Abstract
The variation in manure-amended soil survival capability among 18 Escherichia coli O157 strains (8 animal, 1 food, and 9 human isolates) was studied using a single sandy soil sample and a single sample of cattle manure as the inoculum carrier. The virulence profiles of E. coli O157 strains were characterized by detection of virulence determinants (73 genes, 122 probes in duplicate) by using the Identibac E. coli genotyping DNA miniaturized microarray. Metabolic profiling was done by subjecting all strains to the Biolog phenotypic carbon microarray. Survival times (calculated as days needed to reach the detection limit using the Weibull model) ranged from 47 to 266 days (median, 120 days). Survival time was significantly higher for the group of human isolates (median, 211 days; minimum [min.], 71; maximum [max.], 266) compared to the group of animal isolates (median, 70 days; min., 47; max., 249) (P = 0.025). Although clustering of human versus animal strains was observed based on pulsed-field gel electrophoresis (PFGE) patterns, no relation between survival time and the presence of virulence genes was observed. Principal component analysis on the metabolic profiling data revealed distinct clustering of short- and long-surviving strains. The oxidization rate of propionic acid, α-ketobutyric acid, and α-hydroxybutyric acid was significantly higher for the long-surviving strains than for the short-surviving strains. The oxidative capacity of E. coli O157 strains may be regarded as a phenotypic marker for enhanced survival in manure-amended soil. The large variation observed in survival is of importance for risk assessment models.
INTRODUCTION
Fresh vegetables (especially fresh-cut leafy greens) are increasingly being recognized as important vehicles for the transmission of human pathogens that were traditionally associated with foods of animal origin (12, 43). Produce-associated outbreaks of Escherichia coli O157, Salmonella, and Shigella infections linked to the consumption of leafy green vegetables have been reported in the United States and the European Union (1, 8, 19, 60). E. coli O157 is ubiquitous on farms where healthy cattle and other ruminants harbor the pathogen in their gastrointestinal tracts (14, 32, 46). Consequently, farm animal manure is a source for spreading E. coli O157 into the soil environment and potentially into the fresh vegetable food chain (24).
The observed persistence of E. coli O157 in manure-amended soil can be considered a significant risk factor for the (re)contamination of cattle and food crops and ultimately for human infection (24, 25). The risk of contamination of vegetables grown in soils enriched with contaminated manure will largely depend on the survival capabilities of the pathogen in manure and manure-amended soils. Although the conditions for the survival of enteric pathogens are considered to be unfavorable once pathogens are excreted from the animal gut, pathogens like E. coli O157 are able to survive for extended periods in (manure-amended) soil (23, 27, 33, 47, 50). However, little work has been conducted to investigate key factors determining the survival of E. coli O157 in soil and the mechanisms involved. Generally, nutrient availability is thought to be a key issue in the survival of microbes in soil (34). Although E. coli can potentially exhibit oligotrophic kinetic properties in chemostat cultures (36), a major factor in E. coli die-out in soils is thought to be its inability to lower its metabolic rate to meet the low availability of usable organic carbon and to adjust to conditions of low nutrient availability (24). Recent studies confirm the importance of the nutrient status of the soil with respect to the survival capability of E. coli O157 in manure-amended soil (23, 57, 65). The survival times of a particular E. coli O157 strain in 24 different soils were found to be largely dependent on the level of dissolved organic carbon per unit of soil microbial biomass (23). However, no data are available on E. coli O157 strain variability with respect to survival in soil. It is speculated that variation in E. coli O157 soil survival is a function not only of soil characteristics but also of inherent variability in metabolic capabilities among E. coli O157 strains.
The high prevalence of E. coli O157 in cattle contrasts with the comparative rarity of human infection, despite its highly infectious nature (62). The observed nonrandom distribution of E. coli O157 genotypes observed among human and bovine isolates is indicative of differences in transmissibility and/or virulence among these genotypes (9, 11, 39, 66). Indeed, there is mounting evidence of significant phenotypic differences between human and cattle E. coli O157 isolates (7, 56, 63, 68). However, very limited knowledge of the phenotypic differences between human and animal isolates with respect to environmental survival is available.
