Abstract
The human intestinal microbiota is a complex biological system comprising a vast repertoire of microbes with considerable metabolic activity relevant to both bacterial growth and host health. Greater strides have been made in the analysis of microbial diversity than in the measurement of functional activity, particularly in vivo. Stable isotope probing offers a new approach by coupling measurements of metabolic activity with microbial identification. Using a low-enrichment labeling strategy in vitro, this study has identified metabolically active bacterial groups via magnetic-bead capture methodology and stable isotope ratio analysis. Using five probes (EUB338, Bac303, Bif164, EREC482, and Clep866), changes in the activities of key intestinal microbial groups were successfully measured by exploiting tracers of de novo RNA synthesis. Perturbation of the nutrient source with oligofructose generated changes in the activity of bifidobacteria as expected, but also in the Bacteroides-Prevotella group, the Eubacterium rectale-Clostridium coccoides group, and the Clostridium leptum subgroup. Changes in activity were also observed in response to the medium type. This study suggests that changes in the functional activity of the gut microbiota can be assessed using tracers of de novo nucleic acid synthesis combined with measurement of low isotopic enrichment in 16S rRNA. Such tracers potentially limit substrate bias because they are universally available to bacteria. This low-enrichment labeling approach does not depend on the commercial availability of specific labeled substrates and can be easily translated to in vivo probing experiments of the functional activity of the microbiota in the human gut.
INTRODUCTION
The human gut is inhabited by a large and diverse population of bacteria, which play a major role in human health and well-being. The interaction between the gut bacteria and the host affects many physiological and nutritional host functions, as well as developmental and immunological processes. Great strides have been made toward understanding the diversity of the human gastrointestinal microbiota by using culture-dependent and molecular methods (7, 10, 19). Genomic and metagenomic analyses have revealed an extensive metabolic repertoire encoded in the gut microbiome. However, measurement of the function and metabolic activity of the intestinal bacteria has not kept pace with the growing knowledge of microbial diversity. Although the metabolic activity of some intestinal microbes is well characterized, our understanding of which components of the microbiome's metabolic machinery are activated in response to diet, luminal environment, and ecosystem changes is limited, with insufficient phylogenetic resolution. Increasing our knowledge of the link between microbial phylogeny and metabolic functionality and activity is vital in order to fully understand, and therefore optimize, the role of the intestinal microbiota in human health and disease.
The use of stable-isotope-labeled substrates to probe the microbial utilization of specific substrates has gained much attention in environmental microbial ecology (23) and, more recently, in human microbial ecology (2, 5, 12). This technique, known as stable-isotope probing (SIP), can provide insights into both microbial phylogeny and metabolic activity by tracking stable-isotope-labeled atoms from substrates into microbial biomarkers, such as phospholipid-derived fatty acids (PLFAs), DNA, and RNA. Although PFLA-SIP is potentially a sensitive approach, taxonomic identification of bacteria is difficult because PLFA profiles do not provide resolved phylogenetic information for most uncultivated microorganisms (27). Hence, SIP of nucleic acids has attracted much interest, since it offers the opportunity to link metabolic function with relevant phylogenetic and metabolic genes. Cell division is required to incorporate tracer atoms into DNA and is an important limitation of DNA-SIP, because the relatively low rate of tracer incorporation requires extended incubation times to reach an exploitable amount of isotope in DNA. RNA, with much higher turnover rates than DNA, is more responsive to changes in metabolic conditions and has important advantages over DNA-SIP. The faster turnover of RNA leads to faster incorporation of the isotope, so that the incubation times required to reach a measurable amount of isotope incorporated into RNA are shorter than those for DNA. When in vitro model systems of the human intestinal bacteria are used, rapid isotopic incorporation is highly advantageous, given the changes in both activity and diversity that occur once bacteria are voided from the intestine.
