Abstract
Aerobic granules are dense microbial aggregates with the potential to replace floccular sludge for the treatment of wastewaters. In bubble-column sequencing batch reactors, distinct microbial populations dominated propionate- and acetate-cultivated aerobic granules after 50 days of reactor operation when only carbon removal was detected. Propionate granules were dominated by Zoogloea (40%), Acidovorax, and Thiothrix, whereas acetate granules were mainly dominated by Thiothrix (60%). Thereafter, an exponential increase in enhanced biological phosphorus removal (EBPR) activity was observed in the propionate granules, but a linear and erratic increase was detected in the acetate ones. Besides Accumulibacter and Competibacter, other bacterial populations found in both granules were associated with Chloroflexus and Acidovorax. The EBPR activity in the propionate granules was high and stable, whereas EBPR in the acetate granules was erratic throughout the study and suffered from a deterioration period that could be readily reversed by inducing hydrolysis of polyphosphate in presumably saturated Accumulibacter cells. Using a new ppk1 gene-based dual terminal-restriction fragment length polymorphism (T-RFLP) approach revealed that Accumulibacter diversity was highest in the floccular sludge inoculum but that when granules were formed, propionate readily favored the dominance of Accumulibacter type IIA. In contrast, acetate granules exhibited transient shifts between type I and type II before the granules were dominated by Accumulibacter type IIA. However, ppk1 gene sequences from acetate granules clustered separately from those of propionate granules. Our data indicate that the mere presence of Accumulibacter is not enough to have consistently high EBPR but that the type of Accumulibacter determines the robustness of the phosphate removal process.
INTRODUCTION
In sequencing batch reactors operated under high shear stress and short setting times, microorganisms involved in wastewater treatment can form structures called aerobic granules (1). Aerobic granules are compact quasispherical aggregates formed by self-immobilized mixed microbial communities embedded in a matrix of extracellular biopolymeric substances (35). These microbial aggregates can accommodate a biomass concentration five times higher than that of activated sludge flocs (13) and settle 10 times faster (42, 60). These characteristics translate into small-footprint treatment plants, and therefore the use of aerobic granular sludge-based systems has recently been proposed as a promising alternative technology to conventional activated sludge treatment systems, with annual maintenance costs and land requirements that would be reduced by 17% and 75%, respectively (12).
For a robust application of this novel technology, the granular sludge should be not only physically but also microbiologically stable; that is, granules should contain an appropriate microbial assembly to account for contaminant removal, and ideally this assembly should reveal fitness to environmental changes. Aerobic granules can be used to simultaneously remove carbon, nitrogen, and phosphorus from wastewaters, as has been shown in laboratory sequencing batch reactors (13, 32, 61). However, this technology for combined carbon and nutrient removal still needs to be demonstrated at full scale.
Phosphorus removal is achieved by the Betaproteobacteria species tentatively named “Candidatus Accumulibacter phosphatis” and generally referred to as polyphosphate (poly-P)-accumulating organisms (PAOs) or Accumulibacter (11, 49, 64). When exposed to anaerobic-aerobic/anoxic cycles, PAOs have the metabolic capacity to take up phosphate beyond their anabolic requirements and store it intracellularly as poly-P. This microbial process is called enhanced biological phosphorus removal (EBPR) and has been incorporated in numerous full-scale activated sludge treatment plants (41, 43). However, different reports have indicated that often and for unknown reasons, this process is not stable, and attempts to recover EBPR capacity are not always successful (see reference 43 and references therein). One proposed reason for EBPR failure is that Accumulibacter-related populations are outcompeted by glycogen-accumulating organisms (GAOs). GAOs classed with Gammaproteobacteria are generally named “Candidatus Competibacter phosphatis” (10). Additionally, two GAO lineages that are classed with Alphaproteobacteria have been described; one is related to Sphingomonas and the other to Defluvicoccus vanus (40, 59). GAOs compete for carbon but they do not contribute to EBPR (10, 43). In general, temperatures equal to or lower than 20°C and pH values equal to or higher than 7.5 favor Accumulibacter over GAO populations (36). In addition, propionate provides Accumulibacter with a selective advantage because the propionate uptake rate by Competibacter GAOs is insignificant and because only Alphaproteobacteria GAOs, and not other GAOs, may compete for propionate (44, 45). There is the hypothesis that different PAO types or strains develop when either acetate or propionate is present as the main carbon source (9). Recently, by using the polyphosphate kinase gene (ppk1) as a phylogenetic marker, two major types of ppk1 sequences comprising at least five clades within Accumulibacter were recovered from lab-scale and full-scale EBPR sludges, suggesting that this lineage is more diverse than previously thought (20). Furthermore, a recent study suggests that Accumulibacter belonging to ppk1 clade type I and type II have different denitrification capabilities, with type I being able to couple nitrate reduction with phosphorus uptake, whereas type II cannot (17). It is not clear, however, which reactor operational conditions favor the selection of either Accumulibacter type, as their observed dominance in laboratory-activated sludge systems seems to be random (17). It is also not known whether the propionate-conferred superior phosphorus removal capacity as compared to that for acetate is associated with the selection of a specific Accumulibacter clade.
Because aerobic granular technology is recent, not much information is available in relation to the microbial makeup of the granules (33, 34, 54). In contrast to suspended biomass, strong chemical gradients may develop within the granular environment, leading to the formation of distinct microbial niches and granule architecture where flanking populations may closely interact with PAOs. For full-scale applications, detailed knowledge that guarantees granule robustness concerning efficient nutrient removal is needed. Therefore, the aims of the present study were as follows: (i) to explore the effectiveness of using propionate as the controlling parameter to develop granules with robust phosphate removal activity, (ii) to reveal the microbial community structure and dynamics of aerobic granules cultivated with propionate or acetate and its association to reactor performance over time, and (iii) to explore the associated dynamics of Accumulibacter clades.
MATERIALS AND METHODS
Reactor operating conditions.
Two double-walled glass reactors with working volumes of 2.4 liters each were inoculated with 500 ml of activated sludge obtained from a sewage treatment plant (Uetendorf, Thun, Switzerland) that biologically removes N and P. The reactors were operated at 20°C in sequencing batch mode with cycles of 3 h in a manner similar to that described previously (13). Each cycle consisted of the following phases: (i) a 60-min anaerobic feeding with the introduction of influent from the bottom of the reactor at a flow rate of 1.2 liters h−1, (ii) a 112-min aeration phase with airflow rate at 4 liters min−1, (iii) a 3-min settling phase unless otherwise specified, and (iv) a 5-min withdrawal phase. The reactor pH was controlled at 7.0 to 7.3 during the aerobic phase, and influent allylthiourea (50 mg liter−1) addition, used to prevent nitrification, was stopped at day 15. During the first 10 days, the settling time was gradually reduced from 15 to 3 min; then, at day 33, it was decreased to 2 min; and finally, at day 55, it was increased and maintained at 3 min. The composition of the synthetic wastewater used to feed the reactors was as given previously (13) except that one reactor was fed with acetate and the other with propionate as the sole carbon source. The organic loading rate for both reactors was 1.8 g chemical oxygen demand (COD) liter−1 day−1, and the C/N and C/P ratios were 3.5:1 and 20:1, respectively. Oxygen was maintained at saturation levels during the aerobic phase. To monitor the reactor performance, influent and effluent samples were taken and analyzed for volatile fatty acids (VFAs), PO4–P, NH4+, NO3−, and NO2−. Cycle studies were conducted during the experimental period in which filtered liquid samples were removed from the reactor during the aerobic phase and analyzed for the same components mentioned above.
Analytical procedures.
