding region, pArg5.7, and minigenes derived from it do not support RNA synthesis in a Drosophila Kc cell transcription system. Xenopus and Hela extracts transcribe pArg5.7 albeit at reduced levels compared to the wild-type gene. The tRNA minigene that contained only the D-control region was not able to support RNA synthesis in any of these three transcription systems. A mutant tRNA gene comprising the 3' half of the tRNAArg gene similarly was not able to support RNA synthesis. These experiments show that the DNA sequence from nucleotides 7-58, which contains both intragenic control regions of the tRNA gene, possesses sufficient information to initiate specific transcription by RNA polymerase III in Xenopus and HeLa systems. The transcription efficiency of this tRNA minigene however is reduced to about 20% the transcription level of the wild type tRNA gene. This lowered level of transcriptional efficiency results from deleting the ends of the native tRNA gene and its adjacent flanking sequences. The affects of deleting 5' sequences are most pronounced in the Drosophila transcription system.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Melton D. A., Cortese R. Promoter of a eukaryotic tRNAPro gene is composed of three noncontiguous regions. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1195–1199. doi: 10.1073/pnas.79.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingermann T., Sharp S., Appel B., DeFranco D., Mount S., Heiermann R., Pongs O., Söll D. Transcription of cloned tRNA and 5S RNA genes in a Drosophila cell free extract. Nucleic Acids Res. 1981 Aug 25;9(16):3907–3918. doi: 10.1093/nar/9.16.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Fuhrman S. A., Deininger P. L., LaPorte P., Friedmann T., Geiduschek E. P. Analysis of transcription of the human Alu family ubiquitous repeating element by eukaryotic RNA polymerase III. Nucleic Acids Res. 1981 Dec 11;9(23):6439–6456. doi: 10.1093/nar/9.23.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Guilfoyle R., Weinmann R. Control region for adenovirus VA RNA transcription. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3378–3382. doi: 10.1073/pnas.78.6.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter H., Kressman A., Birnstiel M. L. A split promoter for a eucaryotic tRNA gene. Cell. 1981 May;24(2):573–585. doi: 10.1016/0092-8674(81)90348-2. [DOI] [PubMed] [Google Scholar]
- Kressmann A., Hofstetter H., Di Capua E., Grosschedl R., Birnstiel M. L. A tRNA gene of Xenopus laevis contains at least two sites promoting transcription. Nucleic Acids Res. 1979 Dec 11;7(7):1749–1763. doi: 10.1093/nar/7.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P. P., Gray H. B., Jr Extracellular nucleases of Alteromonas espejiana BAL 31.IV. The single strand-specific deoxyriboendonuclease activity as a probe for regions of altered secondary structure in negatively and positively supercoiled closed circular DNA. Nucleic Acids Res. 1979 Jan;6(1):331–357. doi: 10.1093/nar/6.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ng S. Y., Parker C. S., Roeder R. G. Transcription of cloned Xenopus 5S RNA genes by X. laevis RNA polymerase III in reconstituted systems. Proc Natl Acad Sci U S A. 1979 Jan;76(1):136–140. doi: 10.1073/pnas.76.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Dingermann T., Farrell P., Söll D. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6657–6661. doi: 10.1073/pnas.78.11.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Silberklang M., Hosbach H. A., Schmidt T., Kubli E., Gergen J. P., Wensink P. C., Söll D. The initiator tRNA genes of Drosophila melanogaster: evidence for a tRNA pseudogene. Nucleic Acids Res. 1981 Nov 25;9(22):5867–5882. doi: 10.1093/nar/9.22.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Walseth T. F., Johnson R. A. The enzymatic preparation of [alpha-(32)P]nucleoside triphosphates, cyclic [32P] AMP, and cyclic [32P] GMP. Biochim Biophys Acta. 1979 Mar 28;562(1):11–31. doi: 10.1016/0005-2787(79)90122-9. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]