Abstract
Increasing amounts of livestock manure are being applied to agricultural soil, but it is unknown to what extent this may be associated with contamination of aquatic recipients and groundwater if microorganisms are transported through the soil under natural weather conditions. The objective of this study was therefore to evaluate how injection and surface application of pig slurry on intact sandy clay loam soil cores influenced the leaching of Salmonella enterica serovar Typhimurium bacteriophage 28B, Escherichia coli, and Cryptosporidium parvum oocysts. All three microbial tracers were detected in the leachate on day 1, and the highest relative concentration was detected on the fourth day (0.1 pore volume). Although the concentration of the phage 28B declined over time, the phage was still found in leachate at day 148. C. parvum oocysts and chloride had an additional rise in the relative concentration at a 0.5 pore volume, corresponding to the exchange of the total pore volume. The leaching of E. coli was delayed compared with that of the added microbial tracers, indicating a stronger attachment to slurry particles, but E. coli could be detected up to 3 months. Significantly enhanced leaching of phage 28B and oocysts by the injection method was seen, whereas leaching of the indigenous E. coli was not affected by the application method. Preferential flow was the primary transport vehicle, and the diameter of the fractures in the intact soil cores facilitated transport of all sizes of microbial tracers under natural weather conditions.
INTRODUCTION
Livestock production is increasing worldwide (28), and so is the volume of manure applied to agricultural soil. Fertilization of agricultural land with livestock waste can lead to fecal contamination of waterways through surface water runoff, drainage systems, and groundwater if microorganisms are transported through the soil to groundwater reservoirs (23, 53). Groundwater contamination by microorganisms of fecal origin has been reported over many years (1, 77), and waterborne disease outbreaks have been associated with zoonotic pathogens like Cryptosporidium (29), Escherichia coli O157:H7, and Campylobacter (52, 76), as well as norovirus (12). However, little documentation is available to what extent such contamination was caused by actual transport of pathogens through soil or fecal contamination through boreholes, intruding surface water, etc. Knowledge on transport of bio-colloids (e.g., viruses, bacteria, and protozoa) through soil is therefore important when groundwater supplies are to be protected from contamination with pathogenic microorganisms. Colloids in the subsurface are characterized as mobile abiotic or biotic particles with a diameter less than 10 μm and that possess an electric charge on their surfaces (56).
Livestock waste contains a variety of zoonotic pathogens, including Salmonella, Campylobacter, pathogenic E. coli, enteric viruses, and protozoan parasites (50, 53). Livestock waste is typically applied by injection or surface application on agricultural fields, and the application method may affect transport and survival of pathogens in soil (17, 35). Application of the livestock wastes on the soil surface exposes pathogens to desiccation and UV light, hereby reducing the concentration of fecal microorganisms before the livestock waste is incorporated into the soil, whereas injection directly into the soil protects the microorganisms against these factors and therefore potentially increases the survival of pathogens (40). However, the survival of pathogens in soil depends on many parameters, such as temperature, moisture, pH, soil composition, and competition from indigenous microflora (2, 53). E. coli in manure and slurry-amended soil kept at 16°C was estimated to survive for 14 to 27 days in a greenhouse (72), whereas Avery et al. (8) detected E. coli for 162 days in soil samples contaminated with livestock feces collected from an outdoor pen exposed to natural weather conditions during winter and spring. Survival of E. coli has been reported for at least 40 days in drainage water and feces-amended drainage water (64). Leaching of E. coli from manure-applied intact silt loam soil cores was estimated to continue up to 28 days (31), whereas E. coli was still recovered after 100 days in drainage water from a clay loam field fertilized with slurry (57). The viability of protozoan parasites (e.g., Cryptosporidium parvum oocysts) in leachate after passage through soil is limited. Viable oocysts have been detected in water at 4°C for more than 42 days (14), and in soil chambers, approximately 60% of the oocysts were still viable after 156 days (42). Poliovirus, echovirus, and hepatitis A virus seeded in groundwater showed little inactivation of any of the viruses at 5°C over 8 weeks (75). Gordon and Toze (32) found coxsackievirus survived in unfiltered groundwater at 15°C for at least 35 days. During a normal Danish winter, coxsackievirus was detectable for 23 weeks in soil samples taken from lysimeters amended with sludge (26). During these 23 weeks, air temperatures between −12°C and 26°C were recorded.
Preferential water movement (e.g., through root channels, earthworm channels, and naturally occurring cracks) is probably the primary route for rapid transport of microorganisms through soil (6, 59, 81). Increased transport of microorganisms has been observed in soil with high clay content because water flow in clay-rich soils is usually concentrated in the fractures (3, 11). The ability of microorganisms of different sizes (e.g., viruses, bacteria, or protozoan parasites) to travel quickly through soil fractures has been recognized. Carlander et al. (15) observed a rapid transport of Salmonella bacteriophage 28B in clay soils, with breakthrough (i.e., from the time of application to the detection in the leachate) at a depth of 1.2 m after 2 to 24 h, probably due to the presence of macropores and bypass flow. Salmonella enterica serovar Typhimurium was detected in leachate from intact clay monoliths 24 h after manure application (9), and fecal coliforms originating from dairy shed effluent readily penetrated through 700-mm-deep soil columns within 2 days (43). C. parvum oocysts applied to soil blocks that were subsequently irrigated with artificial rainwater on alternate days were transported within a silty loam soil and detected for up to 9 days in leachate, whereas the oocysts added to clay loam soil blocks could be detected over 21 days in the leachate (54). There is a need to determine the relative leaching potential of microorganisms of different sizes in fractured soil (24, 82) as the infective doses of viruses (one virus particle) and protozoan parasites (ten infective oocysts) are low compared to those of many pathogenic bacteria (27, 47, 80). Leaching of small amounts of pathogenic bacteria would therefore not constitute as great a risk for the public health as would small amounts of virus particles and oocysts of protozoan parasites.
Microbial transport in intact soils has been studied under simulated rain conditions (58, 59, 65) or with a combination of rainfall and irrigation (43, 44), whereas little information is available about microbial transport under natural climate conditions (57, 62). Studies have shown that leaching of bio-colloids such as E. coli is strongly influenced by soil wetness conditions at the time of animal slurry application as well as the time between slurry application and rainfall occurring after spreading: i.e., the wetter the soil, the higher the leaching of bacteria (43, 57). Correspondently, a significant leaching of bacteriophage PRD1 has been observed in connection with rainfall (62).
The main objective of this study was to evaluate how injection and surface application of pig slurry on intact sandy clay loam soil cores influenced the leaching of Salmonella Typhimurium bacteriophage 28B (phage 28B), E. coli, and C. parvum oocysts under natural weather conditions. Differences in the leaching of the different sized microbial tracers were determined, as were the breakthrough and survival time for the tracers in the leachate.
MATERIALS AND METHODS
Soil core excavation.
The soil cores were excavated from the field site Slaeggerup, near the city of Roskilde, Denmark, as described by Bech et al. (10). Briefly, the soil cores were randomly taken within a square of 4 m by 16 m. Fifteen soil cores were collected from the Ap horizon by applying steady-state weight pressure on the top of stainless steel cylinders (15 cm in diameter, 30 cm in height). The steel cylinder was carefully excavated, and the soil core height in the cylinder ranged from 22 cm to 29 cm. The variance in height was due to difficulties in collecting the soil cores. No attempt was made to adjust the soil cores to equal heights in order to avoid the risk of destroying the soil structure or creating a void between the soil core and steel cylinder. The soil is a sandy clay loam (20.1% clay, 19.9% silt, 57.6% sand, and 2.4% organic matter) from glacier deposit classified as Typic Argiudoll with a porosity of 0.37 cm3 cm−3 (49). One pore volume is the amount of space in the soil core occupied by soil pores or fractures. The pore volume for each soil core was determined as the porosity multiplied by the total volume of soil, taking into account the differences in height of the soil cores. A single soil core was destroyed, and the soil revealed no indigenous bacteriophages capable of lysing the Salmonella Typhimurium type 5 host strain. Similarly, E. coli and C. parvum oocysts were not detected.