Concentrations of volatile fatty acids were determined by high-performance liquid chromatography (HPLC) (Jasco Co-2060 Plus, refraction index detector; Omnilab, Germany) with organic acid ion-exclusion column ORH-801 (Transgenomics, United Kingdom). Inorganic anions and cations were measured by ion chromatography (anions: ICS-90, IonPacAS14A column; cations: ICS-3000A, IonPacCS16 column) with electrical conductivity detection (Dionex, Switzerland). The total suspended solids (TSS) and volatile suspended solids (VSS) from the reactor and from the effluent were measured as described in reference 20. The granule mean diameter was estimated from digital images of 300 to 770 granules using the open software ImageJ (http://rsb.info.nih.gov/ij/index.html). The sludge volume index (SVI30), which is the volume occupied by 1 g of activated sludge after 30 min of sedimentation, was determined as described previously (13). For granular sludge, the SVI was determined after 8 min of sedimentation (SVI8) as suggested earlier (13). Polyhydroxyalkanoates (PHAs) were measured by gas chromatography as detailed previously (55).
Fluorescence in situ hybridization (FISH) and flow cytometry.
To determine the relative abundances of PAOs and GAOs, granular sludge samples were fixed overnight at 4°C in 4% formaldehyde in phosphate-buffered saline (PBS), pH 7.4. Then, granules were washed three times with PBS and stored at −20°C in a PBS-ethanol solution (1:1, vol/vol). Fixed granules were mechanically disrupted and the obtained slurry was 5 times diluted (optical density at 600 nm [OD600], ∼0.8) with filter-sterilized PBS. The obtained suspension was then sonicated (Vibra cell 72405) three times for 0.8 s at amplitude 50 with a brief incubation on ice between the sonication events. Samples were then passed through a 30-μm nylon mesh filter, and 0.5-ml aliquots of filtrated cells (OD600, ∼0.3) were centrifuged and hybridized using the protocol described previously (18) with slight modifications. Aliquots of 50 μl of hybridization buffer containing a cocktail of the following probes were used: Cy3-labeled PAO probes PAO462, PAO651, and PAO846 (11) or 6-carboxyfluorescein (FAM)-labeled GAO probes GAOQ431, GAOQ989, and GB_G2 (10, 28). Final concentrations of Cy3-labeled and FAM-labeled probes were 3 ng ml−1 and 5 ng ml−1, respectively, and hybridization conditions were according to references 10, 11, and 28. After the removal of unbound probes with preheated washing buffer for 20 min at 48°C, cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole) (20 μg ml−1) for 20 min. Then, cells were washed and resuspended in 2 ml cold filter-sterilized PBS buffer and filtered through a 7-μm filter. Cells were then analyzed using a CyAn ADP flow cytometer (Dako) equipped with an argon laser (488 nm) and a diode laser (405 nm). FAM- and Cy3-stained cells were excited with the argon laser and detected by the FL-1 (band-pass filter 530/40) and FL-2 (band-pass filter 575/25) detectors, respectively. DAPI-stained cells were excited with the diode laser and detected by the FL-6 (band-pass filter 450/50) detector. The acquisition threshold was set in the side-scatter channel. All parameters were collected as logarithmic signals. For each sample, 100,000 to 200,000 events were recorded, and the obtained data were evaluated using Summit software v4.3 (Dako). Bivariant dot plots of DAPI versus Cy3- or FAM-conferred fluorescence were used to define PAO- or GAO-positive cells in the samples, respectively. As a proof of principle, hybridization results of some samples were checked by fluorescence microscopy.
DNA extraction.
Granules were taken from the reactors during the aerobic phase and immediately stored at −80°C. Before DNA extraction, a slurry of the granules was prepared by mechanical disruption in PBS buffer. Genomic DNA was extracted, in duplicate, from 0.3 to 0.4 ml of slurry using the Power Soil DNA isolation kit (MoBio, Inc.) according to the manufacturer's protocol except that two cycles of bead beating for 30 s (each cycle followed by 30 s on ice) were used. DNA integrity was checked by 0.8% agarose gel electrophoresis using GelRed staining (Biotium, Hayward, CA). DNA concentration and purity were measured using a NanoDrop ND-1000 spectrophotometer.
PCR amplification.
Bacterial 16S rRNA gene fragments were amplified using the primer pair 27f and 1392r (30). The PCR contained 12.5 μl of GoTaq colorless master mix (Promega), 0.4 μM each primer, and 20 to 40 ng of DNA template in a final reaction volume of 25 μl adjusted with nuclease-free water. The thermocycling consisted of an initial step at 94°C for 2 min, followed by 25 cycles at 94°C for 45 s, 50°C for 45 s, and 72°C for 1.5 min, with a final extension at 72°C for 7 min. Accumulibacter-related ppk1 gene fragments were amplified using the primer pair Acc-ppk1-254f and Acc-ppk1-1376r with the two-step PCR protocol as described previously (21). The thermocycling consisted of an initial step at 95°C for 2 min, followed by 25 cycles at 95°C for 45 s, 68°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 7 min. Product sizes of about 1,300 bp for 16S rRNA and about 1,100 bp for ppk1 gene amplicons were confirmed on 1.5% agarose gels. Products of duplicate reactions were pooled and then purified using the MSB spin PCRapace kit (Invitek) according to the manufacturer's protocol.
Cloning and sequencing of bacterial 16S rRNA and Accumulibacter ppk1 genes.
Purified PCR products were ligated into the pGEM-T Easy vector (Promega), which was then used to transform competent E. coli DH5α cells according to the manufacturer's instructions. Transformants were screened by colony PCR in a 25-μl PCR volume with vector-specific primers T7 and SP6. The PCR products from clones containing the correct size, as determined by gel electrophoresis, were digested with the HaeIII enzyme (Promega). Digested samples were separated by electrophoresis in 2.5% agarose gels. Restriction patterns were compared and representative clones were selected for sequencing. Purified PCR products (100 to 150 ng) from the selected clones were sequenced in both directions using the BigDye V3.1 terminator sequencing kit with T7 or SP6 primer. The resulting reaction products were cleaned to remove excess dye by ethanol precipitation and then run in an ABI 3130XL genetic analyzer (Applied Biosystems) according to the manufacturer's instructions. Sequences were assembled using Lasergene software (DNASTAR). Sequences were screened for potential chimeras using the software programs Chimera_Check from Greengenes (15), Bellerophon (23), and Mallard (3); sequences showing anomalies were not considered further. Sequences were matched with GenBank sequences using BLAST (4).
T-RFLP analysis.
Bacterial 16S rRNA gene fragments were amplified as described above except that the forward primer was 5′ terminally labeled with 6-carboxyfluorescein (FAM). Accumulibacter ppk1 gene fragments were amplified as described above except that either the forward or the reverse primer was FAM labeled. Terminal restriction fragments (T-RFs) were obtained by digestion of 50 ng of 16S rRNA or 20 ng of ppk1-amplified gene products with 5 U of HaeIII enzyme at 37°C for 4 h. One microliter of digested sample was mixed with 8.5 μl of Hi-Di formamide and 0.5 μl of internal size standard (GeneScan 1200 Liz Size Standard; Applied Biosystems). This mixture was denatured at 95°C for 2 min and then snap-cooled on ice. To separate the restriction fragments, the denatured samples were loaded on an ABI 3130XL genetic analyzer equipped with 50-cm capillaries filled with POP 7 (Applied Biosystems) as the electrophoresis polymer, and the lengths of the fluorescently labeled T-RFs were determined by comparison with the internal size standard using the GeneMapper software. Resulting T-RF peak sizes were rounded to the nearest integer using the Microsoft Excel macro Treeflap (47), and two peaks were considered identical if their difference was less than 0.5 bp. To avoid detection of primers and uncertainties in size estimation, fragments of less than 50 bp and more than 900 bp were excluded from further analysis. The relative abundance of a detected T-RF within a given terminal-restriction fragment length polymorphism (T-RFLP) profile was estimated from the ratio between its peak area and the total area of all peaks within the profile. T-RFs showing <1% relative abundance were considered not significant and were excluded before percentages of relative abundance were recalculated for the remaining T-RFs. The affiliation of T-RFs to sequences was confirmed using the corresponding clone as the template.