It has been suggested that determinants for infection could also be beneficial for surviving the environment outside the host and vice versa (45). The major E. coli O157 virulence factors intimin (eae) and Shiga toxin (stx) are highly associated with severe disease in humans (10, 21, 26), but their roles with respect to persistence in the cattle reservoir and the environment are marginally understood. The results of a recent study demonstrated the existence of Shiga toxin-producing E. coli (STEC) strains with distinct modes of persistence in the cattle reservoir and the noncattle environment, which are dependent on the capacity to derive fitness benefits from virulence determinants (52). However, the effect of virulence genes on the environmental persistence of STEC has seldom been studied directly. Carriage of Shiga toxin-encoding genes increased survival of E. coli O157 in grazing protozoa and thus may enhance fitness during environmental survival (61).
With the present study it is hypothesized that (i) animal and human E. coli O157 isolates show differential survival capabilities in manure-amended soil, (ii) differences in virulence gene composition between E. coli O157 strains will result in differences in manure-amended soil survival capabilities, and (iii) E. coli O157 strains show differences in carbon nutrition profile which can be related to differences in manure-amended soil survival capability.
MATERIALS AND METHODS
Soil and manure.
Soil was collected from the organic experimental farm Droevendaal (Wageningen University and Research Center, The Netherlands) from a field that had not been cultivated over the last 3 years and was covered with grass. The soil (pH 5.3, C/N 18) was sandy (80% particles of 50 to 2,000 μm and 4% particles of <2 μm) and contained 1,160 mg/kg N, 3.5% organic matter, and 80 mg/kg K, 4.7 mg/kg P (Blgg, The Netherlands). The soil was transported to the laboratory in plastic bags, thoroughly mixed, sieved through 0.5-cm mesh to remove plant parts and earthworms, and stored at 5°C prior to the start of the experiment.
Fresh manure without urine was collected in April 2009 from organically managed Holstein Frisian steers on standard 50% grass/clover silage and 50% dried grass diet from the organic experimental farm Droevendaal (Wageningen University and Research Center, The Netherlands). The manure was stored for 1 week in closed plastic bags at 5°C prior to the start of the experiment.
The collected soil and manure were tested for the natural presence of E. coli O157 by making an overnight enrichment (1:10, vol/vol) in modified tryptone soy broth (mTSB; Oxoid, United Kingdom), followed by plating (50 ml) on CHROMagar O157 (ITK Diagnostics, Uithoorn, The Netherlands) supplemented with 0.025 mg/liter cefixime and 1.25 mg/liter tellurite (1/2 CT CHROMagar O157) (3) and by PCR (54) after DNA extraction (FastDNA spin kit for soil; Bio101 Systems, Qbiogene, Carlsbad, CA) according to the manufacturer's specifications. The CHROMO157 plates were checked for typical mauve E. coli O157 colonies after overnight incubation at 37°C. Both the soil and manure were found negative for E. coli O157.
To ensure standardized water availability, the water content of 10 kg of soil was adjusted to 60% of its maximum water-holding capacity (WHC). To do so, 50 g of field-moist soil sample was taken to determine the actual water content and the maximum WHC. The actual water content was measured by drying approximately 5 g soil of this 50 g for 24 h at 105°C. The maximum WHC of the soil was determined by adding an excess of distilled water to approximately 45 g of remaining field-moist soil. This was left overnight covered with aluminum foil to prevent evaporation. The well-drained soil was filtered in a funnel with filter paper mounted on a collecting flask and again allowed to stand overnight covered with aluminum foil. Subsequently, the WHC was determined by drying 5 g of well-drained soil for 24 h at 105°C. The amount of water to be added was calculated by taking 60% of the WHC minus the water content of the field-moist sample.
Bacteria and genetic characterization.
For this study, 18 E. coli O157 strains were selected from a large collection (National Institute for Public Health and the Environment [RIVM], The Netherlands): 8 veterinary strains (lamb, sheep, cow, and horse feces), 9 human clinical strains (isolated from fecal samples), and 1 food strain (raw-milk cheese). All human clinical strains originated from nonrelated sporadic cases and were not associated with an outbreak. All of these 18 strains were PCR positive for the rfbE gene (O157) (15) and the fliC gene (H7 flagella) (28), although not all strains showed the H7 phenotype (Fig. 1). In addition, all strains were positive for eae (intimin) and hly (hemolysin). Phenotypically, all strains were non-sorbitol-fermenting and β-glucuronidase negative. Detection of pathogenic E. coli virulence genes (73 genes, 122 probes) was done using the Identibac E. coli genotyping DNA miniaturized microarray (Identibac kit; Alere GmbH, Germany) (4). The full Identibac gene list is available at http://identibac.com/fileadmin/Media/Downloads/Genelist__E.coli_03_m.pdf. Analysis was performed as described by the manufacturer. In order to classify the strains into lineages I, I/II, and II, the lineage-specific polymorphism assay (LSPA) was performed as described by Yang and coworkers (66).