So far, RNA-SIP has been used in environmental science to explore plant-microorganism interactions and to identify bacteria responsible for the degradation of individual substrates, e.g., phenol, glucose, or acetate (4, 18, 24). One approach, combining RNA-SIP with magnetic-bead capture, has been developed and optimized to investigate microbial community function by studying the substrate utilization of sulfate-reducing bacteria in marine sediments (15, 16, 18). The selection of the stable-isotope-labeled substrate is a critical determinant of the information obtained from SIP experiments. Many substrates will be selectively utilized by bacteria or bacterial groups whose genes encode proteins that are able to utilize that substrate. Therefore, only bacteria that incorporate labeled atoms into their RNA, with sufficient enrichment, will be identified by this approach, potentially leading to substrate-induced bias in the results. Information about the overall metabolic activity of the microbial community may remain hidden. One major limitation to the wider exploitation of SIP in human microbial ecology is the availability of biologically relevant isotopically labeled substrates. Although monosaccharides such as [U-13C]glucose are readily available and economically viable for SIP experiments, such substrates will induce phylogenetic selectivity (bias) based on the ability of a bacterium to utilize the substrate. In reality, the human intestinal microbiota is fueled by a range of substrates derived mainly from polysaccharides, which are difficult or expensive to procure in sufficient quantities for SIP experiments. Hence, we have explored the use of “generic” tracers that limit phylogenetic selection bias, are economically viable for large-scale experiments, and allow the effects of unlabeled substrates and, importantly, environmental conditions to be investigated.
The objective of this study was therefore to develop a new approach to probing metabolically active bacteria belonging to different phylogenetic groups in humans, while limiting phylogenetic selection bias based on tracer choice. 13C-labeled precursors of de novo RNA synthesis were investigated via in vitro incubation of human gut bacteria. By analyzing the 13C enrichment of the 16S rRNA after using group-specific oligonucleotide probes (which have been established previously for fluorescent in situ hybridization [FISH] analysis) and magnetic-bead capture, this approach has provided information about the metabolic activities of different phylogenetic groups of the human intestinal microbiota in response to a nondigestible carbohydrate.
MATERIALS AND METHODS
Study design.
To determine if a pulse-chase tracer study design resulted in sufficient and reliable incorporation of 13C into bacterial RNA, 13C-labeled tracer precursors of de novo RNA synthesis were incubated with human fecal bacteria and oligofructose as a substrate. To determine which tracer substance leads to the highest level of incorporation of 13C into bacterial RNA, fermentation experiments with 13C-labeled urea, glycine, or NaHCO3 were carried out. [13C]glycine (1.5 mg/ml), NaH13CO3 (0.7 mg/ml), and [13C]urea (0.9 mg/ml) were incubated in fecal fermentation systems, and RNA was harvested and analyzed for isotopic enrichment. [13C]urea was of interest because it is a relatively cheap tracer; it is rapidly hydrolyzed by bacteria to H13CO3− and NH3, thus labeling the inorganic carbon pool; and it can be delivered to the human colon for potential in vivo SIP experiments (15). To explore whether a pulse-chase experiment with an isotopic tracer of de novo nucleic acid synthesis could yield information about bacterial activity, an experiment was conducted that examined the incorporation of an isotope into 16S rRNA in fecal samples incubated in different media with oligofructose as the substrate. The media examined were a mineral medium (MM) and Wilkins-Chalgren (WC) broth, and the carbohydrate investigated was oligofructose (Beneo P95; Beneo-Orafti, Tienen, Belgium). The conditions investigated were, therefore, the medium without oligofructose (MM − OF or WC − OF) and the medium with oligofructose added (MM + OF or WC + OF). All incubations were conducted in triplicate except for that with WC − OF, which was carried out in a single experimental vessel due to the limitations of downstream processing of multiple samples simultaneously.
Fecal-sample incubation and culture conditions.