Pragmatic analysis of T-RFLP data.
The 16S rRNA-based T-RFLP community patterns derived from the reactor samples were compared by nonmetric multidimensional scaling (NMS) based on the distance measure with synthetic axis. The NMS algorithm iteratively searches to place samples as points on k dimensions (axis) while attempting to maintain the relative distances of points as close as possible to the actual rank order of similarities between samples. Hence, sludge samples with similar community structures are plotted together in ordination space. NMS plots were computed using PC-ORD v.4 (39) in a manner similar to that described in reference 8 using the “slow-and-thorough” autopilot mode of the program. A dissimilar distance matrix of Sorensen (Bray-Curtis) was used with 40 runs with random starting configuration using the real data and 50 runs using randomized data. Based on the rules defined in reference 39, the ordination result is considered good when final stress values are <10. The T-RFLP data set was further analyzed following the pragmatic procedure proposed by Marzorati et al. (38), which describes the microbial community based on the three properties described immediately below. Briefly, the range-weighted richness was estimated as the number of peaks in each T-RFLP profile. The dynamics of the community (i.e., rate of change) averages the degree of change between two consecutive profiles over a set time interval and was estimated using the Pearson moment correlation coefficient between day x and day x − 60 days. The functional organization was graphically represented by Pareto-Lorenz evenness curves. Construction of these curves was done as described previously (58). Mathematically, the results yield convex curves. The more a curve deviates from the theoretical perfect evenness line, defined as a 45° diagonal, the less evenness can be observed in structure of the community (58).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences reported in this study are JN090776 to JN090838 (16S rRNA gene) and JN090839 to JN090858 (ppk1 gene).
RESULTS
Initial reactor stage.
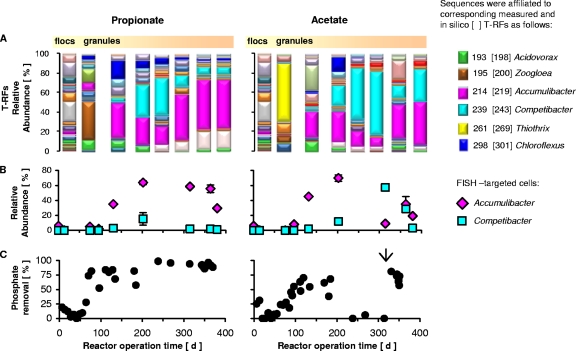
At day 50 to day 65, and in both reactors, most biomass was in granular form (approximately 90% of granules had diameters of >0.5 mm). The average granule sizes of the propionate- and acetate-cultivated granules were 1.8 ± 0.2 mm and 2.0 ± 0.4 mm, respectively. The granules had good settling properties, as reflected by the measured SVI8 values of 48 ml g−1 for the propionate and 63 ml g−1 for the acetate granules. The SVI (sludge volume index) is typically used to monitor sludge settleability and is usually measured after 30 min of sedimentation. The lower the SVI value, the better the sludge sediments. As a reference, the activated sludge used as the inoculum had an SVI30 value of 130 ml g−1, which is common for this type of floccular biomass. At this stage, the microbial community consumed all carbon, mostly during the aerobic phase, and there was no phosphorus removal. Some nitrogen was removed, likely due to assimilation, and neither nitrite nor nitrate was detected in the effluent of the reactors. For both reactors, the 16S rRNA gene T-RFLP data showed that a strong change in the bacterial community composition occurred between days 0 and 65. The number of T-RFs measured for the propionate and acetate granules dropped substantially compared to the number of the T-RFs measured in the inoculum floccular biomass. A strong difference was observed with respect to the dominant T-RFs in the two types of granules at day 65 (Fig. 1A). The dominant T-RF in the propionate granules with about 40% relative abundance was affiliated with the genus Zoogloea, being 99% similar to clones from activated sludge and with the closest isolate being the floc-forming bacterium Zoogloea caeni (50). Other detected important T-RFs were affiliated with Acidovorax-like and Thiothrix-like bacteria, with relative abundances of around 12% each, as well as Accumulibacter-like bacteria, at about 7% abundance. The reported isolate most closely related to the Thiothrix-like sequences from the propionate reactor was Thiothrix lacustris. The Acidovorax-like sequences were 99% to 98% similar, respectively, to clones obtained from denitrifying biofilms (clone HU5) and denitrifying activated sludge (clone KSP1) (25, 27). The closest cultivated representative of the Acidovorax-like sequences is Acidovorax delafieldii (57). In contrast, the T-RFLP profile of the acetate granules was dominated at about 60% by a T-RF affiliated with Thiothrix-like bacteria, the closest cultivated match for which (99%) is Thiothrix fructosivorans ATCC 49748 (22).
Fig. 1.
(A) Microbial community composition and dynamics of propionate- and acetate-cultivated granules expressed in terms of 16S rRNA gene-based T-RFLP patterns. The affiliation of major T-RFs is shown in the legend. (B) Dynamics of abundance of Accumulibacter- and Competibacter-like bacteria in the granules assessed by fluorescence in situ hybridization (FISH). (C) Phosphate removal capacity of the propionate- and acetate-fed sequencing batch reactors. The arrow in the right indicates the day of inducing enhanced release of phosphate from cells.
Mature reactor stage with concurrent P removal.
After day 50, the onset of P removal took place. In the propionate-fed reactor, the establishment of P removal was quasiexponential, reaching 80% by day 80 (Fig. 1C, left). In contrast, the P removal capacity in the acetate-fed reactor increased linearly and erratically, reaching 60% to 70% by day 120 (Fig. 1C, right). Measured chemical profiles for both reactors during cycles were typical for biomass-conducting EBPR. At day 134, the P released per VFA taken up (P-mol/C-mol) during the anaerobic phase was 0.27 for the propionate and 0.26 for the acetate granules. Due to the different carbon sources, the compositions of the polyhydroxyalkanoates (PHAs) stored by the two types of granules were distinct. The proportions of polyhydroxybutyrate, polyhydroxyvalerate, and polyhydroxy-2-methylvalerate (PHB:PHV:PH2MV) were 3:60:37 for the propionate granules and 70:30:0 for the acetate granules (day 134).
Within the time frame of the experiment, P removal in the propionate-fed reactor was high and stable. T-RFLP and FISH analysis showed that the microbial community was dominated by Accumulibacter-like bacteria, with abundances between 20 and 60% (Fig. 1A and B). Competibacter-like bacteria were at all times below 15% based on FISH analysis (Fig. 1B), but T-RFLP analysis showed that on days 200 and 250, T-RFs associated with Competibacter-like bacteria were present at 30 and 35% relative abundances, respectively (Fig. 1A). This, however, did not disturb the P removal activity of the reactor. Other important T-RFs detected in all profiles were affiliated with Chloroflexus-like (18 to 2%) and Acidovorax-like (10 to 2%) bacteria. The Chloroflexus-like clones were 98% similar to Herpetosiphon aurantiacus ATCC 23779 and 97% similar to the filamentous bacterium Herpetosiphon (Ben 15; X86447), previously isolated from activated sludge (6).