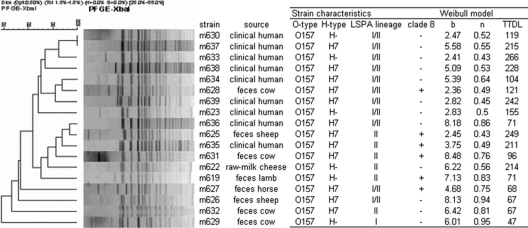
Fig. 1.
Phylogenetic relation between E. coli O157 PFGE pattern, isolation origin, genetic characteristics, and Weibull parameters b (scale parameter), n (shape parameter), and the TTDL (number of days to reach detection limit as calculated by the Weibull model using the estimated parameter values) for survival in manure-amended soil.
Inoculation and sampling.
E. coli O157 strains were stored at −80°C and checked for viability prior to use by growing cultures on tryptic soy agar (TSA) (Oxoid, United Kingdom). From these culture plates, cells were transferred for overnight incubation in mTSB (Oxoid, United Kingdom) at 37°C on an orbital shaker (200 rpm) for 18 h. Liquid cultures were centrifuged at 10,000 × g for 10 min, washed three times, and resuspended in buffered peptone water (BPW). The number of cells per milliliter of suspension was determined using the spectrophotometer, where the optical density (OD) at 630 nm of 0.7 was equal to 1 × 109 CFU ml−1. Cells suspended in BPW were added to half of the manure portions (40 g per portion) with a final density of 1. The amount of inoculum added to the portion was such that the final concentration in the soil-manure mixture was around 107 CFU per gram dry weight, taking into account the water content of the manure and that of the individual soils. Identical volumes of BPW were added to the control treatments. The manure and the inoculum were thoroughly mixed by kneading in a plastic bag from the outside by hand. Subsequently, the inoculated portions of manure were added to and thoroughly mixed with the 460-g portions of soil. The mixture was transferred to 1-liter pots, which were closed (but with the ability of gas exchange) and incubated at 16°C in the dark.
The inoculated soil-manure mixtures were sampled over time (at t = 0, 2, 6, 9, 16, 23, 30, 54, 64, 78, 88, 102, 123, 150, 182, and 212 days) to quantitatively determine the survival of the pathogens. At each sampling time, two samples of approximately 1 g of each replica were removed from the middle of the mixture using a sterile spoon and put in separate preweighed dilution tube with 4.5 ml of 0.1% peptone. Sampling holes were closed. Sample tubes were weighed to determine the exact size of the sample. Samples were vortexed, and appropriate 10-fold serial dilutions were made. From the two highest dilutions, 50 μl was plated in duplicate on petri dishes with CHROMagar O157 agar (ITK Diagnostics, Uithoorn, The Netherlands) supplemented with 0.025 mg/liter cefixime and 1.25 mg/liter tellurite (Oxoid SR 0172) (1/2 CT CHROMagar O157). The inoculated plates were incubated at 37°C for 16 to 20 h. Numbers of viable E. coli O157 cells were determined after incubation for 18 h at 37°C by counting mauve colonies. Two random colonies from two random plates per sampling time were serologically checked for the O157 antigen by latex agglutination (E. coli O157 latex test kit; Oxoid, United Kingdom).
PM.