Fecal samples were collected from a single healthy donor who had no history of gastrointestinal disorders and had not undergone antibiotic therapy within at least 6 months prior to the study. Incubation was carried out in gas-tight 100-ml glass vessels at a total volume of 30 ml under anaerobic conditions. The basal mineral medium (containing, per liter, 8.5 g NaCl, 0.3 g KH2PO4, 0.6 g Na2HPO4, 2.5 g NaCl, 0.24 g MgSO4, 0.011 g CaCl2, 4 g NH4Cl, 1 mg resazurin, and 0.25 mg cysteine-HCl) or Wilkins-Chalgren broth (Oxoid, United Kingdom) was boiled and poured into the incubation vessels while they were flushed with N2. The vessels were immediately sealed with crimp-top-sealed septa and were gassed with N2 for another minute. Following autoclaving, filter-sterilized stock solutions of oligofructose (3.0 g/ml) and the tracer substance were added to yield final concentrations in the medium of 10.0 g/liter oligofructose and either 1.5 mg/ml [13C]glycine, 0.7 mg/ml NaH13CO3, or 0.9 mg/ml [13C]urea. The concentration of the tracer substance used was calculated based on equivalent amounts of 13C for all the different tracer substances. Fresh fecal samples were diluted 10-fold (wt/vol) in prereduced phosphate-buffered saline (PBS; containing, per liter, 8.5 g of NaCl, 0.3 g of KH2PO4, 0.6 g of Na2HPO4, and 0.25 g of cysteine) and were homogenized for 2 min in a stomacher at maximum speed. The samples were subsequently centrifuged at 300 × g for 1 min to remove residual large particles. The supernatant was transferred to the fermentation vessels containing the prereduced medium to yield a 1% (final concentration) fecal suspension in the vessel. The incubation was carried out at 37°C in a shaking water bath. At sampling time points, samples were drawn from the vessels by using a syringe coupled to a Luer Lock tap and needle. For the harvesting of bacterial cells, the fermentation samples were centrifuged at 10,000 × g for 10 min. The pelletized bacterial cells were collected, redissolved in 2 ml of PBS, and stored at −80°C until further use for RNA extraction.
RNA extraction.
Total bacterial RNA was isolated from the fermentation samples according to protocols described previously (28). One volume of acid phenol and 0.3 g of zirconia beads (diameter, 0.1 mm; Thistle Scientific, United Kingdom) were added to 600 μl of the fermentation sample. The sample was treated three times for 1 min at 5,000 rpm in the bead beater (MP Biochemicals, United Kingdom) with cooling on ice for 30 s between treatments. After the addition of 150 μl of chloroform-isoamyl alcohol (24:1) and brief vortexing, the samples were centrifuged at 15,000 × g for 15 min at 4°C. The organic phase was transferred to a new tube, and 1 volume of phenol and 1 volume of chloroform-isoamyl alcohol (24:1) were added. The samples were centrifuged for 5 min at 15,000 × g and 4°C. The last three steps were repeated three times. Then 300 μl of chloroform-isoamyl alcohol (24:1) was added, and the sample was centrifuged at 15,000 × g for 15 min at 4°C. The organic phase was transferred to a new tube. One volume of isopropanol and 0.2 volume of 3 M sodium chloride were added, and the mixture was subsequently centrifuged at 15,000 × g for 30 min at 4°C. The pellet was washed with 500 μl of 70% room temperature ethanol for 30 min at 15,000 × g and 4°C. After air drying, the pellet was resuspended in 500 μl of Tris-EDTA (TE) buffer. Five units of RNase-free DNase was added, and the sample was incubated at 37°C for 30 min. Subsequently, isopropanol precipitation was performed as described above. Finally, the pellet was resolved in 200 μl nuclease-free diethyl pyrocarbonate (DEPC)-treated water and was stored at −80°C until further analysis.
Hybridization and magnetic-bead capture of 16S rRNA.
An appropriate amount of total RNA (30 to 120 μg) was added to the filter-sterilized hybridization buffer (containing, per 50 ml, 50 μl 20% sodium dodecyl sulfate [SDS], 13.3 ml 20× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 500 μl 10% NaCl, 15 ml formamide, and 50 ml Nanopure H2O) to a final volume of 360 μl. The samples were incubated for 10 min at 70°C and for 30 min at room temperature. Subsequently, they were hybridized with 40 nmol of the respective probe at the probe-specific hybridization temperature (Table 1) overnight on a rotator. Streptavidin-coated beads (10 mg/ml; Dynabeads; Invitrogen, United Kingdom) were rinsed three times with 0.5× SSC buffer. One hundred microliters of rinsed beads was added to the samples and was incubated for 2 h at room temperature with occasional agitation by hand. The beads were collected at the wall of the tube using a magnet and were rinsed three times with 7.5× SSC buffer. The 16S rRNA was eluted in Nanopure water at 70°C for 3 min. The RNA was precipitated after the addition of 1 volume of isopropanol and 0.2 volume of 3 M sodium chloride and was subsequently centrifuged for 30 min at 15,000 × g and 4°C. After washing with 70% ethanol at 15,000 × g for 30 min at 4°C, the pellet was air dried overnight and was resuspended in 100 μl Nanopure H2O prior to analysis by liquid chromatography coupled with isotope ratio mass spectrometry (LC-IRMS).