Experimental setup of soil cores.
Soil cores were prepared 2 days before the start of the experiment. The lower 2 cm of soil was replaced with acid-washed filter sand (2 to 3 mm), and an acid-proof steel net was placed at the bottom of each soil core. A total of 14 soil cores were placed randomly in a multicolumn lysimeter together with an additional steel cylinder that contained 2 cm of acid-washed sand. The soil cores were positioned as three rows each containing five soil cores, and the distance between the soil cores was 9 cm. The multicolumn lysimeter was placed outdoor on a roof and therefore exposed to natural variations in temperature and precipitation. A 0.7-m PVC tube was connected to each soil core to give hydraulic suction. The tubes were attached to sterile glass bottles placed in a refrigerator (5°C). The soil cores were equilibrated with tap water to field capacity the day before slurry application. Natural precipitation was the only water applied, except on day 3, when 5 mm distilled water was added to each soil core. The experiment was conducted during the cold season from 31 October 2006 to 28 March 2007. On-site daily logging of precipitation (Fig. 1) and temperature (Fig. 2) data was performed by a TGPR-1201 data logger (Germini, Chichester, United Kingdom) connected to a Rain-O-Matic Professional gauge (Pronamic, Silkeborg, Denmark) and a TGP-4520 with two PT-100 temperature probes for measuring aerial and refrigerator temperatures, respectively.
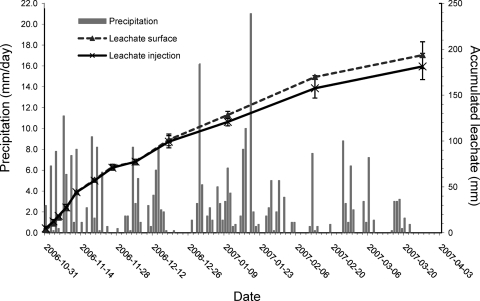
Fig. 1.
Accumulated leachate from soil cores with pig slurry applied to the surface or by injection and daily precipitation during the study period.
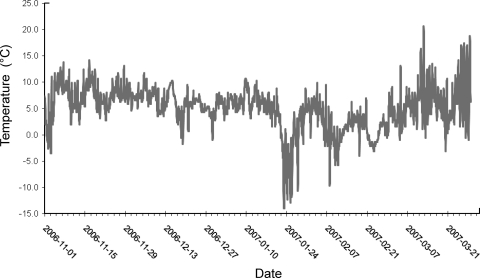
Fig. 2.
Air temperature variation during the study period.
Preparation of pig slurry.
Pig slurry used in the study was collected from a manure storage tank at a pig farm. Initial analyses of the pig slurry showed no indigenous Cryptosporidium oocysts or phages present that could infect the bacterial host strain (Salmonella Typhimurium type 5). The concentration of indigenous E. coli in the slurry was 9.7 × 104 CFU ml−1. The chloride concentration in the pig slurry was initially measured to 28 ppm before additional chloride was added as a tracer. A slurry mixture was prepared for each of the 10 soil cores. Approximately 44 ml of slurry with 8.8% dry matter content was inoculated with phage 28B (48) and C. parvum oocysts. C. parvum oocysts were isolated from a dead calf. The final concentrations of the phage 28B and C. parvum oocysts in the pig slurry were 1.8 × 109 PFU ml−1 and 4.0 × 104 oocysts ml−1, respectively. In addition, chloride was added to the slurry sample as a chemical tracer with a final concentration of 3.14 mg Cl− ml−1. On five replicate soil cores (SA-1 to SA-5), the slurry mixture was applied to the soil surface. To simulate the injection of slurry as applied by farmers, a triangle trench (length, 8 cm; width, 5 cm; depth, 8 cm) was made in five replicate soil cores (IA-1 to IA-5). The thickness of the slurry layer in the triangle trench was approximately 4 cm immediately after slurry application. Three soil cores used as controls (C-1 to C-3) were spiked with 44 ml distilled water with chloride added. One soil core and the steel cylinder with 2 cm of sand were left untouched.
Sample collection.
Leachate was collected in 500-ml sterile glass bottles. Background levels of phage 28B, C. parvum oocysts, and E. coli in leachate collected at the end of the equilibration period (day 0) were below the detection level of 1 PFU ml−1, 1 CFU ml−1, and 1 oocyst per 50 ml, respectively. The chloride concentration in this leachate was 10.4 ppm. Leachate was analyzed on days 1, 4, 6, 9, 13, 20, 26, 36, 49, 72, 106, and 148, totaling 12 sampling times. The days of analysis were chosen during the study when sufficient volumes of leachate were available for the microbial and chemical tracer analyses. A new sterile glass bottle was connected to the PVC tube at each day of analysis and left to collect leachate until the next sampling day. (For example, leachate samples consisted of all percolated water from the soil cores between two successive sampling times.) The leaching periods for microorganisms in intact soil cores were 148 days for phage 28B and E. coli and 36 days for chloride. At day 36, approximately 90% of the chloride had leached, and the analysis therefore ended. Due to sudden changes in trained laboratory staff, concentrations of C. parvum oocysts in leachate were only analyzed during the first 20 days, and viability testing was done on days 1, 4, and 6.
Preparation and enumeration of microbial and chemical tracers.
The phage 28B was used in the study as a model for virus transport as bacteriophages have been suggested as model organisms to predict human enteric viral behaviors and risks for their environmental transmission (37). Phage 28B has a documented resistance toward high temperatures, changes in pH, and high NH3 levels and has not been reported to occur naturally in the environment nor in fecal material (39). Propagation of phage 28B was done according to Höglund et al. (39). Slurry and leachate samples were 10-fold serially diluted in maximum recovery diluent (MRD) (Oxoid, Hampshire, United Kingdom) before phage 28B was enumerated by a double-agar layer method (4). Samples of 10 g of soil were added to 90 ml MRD and treated in an ultrasound bath (Struers, Copenhagen, Denmark) for 30 s to remove microorganisms from the surface of the soil particles. The soil solution was subsequently serially diluted before enumeration. The host strain S. Typhimurium type 5 was grown in nutrient broth at 37°C for 4 h. From the 10-fold-diluted samples, 1 ml was taken and mixed with 1 ml of broth culture of the host strain and 3 ml of soft agar consisting of 70% blood agar base (Oxoid) and 30% nutrient broth (Oxoid). The mixture was spread on a well-dried blood agar base plate (Oxoid), which was incubated at 37°C for 18 h. For each diluted sample, the analysis was done in triplicate. Clear zones (plaques measured in PFU) were counted. When a high level of bacterial background flora was expected (mainly at the lower dilutions), the samples were filtered through 0.45-μm-pore-size filters before being mixed with the soft agar.
E. coli in slurry, soil, and leachate was enumerated on Brilliance E. coli/coliform selective agar (Oxoid), where colonies appear as typical indigo blue colonies after incubation at 37°C for 21 ± 3 h. Samples of 10 g of soil was treated in an ultrasound bath as described for phage 28B. Soil solution, slurry, and leachate were serially diluted in MRD and enumerated for viable E. coli.