The phenotypic microarray (PM) analysis for carbon sources (alcohols, amides, amino acids, carbohydrates, esters, carboxylic acids, and polymers) was performed by Biolog Inc. (Hayward, CA). The PM assay consists of two 96-well PM panels (PM 1 and 2) and tests bacterial growth/survival. Briefly, single colonies from each strain, grown on agar plates, were suspended in inoculating fluid containing a patented redox dye (0.01% tetrazolium violet). Bacterial cell suspensions were transferred into PM panels and incubated in the OmniLog incubator reader. Bacterial growth was assessed by the color change from the reduction of the redox dye and the color intensity was measured for 24 h, resulting in a kinetic response curve. Wells without substrate that theoretically result in no signal were used as negative controls in each PM panel. Data were analyzed with OmniLog PM software. The area under the kinetic response curve was used as a quantitative measure for the kinetic response (i.e., oxidative activity).
Statistical analysis.
The survival of E. coli O157 was modeled by fitting the experimental data to the Weibull survival function with GInaFiT (29) as follows: log10 N/N0 = −(t/b)n, where log10 N/N0 is the log number of the relative population size (CFU g dry weight [gdw]−1) at time t (days), b (scale parameter) represents the time of first decimal reduction (days), and n represents shape parameter. For n > 1, a convex curve is obtained, while for n < 1, a concave curve is obtained. Model performance was assessed by calculating the regression coefficient (R2) and the root mean square error (RMSE). In addition, the residuals were subjected to a test for normality. Based on the model parameters, the time needed to reach the detection limit (TTDL) of log10 1.2 CFU gdw−1 was calculated (TTDL in days). The TTDL values were used for presenting survival times and further statistical analysis. Model parameters and the calculated TTDL were compared between the groups of human and bovine isolates with Student's t test, where a P value of 0.05 was significant. The kinetic response parameters of the Biolog analysis were normalized against the values of the negative control. Differences in normalized kinetic response parameters between human and bovine isolates and between long (>200 days) and short (<200 days) survivors were assessed using Student's t tests. Clustering of isolates based on the presence/absence data of the Identibac genetic data and the kinetic response parameters of the Biolog analysis was assessed by principal component analysis (SAS Stat Studio 3.11). Cluster analysis of the pulsed-field gel electrophoresis (PFGE) and the Identibac microarray results were performed using Bionumerics software (version 6.5). Similarity analysis of the PFGE data was performed using the Dice coefficient (Opt, 1.00%; Tol, 1.00%), and clustering was by unweighted-pair group method using average linkages (UPGMA) means. The similarity coefficients of the Identibac data were calculated using the Pearson correlation and cluster analysis used against UPGMA means.
RESULTS AND DISCUSSION
Diversity in manure-amended soil survival among E. coli O157 isolates.
The application of the Weibull model to the observed E. coli O157 survival data resulted in good fits (average RMSE, 0.45 ± 016; average R2 of 0.94 ± 0.03) with (approximately) normally distributed residuals. A considerable variation in survival times was observed (average TTDL, 145 ± 77; median, 120; minimum [min.], 47; maximum [max.], 266 days) (Fig. 1). The survival time was significantly higher for the group of human isolates (n = 9; average, 179 ± 69; median, 211; min., 71; max., 266) compared to the group of animal isolates (n = 8; average, 98 ± 65; median, 70; min., 47; max., 249) (P = 0.025). The model used is based on the assumption that the resistance of E. coli O157 to stresses, as encountered in the manure-amended soil, follows a Weibull distribution and that the survival curve is the cumulative form of this underlying distribution of individual inactivation kinetics (64). The shape parameter of the Weibull model was significantly lower for the human isolates (0.55 ± 0.13) than for the animal isolates (0.75 ± 0.19) (P = 0.027), resulting in a more concave decline pattern for the human isolates and a more linear decline curve for the animal isolates. This can be interpreted as evidence that for the human isolates a larger fraction of cells are, or become, more resistant to the manure-amended soil conditions encountered (53).
Other studies, mostly using inoculations with single strains, reported E. coli O157 survival times ranging from 25 days (27) to more than 217 days (33). Using a single E. coli O157 strain inoculated into 24 different manure-amended soils, a variation in survival times ranging from 54 to 105 days (average, 80 ± 13 days) was observed (23, 58), which is considerably less than the strain variation observed in the present study. It can therefore be concluded that that strain variation in bacterial characteristics produces at least as much variation in manure-amended soil survival as variations in matrix characteristics. For risk assessment purposes, it would be highly valuable to summarize the observed survival times of the different strains into a single best distribution. However, no acceptable distribution could be fitted through the observed frequency distribution of survival times. Determining a single best distribution for E. coli O157 strain variation in manure-amended soil survival or separate distributions for subgroups likely requires testing a larger collection of strains.