In the acetate-fed reactor, although erratic, P removal was maintained at 60% from day 100 to day 180. Accumulibacter-like bacteria were abundant, as evidenced by T-RFLP (20 to 30%) (Fig. 1A) and FISH (40 to 70%) (Fig. 1B) data. In this reactor, a Chloroflexus-like T-RF was also observed, although at abundances lower (1.5 to 12%) than those found in the propionate reactor, whereas an Acidovorax-like T-RF was detected at values similar to those measured in the propionate granules (Fig. 1A). After about 200 days, the P removal of the acetate reactor deteriorated, and no P removal was detected from day 240 to day 310 (Fig. 1C). Concomitantly, the microbial community structure changed from an Accumulibacter- to a Competibacter-dominated community, as reflected by the T-RFLP patterns (Fig. 1A, right). This population shift was also detected by FISH analysis. Moreover, various clones obtained from the acetate granules on day 201 and 314 were 97% similar to the “Candidatus Competibacter phosphatis” clone SBRH10, which was retrieved from a full-scale activated sludge system by Crocetti et al. (10). In the propionate granules, only one similar Competibacter-related clone was retrieved on day 201.
Although low, the abundance of Accumulibacter during the P deterioration period in the acetate reactor was still at 9 to 14% of the total bacterial community. One possibility for the loss of enhanced phosphorous uptake by the bacteria could be that these became poly-P “saturated.” To verify this hypothesis, a test was conducted on day 330 to enhance the release of phosphate by increasing two times the normal acetate concentration in the influent. This anaerobic feeding phase lasted 2.25 h and was followed by emptying and filling the reactor with carbon-free influent two times before normal operation was reset. The P removal activity by the sludge was readily recovered within 1 day (Fig. 1C, right) and remained at around 60 to 80% until the end of the experiment at day 380. A concomitant increase in Accumulibacter-related T-RFs was also observed (Fig. 1A, right).
Pragmatic analysis of 16S rRNA gene T-RFLP data.
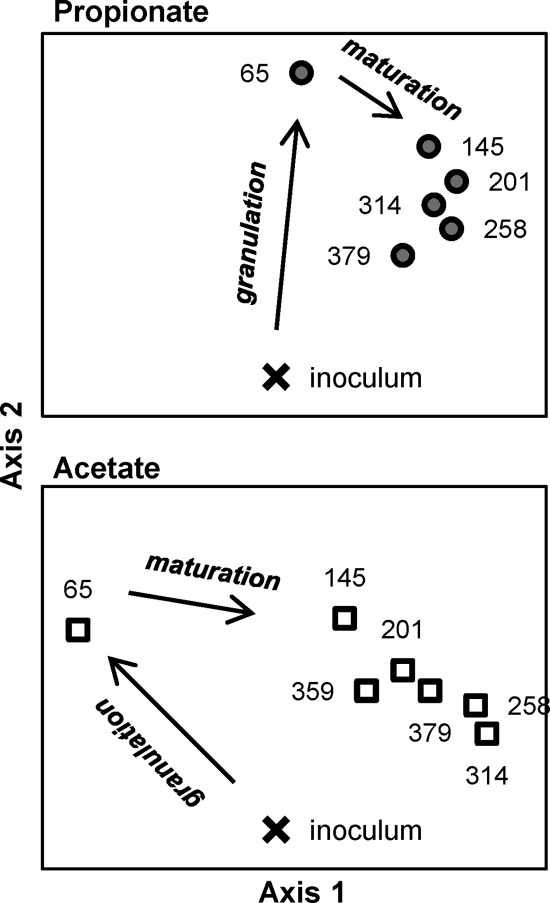
A nonmetric multidimensional scaling analysis (NMS) was used to visualize in two axes the T-RFLP fingerprint patterns from the inoculum and the granule samples over the experimental time. During the development of granules (granulation), there was a marked change in the bacterial community structure. After the granules were formed, a second important change was observed from day 65 to 145. For the propionate reactor, the initial granule community strongly moved along one axis (Fig. 2A, axis 2), whereas for the acetate reactor it moved along both axis (Fig. 2B). Therefore, we define maturation as the second axis movement and clustering of granule communities following about 65 days of reactor operation (Fig. 2).
Fig. 2.
Nonmetric multidimensional scaling (NMS) plots of the 16S rRNA gene-based T-RFLP data derived from the propionate- and acetate-cultivated granules. The NMS final stress was 9.4, which is considered a good value for ordination results. The numbers beside the symbols refer to days of reactor operation.
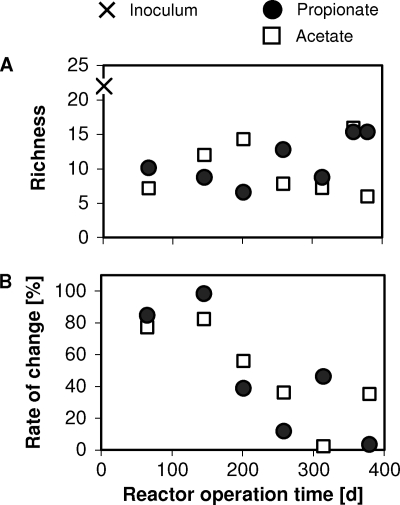
During the transformation of floccular into granular biomass, the microbial community range-weighted richness dropped from 22 to 7 and 10 for the acetate and propionate reactors, respectively. Afterward and throughout the experimental period, the range-weighted richness was lower than 15 for the granular biomass in both reactors (Fig. 3A). To complement the visual inspection of the community dynamics provided by the NMS, the moving window analysis provided a quantitative value of this property. The maximum rate of change at 80% occurred during the first 145 days of reactor operation. Near the end of the experiment, the rate of change fluctuated between 40% and 3% for both propionate and acetate granules (Fig. 3B).
Fig. 3.
(A) Range-weighted richness as a function of reactor operation time. (B) Moving window analysis reflecting the change in the bacterial community composition. Each data point is based on a comparison between 16S rRNA gene-based T-RFLP patterns separated by a time window of 60 days (except for the point at 145 days, where the time window was 80 days).
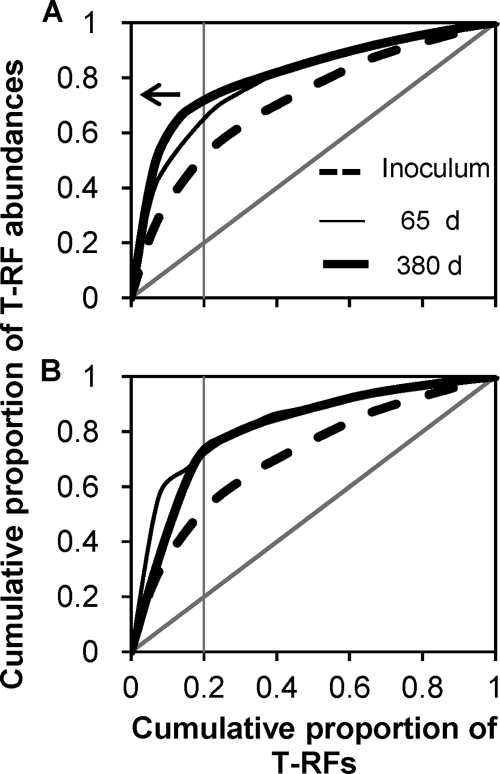
Pareto-Lorenz curves showed that the evenness of the bacterial community notably decreased during the transformation from the inoculum flocs to granules (Fig. 4). In the inoculum, 20% of the T-RFs accounted for 50% of the bacterial relative abundances (i.e., a Pareto value of 0.5), whereas for the granular sludge, 20% of the T-RFs accounted for 70% of the bacterial abundances at day 380, i.e., a Pareto value of 0.7 (Fig. 4).
Fig. 4.
Pareto-Lorenz distribution curves based on 16S rRNA gene T-RFLP patterns showing that, in comparison with the activated sludge used as the inoculum, the evenness of the bacterial community in aerobic granules is lower as the corresponding curve positions further from the perfect evenness represented by the 45° diagonal line. (A) Propionate-cultivated granules. (B) Acetate-cultivated granules. The vertical line is plotted to estimate the Pareto values as indicated by the arrow for the case of granular biomass at day 380 in panel A.
Accumulibacter phylotypes in propionate- and acetate-cultivated granules.