C. parvum oocysts were isolated from fecal material of a dead calf by the method described by Maddox-Hyttel et al. (50). Slurry and soil samples were analyzed correspondently. Briefly, 1.1-g sample was added to 11 ml 0.01% Tween 20 (Sigma), mixed and left standing for 5 min for sedimentation of large particles. Ten milliliters of the supernatant was transferred to 40 ml sucrose solution (density, 0.001 g ml−1) and centrifuged for 10 min at 350 × g. The top 20 ml was mixed with 30 ml distilled water and centrifuged at 1,550 × g for 10 min. The supernatant was removed leaving 2 ml. The oocysts were further concentrated by immunomagnetic separation using Dynabeads GC-Combo kit (Genera Technologies, Newmarket, United Kingdom). The concentration and viability of isolated oocysts were determined by adding fluorescein isothiocyanate (FITC)-conjugated anti-Cryptosporidium monoclonal antibody (Cellabs Pty. Ltd., Australia) and 4′,6′-diamidino-2-phenylindole–propidium iodide (DAPI-PI) (Sigma-Aldrich, Denmark), respectively. The stained oocysts were visualized in a fluorescence microscope. Oocysts to be used in the soil core study were further treated with 70% ethanol for 10 min, washed three times in distilled water, and distributed in 10 Eppendorf tubes each containing 2.2 × 106 oocysts. Approximately 50 ml of leachate was analyzed for oocysts at each sampling time. Concentration of the leachate was done by centrifugation at 3,000 × g for 15 min, carefully removing the supernatant, leaving approximately 2 ml in the tube. The concentration and viability of oocysts in the reduced leachate sample were determined by FITC and DAPI/PI staining.
Chloride concentration in slurry and leachate was determined by ion chromatography (Dionex LC20).
Size determination of E. coli cells.
Analysis of microscope images of E. coli cells was carried out to characterize the variability in size within a population of cells obtained from a pure culture of E. coli originally isolated from the pig slurry used in this study. The E. coli cells were grown on a blood agar plate (Blood Agar Base [Oxoid] with 5% calf blood added) at 37°C for 24 h prior to size determination. Cells were mounted on a microscope slide suspended in MRD at pH 7.0. The size of E. coli cells was determined by analyzing digital images taken in an inverted fluorescence microscope (Zeiss Axioplan2; Germany) operating in phase contrast mode at 1,000× using a DFC340 FX camera (Leica, Germany). The image processing program, ImageJ 1.43 (20), was used to determine the length and diameter of at least 50 cells. A micrometer (2 mm, interval of 0.01 mm; Leica) converted the number of pixels to length in μm. The size of the E. coli cells would represents a maximum size due to the optimal growth conditions.
Statistical methods.
This study is based upon repeated measurements of the same 10 soil cores for a period of 148 days. On day 0, soil cores received pig slurry containing phage 28B, C. parvum oocysts, E. coli, and chloride either applied on the surface (n = 5) or injected (n = 5). The response variable for the study (outcome) was the relative concentration (C/C0) of the microbial and chemical tracers. Here C is the concentration of the tracer in leachate detected at a sampling time and C0 is the concentration of the tracer applied to the slurry sample at the start of the study. The normal distribution of the log-transformed relative concentrations was validated by quantile-quantile plots, and variance homogeneity was validated by residual plots. A model for repeated measurement was selected as results from the same soil cores tend to be positively correlated and that independence between results should not be assumed. Soil cores were set as a random factor. The percentage of soil core variation was used to estimate how large the variation between soil cores was compared to the total variation. If this variation was high, then there would be a large difference between the soil cores.
A statistical analysis of the significance of microbial and chemical tracers and application methods on recovery rate, average run-through time, and removal rate of the tracers was done by analysis of covariance with square root transformation for normalization. The square root transformation was selected based on validation by quantile-quantile and residual plots.
Linear regression analysis was used to find the best-fit slope (i.e., inactivation rate for the phage 28B). Linearity of the inactivation rate was tested within soil cores for each application method as well as between the two application methods. The relative concentration of the phage was normalized by a square root transformation. The soil core was set as a factor, and day and application method were covariates in the model.
All statistical analyses were done in SAS, version 9.2 (SAS Institute, Inc., Cary, NC).
RESULTS
Climate conditions.
A total of 300-mm rain was measured during the 5-month study period (Fig. 1). Rain intensity was highest during the first 20 days of the experiment and in January 2007 with 52% of the water received during these periods. The temperature showed high variations ranging from −14°C during the last part of January to 21°C at the end of the study period (Fig. 2). The mean temperature during the study period was 5°C. On average, a total of 65% of the precipitation leached through soil cores treated by surface application of the slurry, which was similar to soil cores with slurry injected, where 60% of the rainwater was recovered. Some variation was observed in the amount of water leaching through the soil cores as the pore volume was replaced between 1.2 to 2.6 times after 148 days (Fig. 3).
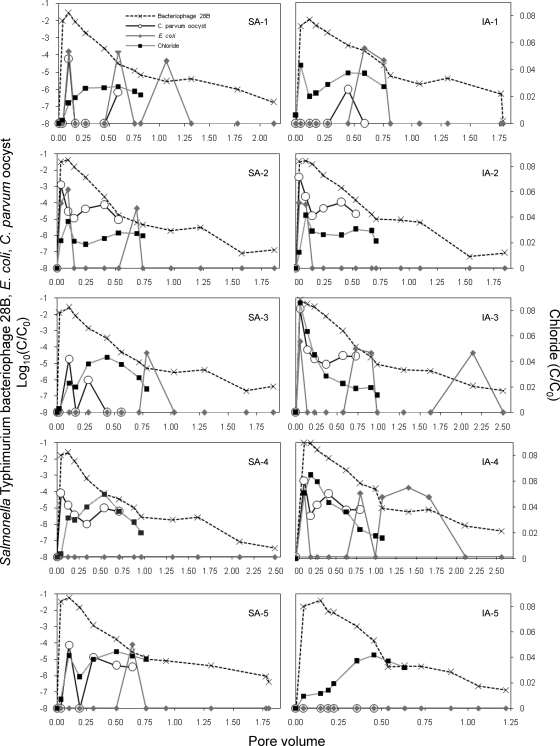
Fig. 3.
Breakthrough curves of Salmonella bacteriophage 28B, E. coli, C. parvum oocysts, and chloride for surface (SA) and injection application (IA) of slurry shown relative to the volume of pore water leached. C is the concentration of tracer in leachate, and C0 is the concentration of tracers in slurry added to the soil cores. In leachate samples, where the tracer concentration was below the detection limit, the value was set at −8.
Sizes of microbial tracers.
The E. coli cells originally isolated from the pig slurry used in this present study were rod shaped, with a mean length of 2.3 μm and mean diameter of 0.9 μm. C. parvum oocysts are spherical, with a mean diameter of 4.9 μm (60), and phage 28B is a double-stranded DNA bacteriophage with a diameter of 60 nm (39). The three microbial tracers used therefore represent organisms with different sizes.
Leaching of chloride.
The leaching of chloride from the soil cores is shown on a linear concentration scale versus the number of pore volumes passed through the soil cores (Fig. 3). A small increase in the concentration of chloride was detected on day 4, while the highest relative concentration was found on day 13, corresponding to approximately 0.5 pore volume. Therefore, leaching of chloride showed both fast transport through macropores as well as transport following the exchange of water in the soil cores. The detection of chloride already at 0.5 pore volume indicated heterogeneity of pore sizes within the soil cores and also that chloride was transported in the smaller pores. The tailing of the breakthrough curve (BTC) showed that zones of immobile water were present in the soil cores and exchange of chloride by diffusion was taking place. The maximum concentration of chloride in leachate varied between 0.086 mg ml−1 and 0.25 mg ml−1, which implies that chloride was diluted 11 to 35 times by the rainwater. Chloride was measured in the leachate for 36 days, and during this period, 51% to 90% of the chloride was recovered. A significant difference in leachate of chloride was seen between injected and surface-applied slurry (P = 0.0034). The largest amount of chloride leached through soil cores where slurry was applied through injection. Variation in chloride leaching could not be attributed to variation between soil cores (0%).