The present study is the first to reveal a significant difference in the capability of manure-amended soil survival among animal and clinical human E. coli O157 isolates. This corresponds to the mounting evidence that suggests there is considerable phenotypic diversity among animal- and human-associated isolates. E. coli O157 strains of animal origin showed much higher colonization of cattle tissue and cells in vitro than human-origin strains (41). The average acid resistance of bovine-biased genotypes was significantly higher than that of human strains (56, 63), and bovine-biased genotypes are equipped with a more efficient nitrogen regulatory response system that enhanced survival under ammonia-limiting conditions (63). In turn, bovine E. coli O157:H7 strains were less virulent in gnotobiotic piglets (7) and produced less Stx2 toxin (55, 68) than strains isolated from clinical human cases. With respect to environmental survival, human-derived E. coli O157 of the H7 phage type that was mainly associated with human disease strains survived significantly better during drying on concrete (6). However, in contrast to our results, the human strains as a group survived significantly less during drying on concrete (6). It is likely that differences in survival capabilities between human- and animal-derived E. coli O157 strains depend on the type of environmental stress encountered.
Relation between virulence gene profile and manure-amended soil survival.
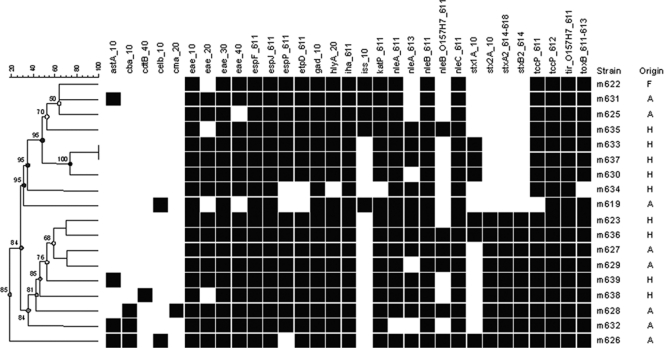
In order to study the effect of virulence genes on manure-amended soil persistence, the virulence profiles of all 18 E. coli O157 isolates were determined using the Identibac microarray. This assay resulted in 17 unique virulence profiles for the 18 E. coli O157 isolates used in the present study (Fig. 2). No clustering of strains according to isolation origin and survival time was observed. These results do not reveal effects of E. coli O157 virulence genes on the capability of manure-amended soil survival. Similarly, no effect of stx1 and/or stx2 gene presence on the ability of E. coli O157 to survive drying on concrete was observed (6). It should be noted that only a selective group of genes is present in the Identibac assay and that there is the possibility of involvement of other (putative) virulence genes.
Fig. 2.
Dendrogram based on the Identibac virulence gene microarray results. Only genes that were present in at least one strain have been included. Black boxes indicate genes present. A, animal origin; F, food origin; H, human origin. The survival time is indicated in days. Gene specifics: astA, heat-stable enterotoxin 1; cba, colicin B pore forming; cdtB, cytolethal distending toxin B; celB, endonuclease colicin E2; cma, colicin M (resembles beta-lactam antibiotics); eae, intimin; espF, type III secretion system; espJ, prophage-encoded type III secretion system effector; espP, putative exoprotein precursor; etpD, type II secretion protein; gad, glutamate decarboxylase; hlyA, hemolysin A; iha, adherence protein; iss, increased serum survival; katP, plasmid-encoded catalase peroxidase; nleA, non-LEE-encoded effector A; nleB, non-LEE-encoded effector B; nleC, non-LEE-encoded effector C; stx1A, Shiga toxin 1 A subunit; stx2A, Shiga toxin 2 A subunit; stxA2, Shiga toxin 2 subunit A; stxB2, Shiga toxin 2 subunit B; tccP, Tir-cytoskeleton-coupling protein; tir, translocated intimin receptor; toxB, toxin B potential adhesion.