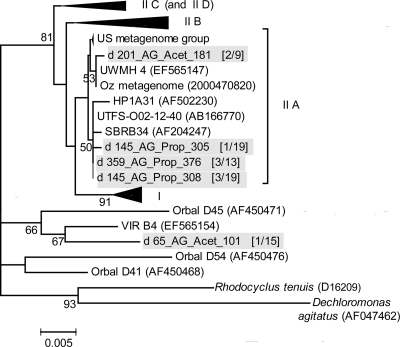
Sequence analysis of 16S rRNA gene fragments showed that in the propionate granules, all the Accumulibacter-related clones were highly similar (99 or 100%) to clone UTFS-O02-12-40. In contrast, the Accumulibacter-related clones of the acetate granules were 97% similar to clone VIR_B4 (21) and 99% similar to clone UWMH_4 (21) (Fig. 5). Although the number of analyzed sequences was small, the phylograms (Fig. 5) showed that Accumulibacter from the propionate granules at days 145 and 359 belonged to clade type IIA. Accumulibacter from the acetate granules on day 201 were also of type IIA, and one sequence on day 65 clustered outside the clades defined previously (21). All these 16S rRNA gene Accumulibacter-related sequences resulted in the same in silico and measured T-RF values. Although the data were used for phylogenetic assignment of T-RFs in the T-RFLP profiles (Fig. 1A), it was not possible to resolve potential differences in Accumulibacter type composition between the two kinds of granules.
Fig. 5.
Unrooted maximum likelihood phylogram of Accumulibacter-related 16S rRNA gene sequences. Bootstrap values of >50% and generated from 1,000 bootstrap resamplings are shown next to nodes. Roman numerals refer to Accumulibacter clades as designated previously (21). Sequences obtained in this study are shaded. The clone names are read as follows: the number following d refers to the reactor's operation day of sampling; AG_Prop refers to the propionate- and AG_Acet to the acetate-cultivated aerobic granules. The numbers in parentheses refer to the frequency of that sequence with respect to the total sequences in the respective clone library. The scale bar indicates changes per site. The phylogram was constructed using version 5 of the MEGA software (53).
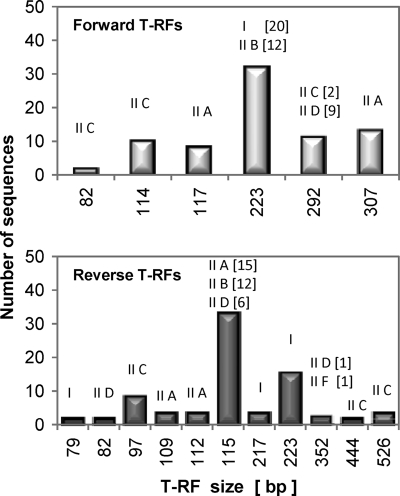
Since the propionate granules exhibited P removal activity higher and more stable than that measured for the acetate granules, one question was whether distinct clades developed in these granules during the long-term reactor operation. Thus, to explore the dynamics of Accumulibacter clades in these two distinct granules in more detail, a ppk1 gene-based approach was used, as this gene is a robust phylogenetic marker to elucidate the Accumulibacter-like community structure at finer scale (21). A high-throughput approach to discriminate between Accumulibacter type I and type II was designed and consisted of obtaining T-RFPL profiles from ppk1 genes by using either the fluorescently labeled forward or reverse primer in two separate analyses. The feasibility of this approach was tested in silico by using previously reported ppk1 gene sequences retrieved from laboratory- and full-scale reactors conducting EBPR (21, 26, 56). The in silico results indicated that type I and type II can be well distinguished (Fig. 6).
Fig. 6.
Forward (top) and reverse (bottom) terminal restriction fragments (T-RFs) generated by in silico digestion (52), using HaeIII endonuclease, of Accumulibacter ppk1 genes reported previously (21, 26, 56) and obtained from the NCBI database. The associated clade types are indicated in roman numerals, and, when required, the number of sequences is given in brackets.
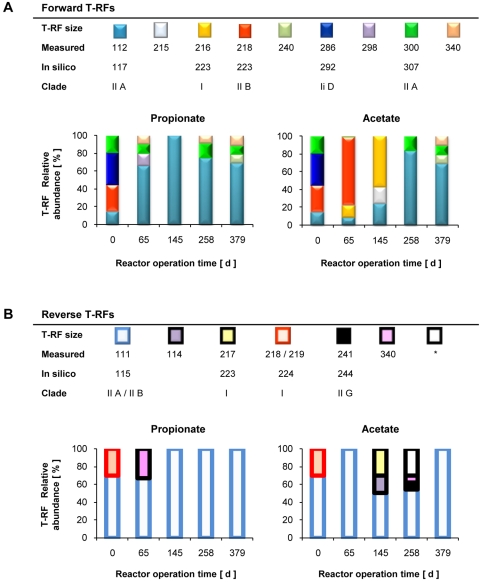
Applying this approach experimentally to the granules revealed that the Accumulibacter community from the inoculum-activated sludge (day 0) was more diverse and more evenly distributed than the ones from the propionate and the acetate granules (Fig. 7). The use of propionate as the carbon source readily selected for type IIA Accumulibacter in the granular sludge. In contrast, acetate stimulated shifts between types I and II. However, type IIA was the dominant clade in both granular sludges at the end of the experiment on day 379 (Fig. 7).
Fig. 7.
Measured Accumulibacter community dynamics of propionate- and acetate-cultivated granules expressed in terms of ppk1 gene-based T-RFLP patterns. *, unassigned T-RFs.
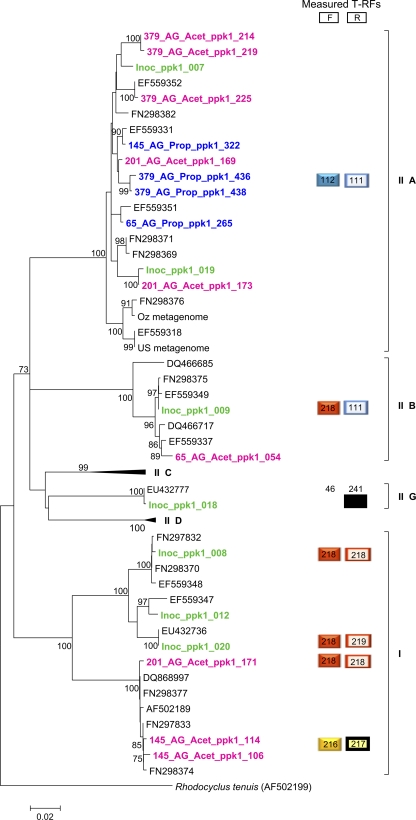
To further explore whether type IIA strains present in the propionate and acetate granules were different, cloning and sequencing of ppk1 gene fragments were conducted. Phylogenetic analysis of retrieved clone sequences revealed that the Accumulibacter community from the activated sludge inoculum was more diverse than that found in the granular sludges, as it contained representatives from nearly all clades (Fig. 8). This supports the observations of the T-RFLP profile for this sample (Fig. 7). On the other hand, and in agreement with the T-RFLP profiles, the phylogenetic analysis indicated that the Accumulibacter strains from the propionate granules were mainly composed of type IIA and that the sequences clustered separately from those of the acetate granules. Furthermore, acetate seemed to induce shifts between different dominant clades as a function of reactor operation time, and the community seemed more diverse than that from the propionate granules (Fig. 8). These observations also agree with the results of the T-RFLP profiles (Fig. 7).
Fig. 8.
Unrooted maximum likelihood phylogram of Accumulibacter-related ppk1 gene sequences. Bootstrap values of >70% and generated from 1,000 bootstrap resamplings are shown next to nodes. Roman numerals refer to Accumulibacter clades as designated previously (21). Sequences obtained in this study are shown in green (inoculum-activated sludge), blue (propionate granules), and pink (acetate granules). For the clone names, the starting numbering refers to the sampling day, and the ending numbering derives from logistics of the clone libraries (i.e., arbitrary). The scale bar indicates changes per site. The phylogram was constructed using version 5 of the MEGA software (53). Forward (F) and reverse (R) T-RFs were measured for all clones.