Leaching of phage 28B.
Phage 28B leaching was expressed in the breakthrough curve (BTC) as the logarithmic relative concentration log10 (C/C0) versus the number of replaced pore volumes (Fig. 3). Phage 28B was detected in the first leachate sample with both application techniques. The average maximum concentrations (Cmax ± standard deviation [SD]) of leached phage 28B for injected and surface applications were 9.4 × 107 ± 6.4 × 107 PFU ml−1 and 6.3 × 107 ± 2.6 × 107 PFU ml−1, respectively. Cmax was found on day 4 to correspond to 0.1 pore volume. Slurry-injected soil cores leached significantly more phage 28B than soil cores with slurry applied to the surface during the study (P = 0.0055). Phage 28B leached from one of the five control soil cores. In this particular soil core, phage 28B was detected on the first day at a concentration of 2.8 × 104 PFU ml−1 with a rapid decline in concentration found during the following sampling times. Phage 28B showed a rapid breakthrough at a pore volume less than 0.1, indicating primarily macropore flow during the first 4 days. A long tailing of the BTC demonstrated inactivation and slow detachment of the phage from manure and soil particles. The inactivation rate of the phage, μs, was expressed as the slope of the tail in the BTC from day 36 to day 148 (Fig. 3). The inactivation rate of the phage was not significantly different within soil cores with slurry applied to the surface (P = 0.79) as well as within the five slurry-injected soil cores (P = 0.42). Thus, our data do not indicate any difference in the inactivation rate of the phage within soil cores that received slurry by the same application method. In addition, the inactivation rate was not affected by application method (P = 0.26). The inactivation rate for phage 28B in soil was estimated to 0.016 day−1 for both soil cores with the slurry surface applied and those with the slurry injected.
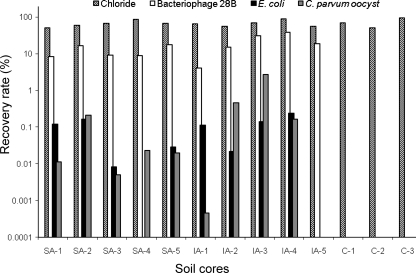
A variance in the leaching pattern of phage 28B was found between the soil cores (39%), indicating that the phage was to some extent affected by the natural heterogeneity within the soil cores as the other, larger, microbial tracers. After 148 days, it was still possible to detect the phage 28B at a concentration of approximately 100 PFU ml−1. The total recovery of phage 28B in leachate ranged from 4% to 38% after 148 days (Fig. 4).
Fig. 4.
Recovery rate of the microbial tracers and chloride in each soil core during the study period. Slurry was applied to the surface of SA-1 to SA-5, and IA-1 to IA-5 had slurry injected. The control soil cores C-1 to C-3 only received chloride. The y axis is in the logarithmic scale.
Leaching of E. coli.
Indigenous E. coli in slurry leached through the soil cores with some variation (7%) between soil cores. E. coli leached at approximately 0.1 pore volume and were detected sporadic at larger pore volumes from three soil cores with surface and injected slurry, respectively. One soil core showed no leaching of E. coli, and another soil core leached E. coli at approximately 0.6 pore volume (Fig. 3). After 106 days, E. coli could no longer be detected in the leachate, and it was not detected in the leachate from the control soil cores. The two slurry application methods were not associated with any significant difference in the leaching of E. coli (P = 0.85). The recovery rates of E. coli in the leachate from soil cores with the slurry injected and surface applied were 0.1% and 0.06%, respectively (Fig. 4).
Recovery of C. parvum oocysts.
Similar to E. coli, C. parvum oocysts leached from the soil cores at approximately 0.1 pore volume but high variation (73%) was seen in the leaching of oocyst between soil cores (Fig. 3). From nine soil cores, a second breakthrough of oocysts was found at 0.3 to 0.6 pore volume, whereas one soil core retained all oocysts (IA-5; Fig. 3). Significantly more oocysts leached from slurry-injected soil cores (P = 0.037) with 10 times more oocysts recovered from injected slurry (0.05%) than from surface-applied slurry (0.66%). The average viability rates for oocysts leached from soil cores injected with slurry were 34%, 52%, and 50% on days 1, 4, and 6, respectively. Mean oocyst viability rates from surface-applied slurry were 83%, 18% and 50% on day 1, 4 and 6, respectively. The viability of the oocysts varied between the soil cores and ranged from no viable oocysts to 100% viability.
Cell size and leaching of microbial tracers.
A significant difference in the recovery rates of the four tracers was found (P < 0.001). The highest recovery rate was found for chloride, followed by phage 28B, E. coli, and C. parvum oocysts. The recovery rate for the phage 28B was significantly higher than those for E. coli and C. parvum oocysts, whereas the recovery rates of E. coli were similar to those of C. parvum oocysts (P = 0.43). There was a 7-cm difference between the longest and shortest soil cores, but the recovery rate was not affected by this difference in height (P = 0.20).
An average run-through time was estimated based on the day that the mean number of the microbial tracer leached through the soil cores compared to the total number of microbial tracer leached during the study period. For chloride, this time was based on the amount of chloride. The average run-through times of tracers leached through the soil cores were 4.4 days for phage 28B, 5.4 days for C. parvum oocysts, 15 days for chloride, and 25 days for E. coli. Soil core height (P = 0.78) did not influence the average run-through time. Phage 28B had a significantly faster run-through time compared to E. coli (P = 0.0054) and chloride (P < 0.0001), but the time was similar to that of C. parvum oocysts (P = 0.70). The average run-through times for E. coli and chloride were not significantly different (P = 0.25).
The microbial removal rate based on mass balance of total amount of microbial tracer leached through the soil cores was estimated. The removal rate was 3.9 log m−1 for phage 28B in surface-applied soil cores and 3.2 log m−1 when slurry was injected. The microbial removal rates for E. coli and C. parvum oocysts were significantly higher than those of phage 28B (P < 0.0001). The mean microbial removal rates for E. coli and C. parvum oocysts were 13.6 and 14.5 log m−1, respectively, and these removal rates were not significantly different (P = 0.81).
DISCUSSION
Flow pattern of tracers.