The loss of the O157 plasmid was shown to increase stress resistance to acid and chemical substances at the expense of cattle colonization efficiency, indicating a possible burden of virulence plasmids with respect to environmental survival (40). When analyzing the genome of E. coli O157 strain TW14359, which was responsible for the 2006 U.S. spinach outbreak characterized by an unusually high hemolytic-uremic syndrome (HUS) rate, seven putative virulence determinants that were not present in the Sakai and EDL933 reference strains were identified (37). These included two putative type III secretion system effector proteins, candidate genes that could result in increased pathogenicity and/or fitness in the plant environment, and an intact anaerobic nitric oxide reductase gene (norV) (37). For future research of the O157 isolates used in this study, it would be worthwhile to investigate whether these genes provide real fitness advantages in the plant and soil environment.
Based on the PFGE pattern of the strains used, a clustering of human versus animal isolates was observed and, although less clear, between short (<200 days) and long (>200 days) survivors (Fig. 1). This clustering indicates the existence of genetic variation between human and animal strains. The food isolate survived for a long time (214 days) but was genetically more related to the animal isolates than to the human isolates. E. coli O157 strains can be divided into three major (LSPA) lineages (I, I/II, and II), and these lineages have a biased distribution among human and animal hosts, suggesting that the lineages could have unique transmissibility or virulence characteristics (35, 66, 67). In addition, several recent studies showed phenotypic differences related to virulence between isolates belonging to these different lineages (17, 18, 41, 68). Although the different LSPA lineages clustered in the PFGE dendrogram, no significant difference in survival times was observed between different LSPA lineages. Likewise, no significant differences in survival in cattle manure were observed between different E. coli O157:H7 lineages (42). It should be noted that the number of LSPA lineage I strains was underrepresented in the current study.
Metabolic profile of E. coli O157 isolates.
The Biolog phenotypic microarray showed some variation among the E. coli O157 isolates with respect to the number of carbon sources that were oxidized at a rate larger than that of the negative control (median = 83/192, min. = 59/192, max. = 93/192). Table S1 in the supplemental material gives a complete overview of the Biolog phenotypic microarray results for all 18 strains. There was not a relation between the number of carbon sources in each metabolic rate group (<1, 1 to 2, 2 to 3, 3 to 4, and >4 times the negative control) and the origin of isolation (human versus animal) or with survival time. Remarkably, the food isolate (raw-milk cheese) showed the highest average oxidative activity when all carbon sources that were oxidized at a rate larger than the negative control were considered.
In total, 60 substrates were oxidized at a rate of at least two times that of the negative control (Table 1). Many of these sugars and organic acids, or their direct derivatives, are common key compounds in several metabolic pathways (like the Krebs cycle), structural components of plant and fungal cell walls, and part of mucous animal secretions. Interestingly, all E. coli O157 isolates were unable to grow on amino acids essential for humans (with the exception of l-threonine), and no growth was observed on amino acids which are mostly absent from intestinal mucins (excepting l-glutamine). Similar results were obtained for commensal E. coli (51). The carbohydrate 3-O-β-d-galactopyranosyl-d-arabinose is an unnatural chemically synthesized compound, which was utilized by all E. coli O157 isolates at a rate 2 times higher than that of the negative control. In contrast, only 18/65 generic E. coli strains could utilize this source (51).
Table 1.