Table 1.
Biotinylated oligonucleotide probes targeting 16S rRNA and hybridization temperatures
| Probe name | Sequence (5′–3′) | Target organisms | Hybridization temp (°C) | Reference |
|---|---|---|---|---|
| EUB338 | Biotin-GCTGCCTCCCGTAGGAGT | Most eubacteria | 46 | 1 |
| Bac303 | Biotin-CCAATGTGGGGGACCTT | Bacteroides-Prevotella group | 46 | 17 |
| Bif164 | Biotin-CATCCGGCATTACCACCC | Bifidobacteria | 45 | 13 |
| EREC482 | Biotin-GCTTCTTAGTCARGTACCG | Eubacterium rectale-Clostridium coccoides group | 50 | 8 |
| Clep866 | Biotin-GGTGGATWACTTATTGTG | Clostridium leptum subgroup | 40 | 14 |
Measurement of 16S rRNA 13C enrichment by LC-IRMS.
The system used for LC-IRMS analysis has been described previously (22). Briefly, the system consists of an ion chromatography system coupled, through an interface, to a gas isotope ratio mass spectrometer (IRMSr). The system has two modes of operation: one for online separation of compounds using ion chromatography and IRMS analysis and one for direct injection (flow injection mode) of single samples into the interface for IRMS analysis. Carbon-containing compounds passing through the interface are oxidized to CO2 with maintenance of isotopic provenance before measurement of the isotope ratio (13C/12C) in the IRMSr. Purified 16S rRNA was injected in flow injection mode using a 20-μl injection loop. The level of 13C enrichment was determined relative to a calibrated laboratory standard that had itself been calibrated against the internationally accepted standard for 13C analysis, Vienna Pee Dee Belemnite (VPDB). Results were expressed as δ13C (‰), which describes the deviation of the measured ratio in parts per thousand from the internationally accepted ratio for VPDB (δ13C = 0‰).
Statistics.
Data were analyzed by analysis of variance (ANOVA) with Bonferroni post hoc analysis to assess significant differences between probes and between media. Statistical analysis was conducted using SPSS, version 18 (IBM United Kingdom Ltd., Middlesex, United Kingdom).
RESULTS AND DISCUSSION
13C enrichment in total and 16S rRNA.
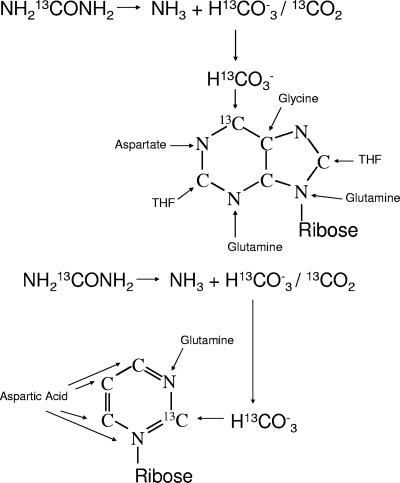
13C-labeled urea was used as the tracer substance for de novo synthesis of precursors for bacterial RNA synthesis in fermentation experiments with human fecal slurries and oligofructose as a substrate. The origin of C atoms in pyrimidines and purines during de novo synthesis and the 13C labeling by use of urea as the tracer substance are shown in Fig. 1. An analogous approach using simple isotope tracers was developed by MacGregor et al. and was further optimized by Miyatake et al. for sulfate-reducing Deltaproteobacteria in marine sediments (15, 16, 18). In the current study, the use of simple tracers has been extended to [13C]urea as a route to labeling RNA for SIP, and this study has demonstrated that [13C]urea labels the 16S rRNA of human gut bacteria.
Fig. 1.
Origin of C atoms in purine and pyrimidine de novo synthesis and 13C labeling by use of [13C]urea as the tracer substance.