The transport of microbial tracers is facilitated by attachment to particles, or the microorganisms may themselves act as mobile colloids through the subsurface environment (2, 56). Colloid transport in fractured porous media is typically characterized by fast preferential flow resulting in an early breakthrough (16, 65). The importance of cell size and shape has been highlighted in studies on transport of microorganisms in soil (24, 38, 82). We observed early breakthrough for all tracers within 1 day, indicating preferential flow through macropores (e.g., naturally occurring cracks, fractures, or earthworm channels). The BTC of chloride showed a rapid water flow through large connective pores resulting in the first peak and additional flow through the fine-grained portion of the soil cores indicated by the second peak at a 0.5 pore volume. The tailing of the BTC indicated diffusive exchange of chloride with immobile water regions. Immobile water has typically been related to thin liquid films around soil particles, dead-end pores, nonmoving intra-aggregate water, or relatively isolated regions associated with unsaturated flow (63). Experiments with transport of tritium in soils with clay content higher than 18% have shown similar double peaks and tailing of BTC (46) corresponding to the 20.1% clay content in the soil cores used in the present study. A second peak in leaching of oocysts corresponding to maximum leaching of chloride at a 0.3 to 0.6 pore volume was observed. The second peak would be a contribution from the fine-grained portion of the soil cores which would be expected to retain microorganisms like the oocysts. An almost similar level of larger-sized bacteria leaching with same double-peaked pattern was explained by an enhanced dispersion of cells to the central canal of coarse sand in the center of a bed of fine sand (30). Increased pH in river water applied to soil columns resulted in an enhanced electrostatic repulsion between attached C. parvum oocysts and soil particles leading to enhanced leaching (38). Furthermore, long-term, low-level elution may also be a potential source of secondary breakthrough of oocysts due to detachment (36). Phage 28B showed correspondingly pronounced tailing of the BTC and colloids with similar sizes as viruses can diffuse into the fine pores of the soil matrix (24). McLeod et al. (58) detected the same preferential flow in intact soil columns for the phage 28B and fecal coliforms, while the bromide lacked the early peak.
Laboratory strains versus indigenous strains.
We found E. coli in leachate during the first sampling and thereafter sporadically for up to 3½ months thereafter. This could indicate the presence of a mixture of planktonic cells as well as cells attached to slurry particles and a subsequent slow detachment of a fraction of these microorganisms into the leachate. Guber et al. (33) concluded based on the kinetic release of indigenous E. coli that E. coli also resided in the liquid part of the manure and was released as the liquid manure fraction was diluted and replaced by rainwater. In addition, the presence of manure colloids reduced the attachment of bacterial cells to clay and silt particles, leading to free-cell transport of manure-borne fecal coliforms (34). E. coli cells present in slurry have been found in drains very rapidly after slurry application on fields and were detectable for 1 to 3 months in low concentrations in the drainage water (79), corresponding well with the observations seen in our study. The microbial elimination rate (MER; i.e., the total log reduction of microorganisms during the study time) for indigenous E. coli (MER, 2.8 log units) from river water in columns was lower than that for spiked E. coli (MER, 4.8 log units), resulting in increased leaching of indigenous E. coli compared to spiked E. coli. This could lead to an overestimation of the elimination of indigenous microorganisms under field conditions if leaching results are based on spiked microorganisms (38). The MER estimated for the indigenous E. coli from slurry in the present study (MER, 3.1 log units) corresponds well with MER values for indigenous E. coli from river water as reported by Hijnen et al. (38). In addition, they reported the sequence of the MER values for Cryptosporidium oocysts > E. coli > phage 28B corresponded to the decrease in size of the organisms with the least elimination of the phage. In our study, the MERs for E. coli and oocysts were similar. This could have been due to the short time oocysts were analyzed in the leachate as well as the use of indigenous E. coli that was adapted to the slurry environment.
Microbial tracers in leachate.
The duration of the study revealed an extended leaching time and a long survival time for both phage 28B and E. coli. The bacteriophage could still be detected in the leachate after 5 months and were able to infect the Salmonella Typhimurium host strain, indicating that infectivity was maintained. E. coli originating from the slurry survived and could be detected in the leachate for 3½ months. E. coli derived from pig slurry has been shown to survive for 30 to 68 days in soils of different textures at low temperatures (22). E. coli originating from livestock feces deposited directly onto pasture survived up to 162 days during the cold season (8). Fecal coliforms from dairy shed effluent were detected in leachate from flood- or spray-irrigated sandy loam soil columns for up to 124 days (44). The extended leaching time for fecal coliforms could have been due to the irrigation practice as preferential flow is more likely to occur under irrigation compared to rainfall (18). McLeod et al. (58) showed the possibility of fast transport of microbial tracers (both bacteria and bacteriophage) through large soil columns, while the survival time was much shorter than that found in the present study.
Breakthrough of bacteriophages MS2 and PRD1 was measured in recharge water for 120 days in an artificial recharge area with an inactivation rate of 0.07 to 0.09 day−1. A 3-log-unit reduction was seen within the first 2.4 m, and an additional 5-log-unit reduction was seen within the following 27 m (71). The inactivation rate for viruses in groundwater has been reviewed by John and Rose (45) to vary from 0.02 to 0.1 log10 day−1. Inactivation of viruses has been shown to be temperature dependent, with greater inactivation at increasing temperatures, but this occurs mainly at temperatures higher than 20°C. We found a similar inactivation rate for phage 28B, as reported for hepatitis A virus and coxsackievirus, despite the fluctuating temperatures recorded during the study period.
C. parvum oocysts leached during the first 20 days, and the recovery rate for surface-applied slurry was significant lower (0.05%) than that found in the soil cores where slurry had been injected. Mawdsley et al. (54) added 108 oocysts on the surface of intact clay loam soil cores and irrigated with 11 mm artificial rainwater on alternate days for a period of 21 days. They recovered 0.045% of the oocysts, which is similar to the result observed in the present study. Ramirez et al. (67) added similar concentrations of oocysts to 1 liter of slurry that was deposited on intact soil blocks, but were able to recover 0.16% of the oocysts in the leachate. This could have been due to a large number of macropores wider than 2 mm, as they also observed undiluted slurry leaching through the soil immediately after application of slurry and before any rain was applied. Cryptosporidium oocysts added to soil blocks following intermitted irrigation under laboratory conditions could move through some soils for more than 70 days (55).
Molecular markers, provided that they are designed to detect viable cells, could have been used to monitor microbial cells in the leachate: e.g., the hsp70 gene encoding heat shock protein 70 in C. parvum oocysts (66). Prolonged exposure to the soil environment for bacteria can induce the viable but nonculturable (VBNC) stage. The bacterial cells cannot be cultured by traditional culture-based methods but only be identified by direct detection, including molecularly based methods (21). VBNC cells introduced to the soil environment did not display enhanced persistence compared to culturable cells (51), and the presence of equal numbers of total and culturable Salmonella cells in the leachate from soil monoliths indicated that most cells leaching were viable (9).
Thus, even under fluctuating temperatures and with natural precipitation, viable microbial tracers leached through intact soil cores with a survival/inactivation time similar to or higher than that reported previously.
Viability of oocysts in leachate.
Little information is available on the viability of the oocysts after passage through soil. A 50% viability of C. parvum oocysts in leachate from both surface-applied and injected slurry was found on day 6 in the present study. In these first 6 days, the temperature ranged from −3.5°C to 13°C. The viability of the oocysts varied between the soil cores and ranged from 33% to 63%. After 156 days at 4°C, 72% of oocysts were viable in a silt loam soil and 27% in distilled water (control), but at 20°C, 59.2% of oocysts were viable in the soil and no viable oocysts could be detected in the water (42). Cool temperatures (10°C) appear to increase viability and maintain infectivity of oocysts (61) as well as increasing the release of oocysts from manure and leaching through karst soil (13). The correlation between methods used to detect viable oocysts and infective oocysts has been questioned, and divergent results have been described in the literature (66, 69). Application of vital dye staining or in vitro excystation provides information on oocyst viability, but the results do not always correlate with the outcome of in vitro and in vivo infectivity assays. Only infectious oocysts are a potential health risk. Based on the few studies reported and results shown here, it seems possible for viable C. parvum oocysts to leach through soil. Whether the oocysts also could be considered infective and thereby able to cause disease in humans is currently unclear.
Effect of rainwater.