DISCUSSION
The microbial community of propionate and acetate granules is less diverse than the community of the activated sludge inoculum flocs.
The transformation from flocs to granules was a rapid process that occurred, in both reactors, within the first 50 days of reactor operation. This agrees with other studies in which full granulation was observed within 12 to 50 days (34) or within 21 days with acetate (16, 33), domestic (14), or industrial (2) wastewater. In all cases, the metabolic activity of these “young” granules was limited to carbon removal. Independent of the microbial assembly, and in relative terms, our pragmatic analysis shows that the microbial weighted richness in the “young” (or early-stage) granules dropped to half of that seen for the inoculum.
The 16S rRNA gene-based T-RFLP patterns clustered the propionate and acetate granule community well apart from that of the floccular biomass. Distinctively, when granules were formed, filamentous-type Thiothrix sp. bacteria in combination with the floc-forming Zoogloea sp. bacteria were predominant. Zoogloea spp. dominated the young propionate granules, whereas Thiothrix spp. dominated the acetate ones. Indeed, theories for the formation of anaerobic (24) and aerobic (5) granules suggest that filamentous microorganisms serve as backbones for granular structure. Young granular sludge comprised hairy granules and smooth granules (not shown) as previously reported for short-term studies on aerobic granulation (5). A key property of these filamentous and floc-forming bacteria is their known capacity to store PHB when exposed to strong transient feast-and-famine conditions, as applied in this study. Eventually, most filaments were outcompeted and washed out from the reactors, as the short settling time applied induced a further selective pressure.
The mature granules range-weighted richness oscillated between 7 and 15 T-RFs. This falls within a low-range value and suggests that granules as the end product, in most anaerobic-aerobic sequencing batch reactors, represent an environment with low carrying capacity and with a tendency to develop dominant-species-based communities (i.e., high functional organization reflected by high Pareto values [38]). We consider that this ecological interpretation of mature aerobic granules applies as well for complex wastewaters, although further research is required (e.g., parallel operation of acetate/propionate-fed floccular versus granular system). In support of this, the microbial community reported for mature aerobic granules treating the complex abattoir wastewater appeared highly dominated by Accumulibacter-like bacteria (approximately 80 to 90%), as revealed by FISH images of entire granule sections (31). In relative terms, it is possible that the intuitively expected high richness values due to more complex wastewaters may be counteracted by the strong physical and redox change selection pressure applied in the anaerobic-aerobic sequencing batch reactors. The mature propionate and acetate granules of this study were also dominated by Accumulibacter- and Competibacter-like bacteria. Additionally, and within the low richness values found in the mature granules, important flanking populations were associated with Acidovorax spp., of which some strains are capable of heterotrophic denitrification of nitrate and may utilize acetate, propionate, and PHB from decomposing cells as the carbon source for denitrification (25, 48). Another important population, belonging to the class the Chloroflexi, was Herpetosiphon spp., filamentous bacteria not associated with bulking sludge that thrive on exopolymers produced by other microorganisms or the cell walls of decaying bacteria (29).
The propionate granules had better and more stable phosphate removal activity than the acetate granules.
The lengthy step in the startup of the granular sludge process is not to obtain granules per se but relates to obtaining granules with the microbial assembly and stability required for a specific treatment process. On this regard, we show that mature granules with robust phosphate removal activity were attained by using propionate. Although fluctuating levels of Competibacter abundances were observed, these did not compromise the granule phosphate removal activity of the microbial community dominated by Accumulibacter-like bacteria. This supports recent model predictions that in 100% propionate-fed systems, Competibacter does not impair Accumulibacter populations (36). Putative GAOs belonging to Alphaproteobacteria and recently reported as responsible for deteriorating phosphate removal in propionate-fed activated sludge (40) were apparently not important in the system of this study, and no T-RF nor clone associated with them was detected.
Conversely, the mature acetate granules sustained phosphate removal capacity for a period of only about 100 days before failure, although the reactor was operated in conditions similar to those for the propionate reactor. During failure, the microbial community predominance shifted from Accumulibacter- to Competibacter-like. It is known that biological phosphorus removal suffers from deterioration events for unexplained reasons. Although reports point to Competibacter as responsible for process deterioration, the parameters that result in their proliferation have not yet been elucidated (51). We hypothesized that Accumulibacter in the acetate reactor channeled large amounts of energy to poly-P formation which eventually “saturated” the cells. Indeed, based on Raman imaging, a recent report shows that poly-P inclusions can occupy as much as 75% of the Accumulibacter cell body (37). This metabolic state of Accumulibacter might in turn have allowed Competibacter to proliferate, as assessed here by two independent methods. After the test providing more substrate to induce hydrolysis of poly-P by the “saturated” cells, the acetate reactor recovered its phosphate removal activity within 1 day, suggesting that the existing Accumulibacter cells in the granules were sufficiently numerous and that recovery was not due to growth.
Differences in the metabolism of acetate and propionate during EBPR may assist in understanding the results obtained. According to metabolic models developed for Accumulibacter, acetate requires more glycogen than propionate to produce sufficient reducing power for anaerobic VFA storage as PHA (36). In addition, glycogen degradation does not provide all the reducing power required for PHA synthesis from acetate, but it may provide it for propionate (63). It is proposed that this extra reducing power is generated by the tricarboxylic acid (TCA) cycle, a split TCA cycle, or the glyoxylate cycle (7, 56, 62). Additionally, on a C-mol basis, the formation of acetyl coenzyme A (acetyl-CoA) from acetate requires more energy than the formation of propyonyl-CoA from propionate (36). Thus, it might be that acetate metabolism by Accumulibacter depends more on poly-P than propionate metabolism does. Possibly, acetate-fed Accumulibacter favored channeling energy for poly-P storage over biomass growth, which in turn resulted in poly-P-“saturated” cells.
In addition to less energy expenditure for polyhydroxyalkanoate formation from propionate as suggested previously (46), contrasting energy efficiencies associated with different Accumulibacter strains utilizing different metabolic pathways may also be responsible for the observed differing functionalities of the acetate- and propionate-cultivated granules.
Contrasting structure and dynamics of Accumulibacter-like community in propionate- and acetate-cultivated granules.
The ppk1 gene T-RFLP data sets and the phylogenetic analysis of retrieved Accumulibacter ppk1 gene sequences revealed that, in our anaerobic-aerobic system, propionate readily favored the dominance of type II Accumulibacter. On the contrary, acetate granules exhibited transient shifts between type I and type II during the first 200 days before the granules were dominated by type II Accumulibacter as well. Interestingly, EBPR sludge samples for metagenome studies were taken after 11 months of reactor operation in the case of an acetate-fed system from the United States (US metagenome) and at a time not specified in the case of a propionate-fed one from Australia (OZ metagenome) (19). Both metagenomes corresponded to type II Accumulibacter and, in accordance with our study, suggest that anaerobic-aerobic EBPR lab-scale systems operated for long periods are apparently mostly enriched in type II Accumulibacter. In the propionate granular sludge system, the initial significant presence of Acidovorax, known to denitrify NO3− utilizing PHA, may have outcompeted type I Accumulibacter. Our data indicate that the mere presence of Accumulibacter is not enough to have consistently high phosphate removal in the reactor system but that the type of Accumulibacter determines the robustness of the phosphate removal process.