The RNA extracted from the cell pellet after fermentation amounted to approximately 600 to 800 μg. Measurement of total RNA by LC-IRMS revealed a high level of enrichment (δ13C = 935.2‰ ± 5.5‰) in total extracted RNA after 24 h of incubation, compared to no enrichment above natural abundance in the control samples at 0 h. The use of 30 μg of total RNA for hybridization with the EUB338 probe resulted in 0.51 ± 0.26 μg C/100 μl, well above the detection limit of the LC-IRMS system. The LC-IRMS measurements showed a high level of enrichment (δ13C = 822.15‰ ± 49.86‰) in the eluted 16S rRNA, confirming that the bacteria had incorporated the 13C-labeled precursor. Analysis of a blank (with water in place of total RNA run through the whole protocol) showed no measurable carbon and no deviation in the isotopic ratio, suggesting little or no contribution from carbon at its natural abundance. Significantly higher enrichment was found for the samples eluted at 70°C than for those eluted at 90°C (P < 0.05); thus, an elution temperature of 70°C was used in further experiments. The elution temperature had no effect on the amount of C in the background samples where water was used instead of RNA, which remained below the detection limit of IRMS analysis (data not shown).
In prior human SIP experiments and in many environmental ecology SIP experiments, 13C-enriched DNA or RNA has been separated from unenriched DNA or RNA by density gradient ultracentrifugation. This technique is limited to highly enriched nucleic acids, which are necessary for the separation of “heavy” and “light” DNA in a buoyant density gradient, and cannot be envisioned as a route to in vivo SIP in humans, simply because of the difficulty of obtaining such high levels of enrichment in gut bacterial nucleic acids. The RNA-SIP procedure coupled with magnetic-bead capture allows low levels of tracer to be used, and the metabolic activity of the bacteria can be monitored after a short incubation with a substrate—a “pulse-chase” experimental design. Only sufficient tracer to produce modest 16S rRNA enrichment above natural abundance is necessary. The natural-abundance 13C signature of intestinal bacterial RNA will reflect the isotopic signature of the carbon inputs to RNA synthesis and therefore will be driven almost exclusively by substrate availability. In areas without extensive maize consumption, such as most Western European countries, the natural-abundance signature of the major dietary inputs will range from ca. −20 to −25‰; with a large maize input—for example, in the United States—the natural-abundance signature of the major dietary inputs will range from ca. −10 to −15‰ (25). The isotopic shifts observed in this study, which exceeded 100‰, are clearly driven by isotopic tracer incorporation and not by dietary or substrate inputs. The extent of isotopic incorporation is also likely to reflect the rate of RNA turnover. Therefore, with IRMS measurement, which has an analytical precision of <0.5‰, very modest amounts of a tracer are needed to enable the detection of isotopic incorporation into RNA.
Testing of different tracer substances.
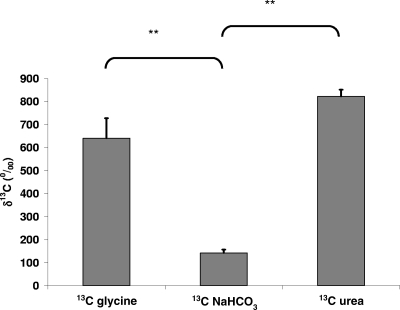
Measurement of 13C enrichment after hybridization with the EUB338 probe showed the highest level of enrichment for the samples incubated with [13C]urea (Fig. 2), so [13C]urea was used as the tracer substance in all further experiments. [13C]glycine and [13C]bicarbonate, however, also appeared to be viable SIP tracers and followed known routes for carbon sequestration into nucleic acid synthesis. Our particular interest in [13C]urea is based on previous work demonstrating that [13C]urea can be delivered to the colon in humans using the synthetic compound lactose [13C]ureide, which resists digestion and absorption but releases [13C]urea upon fermentation by bacteria in the colon (20). These data suggest at least the potential to label bacterial nucleic acids in vivo using lactose [13C]ureide.
Fig. 2.