Soil cores in this study received primarily rainwater, and all of the microbial tracers were able to leach through the soil cores. Even though the soil cores were placed close together in the multicolumn lysimeter, variation in the amount of leachate was observed. This may indicate variation in soil structure between each of the soil cores and also variation in the amount of rain within the very small area where the soil cores were placed. The variation in the amount and intensity of natural rain could differ compared to artificial irrigation where the amount and intensity of irrigation water can be controlled. Natural raindrops would have reach terminal velocities when hitting the ground, and the kinetic energy of the drops would be high, with a greater chance to erode the manure compared to drops from irrigation (13). Schijven et al. (70) observed that drops exerted greater mechanical forces on manure than mist did, resulting in higher oocyst release rates from manure under simulated rain conditions. The oocyst recovery rates of Boyer et al. (13) from surface-applied manure were greater (0.12 to 0.27%) than those of the present study (0.05%), but that might have been a result of rainfall intensities of 2.8 cm h−1, as opposed to this study's maximum rainfall intensity of 0.48 cm h−1. The greater finding of oocysts from the injected soil cores in leachate could have been the result of lower exposure to inactivation factors such as desiccation or UV light (40). One control soil core in the study leached phage 28B. This contamination could have been the result of splashing from soil cores with surface-applied slurry onto this control soil core due to raindrop intensity as the soil cores were situated close together (9 cm apart) to maintain similar climate conditions for all soil cores.
Method of slurry application.
Injection of slurry is practiced on both grasslands and arable fields to reduce ammonia emission and odor as well as limiting the direct contamination of crops and the external environment with fecal microorganisms (19, 41). This practice is associated with risks of percolation and survival of enteric pathogens in soil (53) as compared to surface-applied slurry, where pathogens are exposure to high temperatures, desiccation, and UV light (40). Semenov et al. (72) suggested that surface application of slurry can decrease the risk of groundwater contamination as this led to shorter survival times for Salmonella Typhimurium in soil, which were in contrast to the results for E. coli O157:H7, for which no difference in the survival time was observed. In addition, higher concentrations of E. coli O157:H7 and Salmonella Typhimurium were detected in soil sampled at depth of 10 to 40 cm after 21 days when slurry was injected. On the contrary, Avery et al. (7) reported a higher concentration of E. coli O157:H7 in soil cores with surface-spread slurry compared to slurry injected at a depth of 25 cm after 8 weeks. Cryptosporidium oocysts were isolated at all depths (0 to 90 cm) without a clear pattern of distribution (0 to 640 oocysts g−1) in a surface- and subsurface-irrigated field study (5). Slurry broadcasting can lead to higher contamination with fecal coliforms and somatic coliphages in surface runoff waters, and injection of slurry only partially prevented microbial losses of runoff waters compared with broadcasting (78). We observed a significantly higher recovery rate of phage 28B and C. parvum oocysts in the leachate when slurry was injected, whereas the difference was insignificant for E. coli. This supports the conclusion made by Semenov et al. (72) that there could be higher risk for contamination of groundwater by injection of slurry.
In feces, concentrations as high as 1011 virus particles g−1 (68) and 1010 oocysts g−1 (73) can be excreted, but concentrations of 10 enterovirus particles ml−1 (25) and 1 oocyst ml−1 (74) have been reported in different wastes. In the present study, the slurry that was applied had 1.8 × 109 phage ml−1 and 4.0 × 104 oocysts ml−1, so the study is illustrating a worst case scenario in relation to virus and oocyst contamination. With the recovery and removal rates found in the present study, there would be a possible health risk, especially related to leaching of viruses, while infective oocysts and bacteria with low infective dose (e.g., E. coli O157:H7) had a low recovery, and a very large removal rate may constitute a minor health risk. Further epidemiological risk factor studies are needed for drinking-waterborne-associated diseases to assess whether such potential risks for pathogen breakthrough to groundwater represent actual human health risks.
Overall, this study showed that the risk of groundwater contamination was especially related to viruses and protozoan parasites by injection of slurry. The transport through intact soil does not seem to be related only to the sizes of the microorganisms but also initially adaptation to the organic waste environment and possible attachment to slurry particles may play an important role.
ACKNOWLEDGMENTS
We thank Cynthia D. Juel at the National Veterinary Institute, Technical University of Denmark, for excellent guidance and technical support regarding the parasitological analyses. The technical assistance of Dang Thi Thanh Son was very much appreciated. We thank Jakob Ottoson and Annika Holmqvist for the provision of S. Typhimurium bacteriophage 28B and the host strain S. Typhimurium type 5.
This study was supported by the “Safe and High Quality Food Production using Low Quality Waters and Improved Irrigation Systems and Management” project (SAFIR, EU, FOOD-CT-2005-023168) (www.safir4eu.org) funded by European Commission, the PATHOS project (“From Manure to Freshwater—Technology Avoiding Contamination with Pathogens, Hormones and Pharmaceuticals”) (www.pathos.geus.net) supported by the Danish Council for Strategic Research (ENV 2104-07-0015), and the EU-funded PathOrganic under the CoreOrganic ERA-net (project no. 1888) on risks and recommendations regarding human pathogens in the organic vegetable production chain (www.pathorganic.coreportal.org).
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Abbaszadegan M., LeChevallier M., Gerba C. 2003. Occurrence of viruses in U.S. groundwaters. J. Am. Water Works Assoc. 95:107–120 [Google Scholar]
- 2. Abu-Ashour J., Joy D. M., Lee H., Whiteley H. R., Zelin S. 1994. Transport of microorganisms through soil. Water Air Soil Pollut. 75:141–158 [Google Scholar]
- 3. Abu-Ashour J., Joy D. M., Lee H., Whiteley H. R., Zelin S. 1998. Movement of bacteria in unsaturated soil columns with macropores. Trans. ASAE 41:1043–1050 [Google Scholar]
- 4. Adams M. H. 1959. Bacteriophages. Interscience Publishers Inc., New York, NY [Google Scholar]
- 5. Armon R., Gold D., Brodsky M., Oron G. 2002. Surface and subsurface irrigation with effluents of different qualities and presence of Cryptosporidium oocysts in soil and on crops. Water Sci. Technol. 46:115–122 [PubMed] [Google Scholar]
- 6. Artz R. R. E., Townend J., Brown K., Towers W., Killham K. 2005. Soil macropores and compaction control the leaching potential of Escherichia coli O157:H7. Environ. Microbiol. 7:241–248 [DOI] [PubMed] [Google Scholar]
- 7. Avery L. M., Hill P., Killham K., Jones D. L. 2004. Escherichia coli O157 survival following the surface and sub-surface application of human pathogen contaminated organic waste to soil. Soil Biol. Biochem. 36:2101–2103 [Google Scholar]