Since previous research points to contrasting denitrifying functions for type I (NO3−/NO2− reduction) and type II Accumulibacter (NO2− reduction) and highlights that these different functions have important process applications, recent efforts have been directed toward manipulating these clades at discretion. To aid on this effort, we propose a distinct dual T-RFLP approach based on ppk1 genes to readily monitor changes in those clades under specified conditions. The T-RF profiles obtained with the reverse primer distinguish between type I and type II, whereas T-RF profiles obtained with the forward primer give resolution within the type II clade by separating IIA, IIC, and IID (Fig. 6). Combining forward and reverse T-RF profiles further resolves clade IIB. In addition, potential differences within type I clusters from the phylograms might be resolved, as we observed an ∼2-bp difference in forward T-RFs within type I members (Fig. 8). Our approach contrasts with the recently reported multiple reactions-various restriction enzyme approach (51), as it requires a single restriction enzyme, and if the forward and reverse primers are labeled with two different fluorophores, there is an option to conduct a single-reaction run for analysis.
ACKNOWLEDGMENTS
This research was partially funded by the Swiss National Science Foundation, grant no. 205321-120536.
We thank S. Lochmatter, D. Weissbrodt, and O. Zanoletti for reactor operation and laboratory assistance and Matthias Noll for assistance with NMS analysis. Helpful suggestions by anonymous reviewers were highly appreciated.
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Adav S. S., Lee D.-J., Show K.-Y., Tay J.-H. 2008. Aerobic granular sludge: recent advances. Biotechnol. Adv. 26:411–423 [DOI] [PubMed] [Google Scholar]
- 2. Arrojo B., Mosquera-Corral A., Garrido J. M., Mendez R. 2004. Aerobic granulation with industrial wastewater in sequencing batch reactors. Water Res. 38:3389–3399 [DOI] [PubMed] [Google Scholar]
- 3. Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J., Weightman A. J. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W. 2009. GenBank. Nucleic Acids Res. 37:D26–D31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beun J. J., et al. 1999. Aerobic granulation in a sequencing batch reactor. Water Res. 33:2283–2290 [DOI] [PubMed] [Google Scholar]
- 6. Bradford D., et al. 1996. 16S rRNA analysis of isolates obtained from Gram-negative filamentous bacteria micromanipulated from activated sludge. Syst. Appl. Microbiol. 19:334–343 [Google Scholar]
- 7. Burow L. C., Mabbett A. N., Blackall L. L. 2008. Anaerobic glyoxylate cycle activity during simultaneous utilization of glycogen and acetate in uncultured Accumulibacter enriched in enhanced biological phosphorus removal communities. ISME J. 2:1040–1051 [DOI] [PubMed] [Google Scholar]
- 8. Butler J. L., Williams M. A., Bottomley P. J., Myrold D. D. 2003. Microbial community dynamics associated with rhizosphere carbon flow. Appl. Environ. Microbiol. 69:6793–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carvalho G., Lemos P. C., Oehmen A., Reis M. A. M. 2007. Denitrifying phosphorus removal: linking the process performance with the microbial community structure. Water Res. 41:4383–4396 [DOI] [PubMed] [Google Scholar]
- 10. Crocetti G. R., Banfield J. F., Keller J., Bond P. L., Blackall L. L. 2002. Glycogen-accumulating organisms in laboratory-scale and full-scale wastewater treatment processes. Microbiology 148:3353–3364 [DOI] [PubMed] [Google Scholar]
- 11. Crocetti G. R., et al. 2000. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 66:1175–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Bruin L. M. M., de Kreuk M. K., van der Roest H. F. R., Uijterlinde C., van Loosdrecht M. C. M. 2004. Aerobic granular sludge technology: an alternative to activated sludge? Water Sci. Technol. 49:1–7 [PubMed] [Google Scholar]
- 13. de Kreuk M., Heijnen J. J., van Loosdrecht M. C. M. 2005. Simultaneous COD, nitrogen, and phosphate removal by aerobic granular sludge. Biotechnol. Bioeng. 90:761–769 [DOI] [PubMed] [Google Scholar]
- 14. de Kreuk M. K., van Loosdrecht M. C. M. 2006. Formation of aerobic granules with domestic sewage. J. Environ. Eng. 132:694–697 [Google Scholar]
- 15. DeSantis T. Z., et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ebrahimi S., et al. 2010. Performance and microbial community composition dynamics of aerobic granular sludge from sequencing batch bubble column reactors operated at 20°C, 30°C and 35°C. Appl. Microbiol. Biotechnol. 87:1555–1568 [DOI] [PubMed] [Google Scholar]
- 17. Flowers J., He J. S., Yilmaz S., Noguera D. R., McMahon K. D. 2009. Denitrification capabilities of two biological phosphorus removal sludges dominated by different ‘Candidatus Accumulibacter’ clades. Environ. Microbiol. Rep. 1:583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fuchs B. M., et al. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. García Martín H., et al. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24:1263–1269 [DOI] [PubMed] [Google Scholar]
- 20. Greenberg A. E. (ed.). 1985. Standard methods for the examination of water and wastewater, 16th ed American Public Health Association, Washington, DC [Google Scholar]
- 21. He S., Gall D. L., McMahon K. D. 2007. “Candidatus Accumulibacter” population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl. Environ. Microbiol. 73:5865–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Howarth R., et al. 1999. Phylogenetic relationships of filamentous sulfur bacteria (Thiothrix spp. and Eikelboom type 021N bacteria) isolated from wastewater treatment plants and description of Thiothrix eikelboomii sp. nov., Thiothrix unzii sp. nov., Thiothrix fructosivorans sp. nov. and Thiothrix defluvii sp. nov. Int. J. Syst. Bacteriol. 49:1817–1827 [DOI] [PubMed] [Google Scholar]
- 23. Huber T., Faulkner G., Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 24. Hulshoff Pol L. W., de Castro Lopes S. I., Lettinga G., Lens P. N. L. 2004. Anaerobic sludge granulation. Water Res. 38:1376–1389 [DOI] [PubMed] [Google Scholar]
- 25. Khan S. T., Horiba Y., Yamamoto M., Hiraishi A. 2002. Members of the family Comamonadaceae as primary poly(3-hydroxybutyrate-co-3-hydroxyvalerate)-degrading denitrifiers in activated sludge as revealed by a polyphasic approach. Appl. Environ. Microbiol. 68:3206–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim J. M., et al. 2010. Analysis of the fine-scale population structure of “Candidatus Accumulibacter phosphatis” in enhanced biological phosphorus removal sludge, using fluorescence in situ hybridization and flow cytometric sorting. Appl. Environ. Microbiol. 76:3825–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kindaichi T., et al. 2007. In situ activity and spatial organization of anaerobic ammonium-oxidizing (anammox) bacteria in biofilms. Appl. Environ. Microbiol. 73:4931–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kong Y. H., Ong S. L., Ng W. J., Liu W. T. 2002. Diversity and distribution of a deeply branched novel proteobacterial group found in anaerobic-aerobic activated sludge processes. Environ. Microbiol. 4:753–757 [DOI] [PubMed] [Google Scholar]
- 29. Kragelund C., et al. 2007. Identity, abundance and ecophysiology of filamentous Chloroflexi species present in activated sludge treatment plants. FEMS Microbiol. Ecol. 59:671–682 [DOI] [PubMed] [Google Scholar]