13C enrichment of the 16S rRNA of total bacteria (measured using probe EUB338) after 24 h of incubation of 1% human fecal sample with either [13C]glycine (1.5 mg/ml), NaH13CO3 (0.679 mg/ml), or [13C]urea (0.939 mg/ml) in mineral medium with 10 g/liter oligofructose. Values are given as means ± standard deviations; **, P < 0.01.
13C enrichment of 16S rRNAs from different bacterial groups incubated in different media.
After 6 h, total RNA was extracted and used for hybridization with five different biotinylated oligonucleotide probes targeting the 16S rRNAs of different dominant bacterial groups of the human gut microbiota. Thirty micrograms of total RNA, which had yielded sufficient 16S rRNA when hybridized with EUB338, did not lead to measurable amounts of carbon in 16S rRNA when hybridized with the group-specific probes. Therefore, the amount of RNA used was increased to 90 to 120 μg, which resulted in 0.34 ± 0.14 ng C in captured 16S rRNA, which is close to the limit of quantitation of the LC-IRMS.
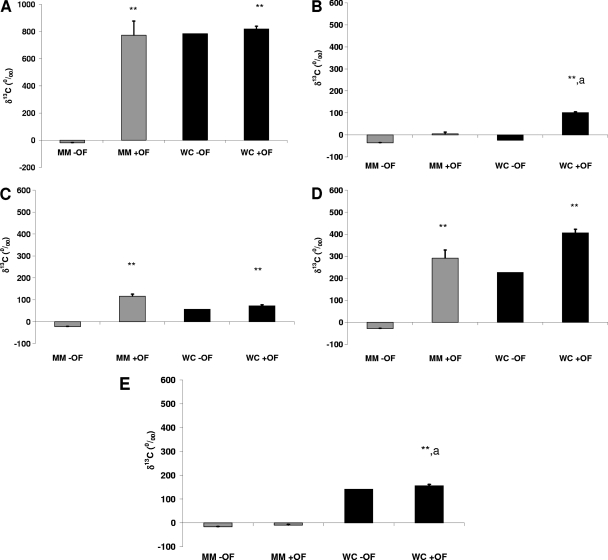
Figure 3 shows the 13C enrichment of the 16S rRNAs of Eubacteria (Fig. 3A), the Bacteroides-Prevotella group (Fig. 3B), bifidobacteria (Fig. 3C), the Eubacterium rectale-Clostridium coccoides group (Fig. 3D), and the Clostridium leptum subgroup (Fig. 3E) after 6 h of incubation in MM or WC broth with or without oligofructose (+ OF or − OF, respectively). The incubation of human gut bacteria in MM − OF did not result in any incorporation of 13C into the 16S rRNA of any of the bacterial groups. When oligofructose had been added as a substrate to the minimal medium (MM + OF), a significantly higher level of 13C enrichment than that with MM − OF was measured for total bacteria (P = 0.006). Significantly higher enrichment was also measured for the Eubacterium rectale-Clostridium coccoides group (P < 0.001) and bifidobacteria (P < 0.001). No significant enrichment was measured for the Bacteroides-Prevotella group (P = 0.40) or the Clostridium leptum subgroup (P = 0.87). When the fermentation was carried out in WC broth, which, in contrast to the mineral medium, is a rich, nutrient-containing medium for bacteria, both WC − OF and WC + OF showed high 13C enrichment in eubacterial 16S rRNA (P, 0.004 for WC + OF versus MM − OF). When oligofructose was added to WC broth (WC + OF), all groups showed highly significant enrichment compared with the MM − OF control (P, <0.005 for all groups). Again at the group level, the highest level of 13C enrichment was measured for the Eubacterium rectale-Clostridium coccoides group. Although the WC − OF data were from single observations, the different groups showed different patterns of enrichment with WC − OF relative to that with either MM − OF or WC + OF. Eubacteria, the Eubacterium rectale-Clostridium coccoides group, the Clostridium leptum subgroup, and bifidobacteria appeared to respond to the WC medium but showed levels of enrichment similar to that with WC + OF. The Bacteroides-Prevotella group appeared to respond only to the WC medium and oligofructose in combination.
Fig. 3.