- 8. Avery S. M., Moore A., Hutchison M. L. 2004. Fate of Escherichia coli originating from livestock faeces deposited directly onto pasture. Lett. Appl. Microbiol. 38:355–359 [DOI] [PubMed] [Google Scholar]
- 9. Bech T. B., et al. 2010. Transport and distribution of Salmonella enterica serovar Typhimurium in loamy and sandy soil monoliths with applied liquid manure. Appl. Environ. Microbiol. 76:710–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bech T. B., Dalsgaard A., Jacobsen O. S., Jacobsen C. S. 2011. Leaching of Salmonella enterica in clay columns comparing two manure application methods. Ground Water 49:32–42 [DOI] [PubMed] [Google Scholar]
- 11. Beven K., Germann P. 1982. Macropores and water flows in soil. Water Resour. Res. 18:1311–1325 [Google Scholar]
- 12. Borchardt M. A., et al. 2010. Norovirus outbreak caused by a new septic system in a dolomite aquifer. Ground Water 49:85–97 [DOI] [PubMed] [Google Scholar]
- 13. Boyer D. G., Kuczynska E., Fayer R. 2009. Transport, fate, and infectivity of Cryptosporidium parvum oocysts released from manure and leached through macroporous soil. Environ. Geol. 58:1011–1019 [Google Scholar]
- 14. Brasseur P., Uguen C., Moreno-Sabater A., Favennec L., Ballet J. J. 1998. Viability of Cryptosporidium parvum oocysts in natural waters. Folia Parasitol. 45:113–116 [PubMed] [Google Scholar]
- 15. Carlander A., Aronsson P., Allestam G., Stenström T. A., Perttu K. 2000. Transport and retention of bacteriophages in two types of willow-cropped lysimeters. J. Environ. Sci. Health A 35:1477–1492 [Google Scholar]
- 16. Casey F. X. M., Logsdon S. D., Horton R., Jaynes D. B. 1998. Measurement of field soil hydraulic and solute transport parameters. Soil Sci. Soc. Am. J. 62:1172–1178 [Google Scholar]
- 17. Chambers B., et al. 2001. Managing livestock manures, booklet 3. Spreading systems for slurries and solid manures, ADAS, Ltd., Wolverhampton, United Kingdom [Google Scholar]
- 18. Chen C., Roseberg R. J., Selker J. S. 2002. Using microsprinkler irrigation to reduce leaching in a shrink/swell clay soil. Agric. Water Manag. 54:159–171 [Google Scholar]
- 19. Chen Y., Zhang Q., Petkau D. S. 2001. Evaluation of different techniques for liquid manure application on grassland. Appl. Eng. Agric. 17:489–496 [Google Scholar]
- 20. Collins T. J. 2007. ImageJ for microscopy. Biotechniques 43:S25–S30 [DOI] [PubMed] [Google Scholar]
- 21. Colwell R. R. 2000. Viable but nonculturable bacteria: a survival strategy. J. Infect. Chemother. 6:121–125 [DOI] [PubMed] [Google Scholar]
- 22. Cools D., Merckx R., Vlassak K., Verhaegen J. 2001. Survival of E. coli and Enterococcus spp. derived from pig slurry in soils of different texture. Appl. Soil Ecol. 17:53–62 [Google Scholar]
- 23. Crane S. R., Moore J. A. 1984. Bacterial pollution of groundwater: a review. Water Air Soil Pollut. 22:67–83 [Google Scholar]
- 24. Cumbie D. H., McKay L. D. 1999. Influence of diameter on particle transport in a fractured shale saprolite. J. Contam. Hydrol. 37:139–157 [Google Scholar]
- 25. Dahling D. R., Stafferman R. S., Wright B. A. 1989. Isolation of enterovirus and reovirus from sewage and treated effluents in selected Puerto Rican communities. Appl. Environ. Microbiol. 55:503–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Damgaard-Larsen S., Jensen K. O., Lund E., Nissen B. 1977. Survival and movement of enterovirus in connection with land disposal of sludges. Water Res. 11:503–508 [Google Scholar]
- 27. DuPont H. L., et al. 1995. The infectivity of Cryptosporidium parvum in healthy volunteers. N. Engl. J. Med. 332:855–859 [DOI] [PubMed] [Google Scholar]
- 28. FAO 2006. Lifestock's long shadow—environmental issues and options. Food and Agriculture Organization of the United Nations; ftp://ftp.fao.org/docrep/fao/010/a0701e/A0701E00.pdf [Google Scholar]
- 29. Fayer R., Morgan U., Upton S. J. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305–1322 [DOI] [PubMed] [Google Scholar]
- 30. Fontes D. E., Mills A. L., Hornberger G. M., Herman J. S. 1991. Physical and chemical factors influencing transport of microorganisms through porous media. Appl. Environ. Microbiol. 57:2473–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gagliardi J. V., Karns J. S. 2000. Leaching of Escherichia coli O157:H7 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gordon C., Toze S. 2003. Influence of groundwater characteristics on the survival of enteric viruses. J. Appl. Microbiol. 95:536–544 [DOI] [PubMed] [Google Scholar]
- 33. Guber A. K., et al. 2007. Comparison of release and transport of manure-borne Escherichia coli and enterococci under grass buffer conditions. Lett. Appl. Microbiol. 44:161–167 [DOI] [PubMed] [Google Scholar]
- 34. Guber A. K., Pachepsky Y. A., Shelton D. R., Yu O. 2007. Effect of bovine manure on fecal coliform attachment to soil and soil particles of different sizes. Appl. Environ. Microbiol. 73:3363–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hahesy T., Scanlon M., Carton O. T., Quinn P. J., Lenejan J. J. 1992. The effect of spreading methods on the dispersal of bacteria in slurry. Irish J. Agric. Food Res. 31:98 [Google Scholar]
- 36. Harter T., Wagner S. 2000. Colloid transport and filtration of Cryptosporidium parvum in sandy soils and aquifer sediments. Environ. Sci. Technol. 34:62–70 [Google Scholar]
- 37. Havelaar A. H. 1991. Bacteriophages as model viruses in water quality control. Water Res. 25:529–545 [Google Scholar]
- 38. Hijnen W. A. M., Brouwer-Hanzens A. J., Charles K. J., Medema G. J. 2005. Transport of MS2 phage, Escherichia coli, Clostridium perfringens, Cryptosporidium parvum and Giardia intestinalis in a gravel and a sandy soil. Environ. Sci. Technol. 39:7860–7868 [DOI] [PubMed] [Google Scholar]
- 39. Höglund C., Ashbolt N., Stenström T. A., Svensson L. 2002. Viral persistence in source-separated humane urine. Adv. Environ. Res. 6:265–275 [Google Scholar]
- 40. Hutchison M. L., Walters L. D., Moore A., Crookes K. M., Avery S. M. 2004. Effect of length of time before incorporation on survival of pathogenic bacteria present in livestock wastes applied to agricultural soil. Appl. Environ. Microbiol. 70:5111–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Islam M., et al. 2004. Persistence of Salmonella enterica serovar Typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathog. Dis. 1:27–35 [DOI] [PubMed] [Google Scholar]
- 42. Jenkins M. B., Bowman D. D., Fogarty E. A., Ghiorse W. C. 2002. Cryptosporidium parvum oocyst inactivation in three soil types at various temperatures and water potentials. Soil Biol. Biochem. 34:1101–1109 [Google Scholar]
- 43. Jiang S., et al. 2008. Bacterial leaching from dairy shed effluent applied to a fine sandy loam under irrigated pasture. Aust. J. Soil. Res. 46:552–564 [Google Scholar]
- 44. Jiang S., et al. 2010. Modeling water flow and bacterial transport in undisturbed lysimeters under irrigations of dairy shed effluent and water using HYDRUS-1D. Water Res. 44:1050–1061 [DOI] [PubMed] [Google Scholar]
- 45. John D. E., Rose J. B. 2005. Review of factors affecting microbial survival in groundwater. Environ. Sci. Technol. 39:7345–7356 [DOI] [PubMed] [Google Scholar]
- 46. Kjaergaard C., Poulsen T. G., Moldrup P., de Jonge L. W. 2004. Colloid mobilization and transport in undisturbed soil columns. I. Pore structure characterization and tritium transport. Vadose Zone J. 3:413–423 [Google Scholar]