- 30. Lane D. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 31. Lemaire R., Webb R. I., Yuan Z. 2008. Micro-scale observations of the structure of aerobic microbial granules used for the treatment of nutrient-rich industrial wastewater. ISME J. 2:528–541 [DOI] [PubMed] [Google Scholar]
- 32. Lemaire R., Yuan Z., Blackall L. L., Crocetti G. R. 2008. Microbial distribution of Accumulibacter spp. and Competibacter spp. in aerobic granules from a lab-scale biological nutrient removal system. Environ. Microbiol. 10:354–363 [DOI] [PubMed] [Google Scholar]
- 33. Li A.-J., Li X.-Y. 2009. Selective sludge discharge as the determining factor in SBR aerobic granulation: numerical modelling and experimental verification. Water Res. 43:3387–3396 [DOI] [PubMed] [Google Scholar]
- 34. Li A.-J., Yang S.-F., Li X.-Y., Gu J.-D. 2008. Microbial population dynamics during aerobic sludge granulation at different organic loading rates. Water Res. 42:3552–3560 [DOI] [PubMed] [Google Scholar]
- 35. Liu X.-W., Sheng G.-P., Yu H.-Q. 2009. Physicochemical characteristics of microbial granules. Biotechnol. Adv. 27:1061–1070 [DOI] [PubMed] [Google Scholar]
- 36. Lopez-Vazquez C. M., et al. 2009. Modeling the PAO-GAO competition: effects of carbon source, pH and temperature. Water Res. 43:450–462 [DOI] [PubMed] [Google Scholar]
- 37. Majed N., Matthäus C., Diem M., Gu A. Z. 2009. Evaluation of intracellular polyphosphate dynamics in enhanced biological phosphorus removal process using Raman microscopy. Environ. Sci. Technol. 43:5436–5442 [DOI] [PubMed] [Google Scholar]
- 38. Marzorati M., Wittebolle L., Boon N., Daffonchio D., Verstraete W. 2008. How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ. Microbiol. 10:1571–1581 [DOI] [PubMed] [Google Scholar]
- 39. McCune B., Mefford M. J. 1999. PC-ORD, multivariate analysis of ecological data. MJM Software, Gleneden Beach, OR [Google Scholar]
- 40. Meyer R. L., Saunders A. M., Blackall L. L. 2006. Putative glycogen-accumulating organisms belonging to the Alphaproteobacteria identified through rRNA-based stable isotope probing. Microbiology 152:419–429 [DOI] [PubMed] [Google Scholar]
- 41. Mino T., Van Loosdrecht M. C. M., Heijnen J. J. 1998. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32:3193–3207 [Google Scholar]
- 42. Ni B.-J., et al. 2009. Granulation of activated sludge in a pilot-scale sequencing batch reactor for the treatment of low-strength municipal wastewater. Water Res. 43:751–761 [DOI] [PubMed] [Google Scholar]
- 43. Oehmen A., et al. 2007. Advances in enhanced biological phosphorus removal: from micro to macro scale. Water Res. 41:2271–2300 [DOI] [PubMed] [Google Scholar]
- 44. Oehmen A., Saunders A. M., Vives M. T., Yuan Z. G., Keller H. 2006. Competition between polyphosphate and glycogen accumulating organisms in enhanced biological phosphorus removal systems with acetate and propionate as carbon sources. J. Biotechnol. 123:22–32 [DOI] [PubMed] [Google Scholar]
- 45. Oehmen A., Yuan Z. G., Blackall L. L., Keller J. 2005. Comparison of acetate and propionate uptake by polyphosphate accumulating organisms and glycogen accumulating organisms. Biotechnol. Bioeng. 91:162–168 [DOI] [PubMed] [Google Scholar]
- 46. Oehmen A., Zeng R. J., Yuan Z. G., Keller J. 2005. Anaerobic metabolism of propionate by polyphosphate-accumulating organisms in enhanced biological phosphorus removal systems. Biotechnol. Bioeng. 91:43–53 [DOI] [PubMed] [Google Scholar]
- 47. Rees G., Baldwin D., Watson G., Perryman S., Nielsen D. 2004. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie Van Leeuwenhoek 86:339–347 [DOI] [PubMed] [Google Scholar]
- 48. Schloe K., et al. 2000. Polyphasic characterization of poly-3-hydroxybutyrate-co-3-hydroxyvalerate (P(HB-co-HV)) metabolizing and denitrifying Acidovorax sp. strains. Syst. Appl. Microbiol. 23:364–372 [DOI] [PubMed] [Google Scholar]
- 49. Seviour R. J., McIlroy S. 2008. The microbiology of phosphorus removal in activated sludge processes—the current state of play. J. Microbiol. 46:115–124 [DOI] [PubMed] [Google Scholar]
- 50. Shao Y., et al. 2009. Zoogloea caeni sp. nov., a floc-forming bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 59:526–530 [DOI] [PubMed] [Google Scholar]
- 51. Slater F. R., Johnson C. R., Blackall L. L., Beiko R. G., Bond P. L. 2010. Monitoring associations between clade-level variation, overall community structure and ecosystem function in enhanced biological phosphorus removal (EBPR) systems using terminal-restriction fragment length polymorphism (T-RFLP). Water Res. 44:4908–4923 [DOI] [PubMed] [Google Scholar]
- 52. Stres B., Tiedje J. M., Murovec B. 2009. BEsTRF: a tool for optimal resolution of terminal-restriction fragment length polymorphism analysis based on user-defined primer-enzyme-sequence databases. Bioinformatics 25:1556–1558 [DOI] [PubMed] [Google Scholar]
- 53. Tamura K., et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weber S. D., Ludwig W., Schleifer K. H., Fried J. 2007. Microbial composition and structure of aerobic granular sewage biofilms. Appl. Environ. Microbiol. 73:6233–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Werker A., Lind P., Bengtsson S., Nordström F. 2008. Chlorinated-solvent-free gas chromatographic analysis of biomass containing polyhydroxyalkanoates. Water Res. 42:2517–2526 [DOI] [PubMed] [Google Scholar]
- 56. Wexler M., Richardson D. J., Bond P. L. 2009. Radiolabelled proteomics to determine differential functioning of Accumulibacter during the anaerobic and aerobic phases of a bioreactor operating for enhanced biological phosphorus removal. Environ. Microbiol. 11:3029–3044 [DOI] [PubMed] [Google Scholar]
- 57. Willems A., et al. 1990. Acidovorax, a new genus for Pseudomonas facilis, Pseudomonas delafieldii, E. Falsen (EF) Group 13, EF Group 16, and several clinical isolates, with the species Acidovorax facilis comb. nov., Acidovorax delafieldii comb. nov., and Acidovorax temperans sp. nov. Int. J. Syst. Bacteriol. 40:384–398 [DOI] [PubMed] [Google Scholar]
- 58. Wittebolle L., Vervaeren H., Verstraete W., Boon N. 2008. Quantifying community dynamics of nitrifiers in functionally stable reactors. Appl. Environ. Microbiol. 74:286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wong M. T., Tan F. M., Ng W. J., Liu W. T. 2004. Identification and occurrence of tetrad-forming Alphaproteobacteria in anaerobic-aerobic activated sludge processes. Microbiology 150:3741–3748 [DOI] [PubMed] [Google Scholar]
- 60. Xiao F., Yang S. F., Li X. Y. 2008. Physical and hydrodynamic properties of aerobic granules produced in sequencing batch reactors. Sep. Purif. Technol. 63:634–641 [Google Scholar]
- 61. Yilmaz G., Lemaire R., Keller J., Yuan Z. 2008. Simultaneous nitrification, denitrification, and phosphorus removal from nutrient-rich industrial wastewater using granular sludge. Biotechnol. Bioeng. 100:529–541 [DOI] [PubMed] [Google Scholar]
- 62. Zhou Y., Pijuan M., Oehmen A., Yuan Z. 2010. The source of reducing power in the anaerobic metabolism of polyphosphate accumulating organisms (PAOs) – a mini-review. Water Sci. Technol. 61. 7:1653–1662 [DOI] [PubMed] [Google Scholar]
- 63. Zhou Y., Pijuan M., Zeng R. J., Yuan Z. 2009. Involvement of the TCA cycle in the anaerobic metabolism of polyphosphate accumulating organisms (PAOs). Water Res. 43:1330–1340 [DOI] [PubMed] [Google Scholar]
- 64. Zilles J. L., Peccia J., Kim M. W., Hung C. H., Noguera D. R. 2002. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl. Environ. Microbiol. 68:2763–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]