13C enrichment of the 16S rRNAs of total bacteria (A), the Bacteroides-Prevotella group (B), bifidobacteria (C), the Eubacterium rectale-Clostridium coccoides group (D), and the Clostridium leptum subgroup (E) after 6 h of incubation of 1% human fecal sample with [13C]urea (0.939 mg/ml) in mineral medium without oligofructose (MM − OF) or with 10 g/liter oligofructose (MM + OF) and in Wilkins-Chalgren broth without oligofructose (WC − OF) or with 10 g/liter oligofructose (WC + OF). **, P < 0.01 versus the control (MM − OF); a, P < 0.05 versus MM + OF.
In WC broth, total bacteria and all the bacterial groups investigated except for the Bacteroides-Prevotella group showed high metabolic activity even without the addition of oligofructose, likely due to the presence of nutrients (tryptone, peptone, yeast, and glucose) capable of providing substrates for growth in this medium. The lack of activity of the Bacteroides-Prevotella group in the absence of oligofructose could be due to the short incubation time of 6 h, since it has been observed previously that members of this group are able to grow in this medium (data not shown). When oligofructose had been added to WC medium, the overall activity of total bacteria remained in a range similar to that without oligofructose, as did the activities of bifidobacteria and the Clostridium leptum subgroup. However, the Bacteroides-Prevotella group showed higher activity than that in WC broth without oligofructose, and the activity of the Eubacterium rectale-Clostridium coccoides group also appeared higher than that without oligofructose. The similar activities of bifidobacteria in WC broth with and without oligofructose could mean either that oligofructose does not induce the activity of bifidobacteria in a complex medium or that their metabolism switched from using medium components to using their preferred substrate, oligofructose, which then resulted in similar activity. Many in vitro and in vivo studies have investigated and established the bifidogenic effect of oligofructose (9, 11, 26), so the latter alternative is more likely. The changes in metabolic activity in the Bacteroides-Prevotella group and the Eubacterium rectale-Clostridium coccoides group suggest that oligofructose fermentation is complex and involves many bacterial groups, possibly acting in consort. This complexity has been demonstrated by tracing the flux of products of oligofructose metabolism (3, 6, 21), but SIP provides the key information about which groups are involved in this complex process. Combining such metabolic tracing methodologies with SIP has the potential to give a global and dynamic view of substrate utilization by the intestinal microbial community, which is a dynamic process. The bacterial species that contribute to these changes in activity merit further investigation, because they may also be important contributors to the observed beneficial effects of oligofructose.
Conclusions.
A new approach to investigating the metabolic activity in relation to the phylogeny of human gut bacteria in vitro has been established by this study. By the use of 13C-labeled precursors of de novo RNA synthesis, changes in the metabolic activities of different bacterial groups under different growth conditions on the group level could be monitored. By using species-specific probes, this method could provide more insight into the metabolic roles of single bacterial species in in vitro incubation experiments. A further development of this method would be in vivo SIP, which could lead to a better understanding of the contributions of the metabolic activities of different bacterial groups in health and disease under true physiological conditions, which are almost impossible to reproduce in vitro.
ACKNOWLEDGMENTS
This work was funded by the Crohn's in Childhood Research Association (CICRA) and the Yorkhill Children's Foundation. We gratefully acknowledge their financial support.
We thank Tom Preston for helpful discussions and Martin McMillan for technical assistance.