- 47. Kothary M. H., Babu U. S. 2001. Infective dose of foodborne pathogens in volunteers: a review. J. Food Saf. 21:49–73 [Google Scholar]
- 48. Lilleengen K. 1948. Typing of Salmonella typhimurium by means of a bacteriophage. Ph.D. thesis Bacteriological and Hygienical Department of the Royal Veterinary College, Stockholm, Sweden [Google Scholar]
- 49. Lindhardt B., et al. 2001. The Danish Pesticide Leaching Assessment Programme—site characterization and monitoring design. Geological Survey of Denmark and Greenland, Copenhagen, Denmark [Google Scholar]
- 50. Maddox-Hyttel C., Langkjær R. B., Enemark H. L., Vigre H. 2006. Cryptosporidium and Giardia in different age groups of Danish cattle and pigs—occurrence and management associated risk factors. Vet. Parasitol. 141:48–59 [DOI] [PubMed] [Google Scholar]
- 51. Mascher F., Hase C., Moënne-Loccoz Y., Défago G. 2000. The viable-but-nonculturable state induced by abiotic stress in biocontrol agent Pseudomonas fluorescens CHA0 does not promote strain persistence in soil. Appl. Environ. Microbiol. 66:1662–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maurer A. M., Sturchler D. 2000. A waterborne outbreak of small round structured virus, campylobacter and Shigella co-infections in La Neuveville, Switzerland, 1998. Epidemiol. Infect. 125:325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mawdsley J. L., Bardgett R. D., Merry R. J., Pain B. F., Theodorou M. K. 1995. Pathogens in livestock waste, their potential for movement through soil and environmental pollution. Appl. Soil Ecol. 2:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mawdsley J. L., Brooks A. E., Merry R. J. 1996. Movement of the protozoan pathogen Cryptosporidium parvum through three contrasting soil types. Biol. Fertil. Soils 21:30–36 [Google Scholar]
- 55. Mawdsley J. L., Brooks A. E., Merry R. J., Pain B. F. 1996. Use of a novel soil tilting table apparatus to demonstrate the horizontal and vertical movement of the protozoan pathogen Cryptosporidium parvum in soil. Biol. Fertil. Soils 21:215–220 [Google Scholar]
- 56. McCarthy J. F., Zachara J. M. 1989. Subsurface transport of contaminants. Environ. Sci. Technol. 23:496–502 [Google Scholar]
- 57. McGechan M. B., Vinten A. J. A. 2003. Simulation of transport through soil of E. coli derived from livestock slurry using the MACRO model. Soil Use Manag. 19:321–330 [Google Scholar]
- 58. McLeod M., Aislabie J., Ryburn J., McGill A., Taylor M. 2003. Microbial and chemical tracer movement through two Southland soils, New Zealand. Aust. J. Soil Res. 41:1163–1169 [Google Scholar]
- 59. McMurry S. W., Coyne M. S., Perfect E. 1998. Fecal coliform transport through intact soil blocks amended with poultry manure. J. Environ. Qual. 27:86–92 [Google Scholar]
- 60. Medema G. J., Schets F. M., Teunis P. F. M., Havelaar A. H. 1998. Sedimentation of free and attached Cryptosporidium oocysts and Giardia cysts in water. Appl. Environ. Microbiol. 64:4460–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nasser A. M., Tweto E., Nitzan Y. 2007. Die-off of Cryptosporidium parvum in soil and wastewater effluent. J. Appl. Microbiol. 102:169–176 [DOI] [PubMed] [Google Scholar]
- 62. Nicosia L. A., Rose J. B., Stark L., Stewart M. Y. 2001. A field study of virus removal in septic tank drainfields. J. Environ. Qual. 30:1933–1939 [DOI] [PubMed] [Google Scholar]
- 63. Nielsen D. R., van Genuchten M. T., Biggar J. W. 1986. Water flow and solute transport processes in the unsaturated zone. Water Resour. Res. 22:89–108 [Google Scholar]
- 64. Oliver D. M., Haygarth P. M., Clegg C. D., Heathwaite A. L. 2006. Differential E. coli die-off patterns associated with agricultural matrices. Environ. Sci. Technol. 40:5710–5716 [DOI] [PubMed] [Google Scholar]
- 65. Pang L., McLeod M., Aislabie J., Šimùnek J., Close M., Hector R. 2008. Modeling transport of microbes in ten undisturbed soils under effluent irrigation. Vadose Zone J. 7:97–111 [Google Scholar]
- 66. Quintero-Betancourt W., Peele E. R., Rose J. B. 2002. Cryptosporidium parvum and Cyclospora cayetanensis: a review of laboratory methods for detection of these waterborne parasites. J. Microbiol. Methods 49:209–224 [DOI] [PubMed] [Google Scholar]
- 67. Ramirez N. E., et al. 2009. Effect of tillage and rainfall on transport of manure-applied Cryptosporidium parvum oocysts through soil. J. Environ. Qual. 38:2394–2401 [DOI] [PubMed] [Google Scholar]
- 68. Reynolds K. A., Mena K. D., Gerba C. P. 2008. Risk of waterborne illness via drinking water in the United States. Rev. Environ. Contam. Toxicol. 192:117–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schets F. M., Engels G. B., During M., de Roda Husman A. M. 2005. Detection of infectious Cryptosporidium oocysts by cell culture immunofluorescence assay: application to environmental samples. Appl. Environ. Microbiol. 71:6793–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schijven J. F., Bradford S. A., Yang S. 2004. Release of Cryptosporidium and Giardia from dairy cattle manure: physical factors. J. Environ. Qual. 33:1499–1508 [DOI] [PubMed] [Google Scholar]
- 71. Schijven J. F., Hoogenboezem W., Hassamizadeh S. M., Peters J. H. 1999. Modeling removal of bacteriophage MS2 and PRD1 by dune recharge at Castricum, Netherlands. Water Resour. Res. 35:1101–1111 [Google Scholar]
- 72. Semenov A. V., van Overbeek L., van Bruggen A. H. C. 2009. Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Appl. Environ. Microbiol. 75:3206–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Smith H. V. 1992. Cryptosporidium and water: a review. Water Environ. J. 6:443–451 [Google Scholar]
- 74. Smith H. V., et al. 1989. An outbreak of waterborne cryptosporidiosis caused by post-treatment contamination. Epidemiol. Infect. 103:703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sobsey M. D., Shields P. A., Hauchman F. H., Hazard R. L., Caton L. W., III 1989. Survival and transport of hepatitis A virus in soils, groundwater and wastewater. Water Sci. Technol. 10:97–106 [Google Scholar]
- 76. Unc A., Goss M. J. 2004. Transport of bacteria from manure and protection of water resources. Appl. Soil Ecol. 25:1–18 [Google Scholar]
- 77. US EPA 2006. Occurrence and monitoring document for the final ground water rule. EPA 815-R-06-012. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 78. Uusi-Kämppä J., Heinonen-Tanski H. 2008. Evaluating slurry broadcasting and injection to ley for phosphorus losses and fecal microorganisms in surface runoff. J. Environ. Qual. 37:2339–2350 [DOI] [PubMed] [Google Scholar]
- 79. Vinten A. J. A., et al. 2002. Fate of Escherichia coli and Escherichia coli O157 in soils and drainage water following cattle slurry application at three sites in southern Scotland. Soil Use Manag. 18:223–231 [Google Scholar]
- 80. Ward R. L., et al. 1986. Human rotavirus studies in volunteers: determination of infectious dose and serological response to infection. J. Infect. Dis. 154:871–880 [DOI] [PubMed] [Google Scholar]
- 81. Wildenschild D., Jensen K. H., Villholth K., Illangasekare T. H. 1994. A laboratory analysis of the effect of macropores on solute transport. Ground Water 32:381–389 [Google Scholar]
- 82. Zhuang J., Qui J., Jin A. 2005. Retention and transport of amphiphilic colloids under unsaturated flow conditions: effect of particle size and surface property. Environ. Sci. Technol. 39:7853–7859 [DOI] [PubMed] [Google Scholar]