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Amann R. I., et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barclay A. R., Morrison D. J., Weaver L. T. 2008. What is the role of the metabolic activity of the gut microbiota in inflammatory bowel disease? Probing for answers with stable isotopes. J. Pediatr. Gastroenterol. Nutr. 46:486–495 [DOI] [PubMed] [Google Scholar]
- 3. Bourriaud C., et al. 2005. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J. Appl. Microbiol. 99:201–212 [DOI] [PubMed] [Google Scholar]
- 4. Degelmann D. M., Kolb S., Dumont M., Murrell J. C., Drake H. L. 2009. Enterobacteriaceae facilitate the anaerobic degradation of glucose by a forest soil. FEMS Microbiol. Ecol. 68:312–319 [DOI] [PubMed] [Google Scholar]
- 5. de Graaf A. A., et al. 2010. Profiling human gut bacterial metabolism and its kinetics using [U-13C]glucose and NMR. NMR Biomed. 23:2–12 [DOI] [PubMed] [Google Scholar]
- 6. Duncan S. H., et al. 2004. Contribution of acetate to butyrate formation by human faecal bacteria. Br. J. Nutr. 91:915–923 [DOI] [PubMed] [Google Scholar]
- 7. Eckburg P. B., et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franks A. H., et al. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gibson G. R., Beatty E. R., Wang X., Cummings J. H. 1995. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 108:975–982 [DOI] [PubMed] [Google Scholar]
- 10. Hayashi H., Sakamoto M., Benno Y. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46:535–548 [DOI] [PubMed] [Google Scholar]
- 11. Kleessen B., Hartmann L., Blaut M. 2001. Oligofructose and long-chain inulin: influence on the gut microbial ecology of rats associated with a human faecal flora. Br. J. Nutr. 86:291–300 [DOI] [PubMed] [Google Scholar]
- 12. Kovatcheva-Datchary P., et al. 2009. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ. Microbiol. 11:914–926 [DOI] [PubMed] [Google Scholar]
- 13. Langendijk P. S., et al. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lay C., et al. 2005. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ. Microbiol. 7:933–946 [DOI] [PubMed] [Google Scholar]
- 15. MacGregor B. J., Boschker H. T., Amann R. 2006. Comparison of rRNA and polar-lipid-derived fatty acid biomarkers for assessment of 13C-substrate incorporation by microorganisms in marine sediments. Appl. Environ. Microbiol. 72:5246–5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. MacGregor B. J., Bruchert V., Fleischer S., Amann R. 2002. Isolation of small-subunit rRNA for stable isotopic characterization. Environ. Microbiol. 4:451–464 [DOI] [PubMed] [Google Scholar]
- 17. Manz W., Amann R., Ludwig W., Vancanneyt M., Schleifer K. H. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142(Pt. 5):1097–1106 [DOI] [PubMed] [Google Scholar]
- 18. Miyatake T., MacGregor B. J., Boschker H. T. 2009. Linking microbial community function to phylogeny of sulfate-reducing Deltaproteobacteria in marine sediments by combining stable isotope probing with magnetic-bead capture hybridization of 16S rRNA. Appl. Environ. Microbiol. 75:4927–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moore W. E., Holdeman L. V. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morrison D. J., Dodson B., Preston T., Weaver L. T. 2003. Gastrointestinal handling of glycosyl [13C]ureides. Eur. J. Clin. Nutr. 57:1017–1024 [DOI] [PubMed] [Google Scholar]
- 21. Morrison D. J., et al. 2006. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br. J. Nutr. 96:570–577 [PubMed] [Google Scholar]
- 22. Morrison D. J., Taylor K., Preston T. 2010. Strong anion-exchange liquid chromatography coupled with isotope ratio mass spectrometry using a Liquiface interface. Rapid Commun. Mass Spectrom. 24:1755–1762 [DOI] [PubMed] [Google Scholar]
- 23. Radajewski S., Ineson P., Parekh N. R., Murrell J. C. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649 [DOI] [PubMed] [Google Scholar]
- 24. Sueoka K., Satoh H., Onuki M., Mino T. 2009. Microorganisms involved in anaerobic phenol degradation in the treatment of synthetic coke-oven wastewater detected by RNA stable-isotope probing. FEMS Microbiol. Lett. 291:169–174 [DOI] [PubMed] [Google Scholar]
- 25. Tanis A., Rietveld A. T., Van den Berg J. W., Wattimena J. L., Swart G. R. 2000. Influence of the 13C-enrichment of the habitual diet on a 13CO2 breath test used as an index of liver glycogen oxidation: a validation study in western Europe and Africa. Nutrition 16:6–10 [DOI] [PubMed] [Google Scholar]
- 26. Wang X., Gibson G. R. 1993. Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine. J. Appl. Bacteriol. 75:373–380 [DOI] [PubMed] [Google Scholar]
- 27. Webster G., et al. 2006. A comparison of stable-isotope probing of DNA and phospholipid fatty acids to study prokaryotic functional diversity in sulfate-reducing marine sediment enrichment slurries. Environ. Microbiol. 8:1575–1589 [DOI] [PubMed] [Google Scholar]
- 28. Zoetendal E. G., Akkermans A. D., De Vos W. M. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]