Average E. coli O157 oxidative activity on various carbon sources as measured by Biolog phenotypic microarraysa
| Carbon source | Oxidative activity |
|---|---|
| d-Glucose-6-phosphate | 4.75 ± 1.21 |
| d-Galacturonic acid | 4.58 ± 1.22 |
| d-Fructose-6-phosphate | 4.46 ± 1.21 |
| Pyruvic acid | 4.32 ± 1.05 |
| d-Glucose-1-phosphate | 4.28 ± 1.12 |
| Inosine | 4.29 ± 1.06 |
| d-Gluconic acid | 4.22 ± 1.12 |
| Adenosine | 4.20 ± 1.08 |
| Thymidine | 4.19 ± 1.11 |
| d-Ribose | 4.13 ± 1.05 |
| 2′-Deoxyadenosine | 4.09 ± 1.00 |
| d-Glucuronic acid | 4.08 ± 1.11 |
| Uridine | 3.95 ± 1.04 |
| d-Trehalose | 3.92 ± 1.00 |
| l-Galactonic acid-γ-lactone | 3.91 ± 1.11 |
| d-Mannose | 3.85 ± 1.02 |
| d-Galactose | 3.80 ± 1.08 |
| Maltose | 3.73 ± 1.09 |
| l-Lactic acid | 3.72 ± 0.86 |
| Maltotriose | 3.69 ± 0.91 |
| Methylpyruvate | 3.69 ± 0.97 |
| Glycerol | 3.67 ± 1.03 |
| d-Melibiose | 3.66 ± 1.14 |
| α-Methyl-d-galactoside | 3.61 ± 1.07 |
| l-Serine | 3.59 ± 0.96 |
| d-Fructose | 3.59 ± 0.97 |
| d-Xylose | 3.49 ± 0.93 |
| α-d-Lactose | 3.41 ± 0.82 |
| l-Malic acid | 3.38 ± .95 |
| d-Mannitol | 3.34 ± 0.93 |
| l-Arabinose | 3.32 ± 0.81 |
| l-Asparagine | 3.32 ± 1.02 |
| N-Acetyl-d-glucosamine | 3.30 ± 0.91 |
| l-Fucose | 3.26 ± 0.94 |
| d,l-α-Glycerol phosphate | 3.22 ± 0.84 |
| Succinic acid | 3.12 ± 0.84 |
| l-aspartic acid | 3.07 ± 1.00 |
| d-Malic acid | 3.06 ± 0.84 |
| d,l-Malic acid | 3.04 ± 0.86 |
| α-d-Glucose | 3.02 ± 0.82 |
| Acetic acid | 3.00 ± 0.91 |
| l-Alanine | 2.93 ± 0.94 |
| d-Alanine | 2.89 ± 0.81 |
| Ala-Gly | 2.88 ± 0.98 |
| N-Acetyl-d-mannosamine | 2.85 ± 0.92 |
| Sucrose | 2.70 ± 1.40 |
| l-Glutamine | 2.60 ± 0.91 |
| Fumaric acid | 2.54 ± 0.78 |
| Mono-methyl succinate | 2.46 ± 0.84 |
| d-Raffinose | 2.39 ± 0.43 |
| d-Glucosamine | 2.38 ± 0.15 |
| Mucic acid | 2.38 ± 1.58 |
| Bromosuccinic acid | 2.34 ± 0.88 |
| l-Threonine | 2.25 ± 1.00 |
| Gly-Pro | 2.24 ± 0.84 |
| Dextrin | 2.20 ± 0.21 |
| l-Rhamnose | 2.13 ± 1.20 |
| 3-O-β-d-galactopyranosyl-d-arabinose | 2.11 ± 0.33 |
| N-Acetyl-d-Galactosamine | 2.10 ± 0.51 |
| l-Proline | 2.08 ± 1.21 |
Only those carbon sources that were oxidized at an average rate of at least 2 times the negative control value (60 out of 192 carbon sources) are shown. Numbers in boldface indicate a standard deviation of catabolic activity larger then 1, indicating considerable diversity among the E. coli O157 isolates.
Relation between metabolic profile and strain origin.
Only two carbon substrates were oxidized at significantly different rates in human and animal E. coli O157 isolates. Human isolates oxidized N-acetyl-d-mannosamine at a high rate compared to the animal isolates (P = 0.041). N-Acetyl-d-mannosamine is a precursor for the biosynthesis of N-acetyl-neuraminic acid (glucuronate), which was together with gluconate identified as a carbon source preferred by generic E. coli but not by E. coli O157:H7 (22). Also, β-methyl-d-glucoside was oxidized at a high rate by the human isolates compared to the bovine isolates (P = 0.044). The relevance of the higher oxidative activity of human clinical strains for these carbon sources remains unclear, but it may contribute to metabolic adaptation to the human digestive tract.
The E. coli O157 isolates belonging to the intermediate LSPA lineage I/II oxidized l-rhamnose and bromosuccinic acid at a significantly high rate compared to lineage II (P = 0.027 and 0.013, respectively). Interestingly, 4 of the 18 strains (1 lineage I/II, 3 lineage II) were not able to ferment l-rhamnose. In contrast, all E. coli O157 isolates from the United States (n = 20) and Japan (n = 100) were found to be rhamnose positive (31, 49), while the E. coli O157 isolates from the United Kingdom were all rhamnose negative (13). Clearly, considerable geographic difference exists with respect to E. coli O157 metabolic capacities.
Relation between metabolic profile and soil survival.
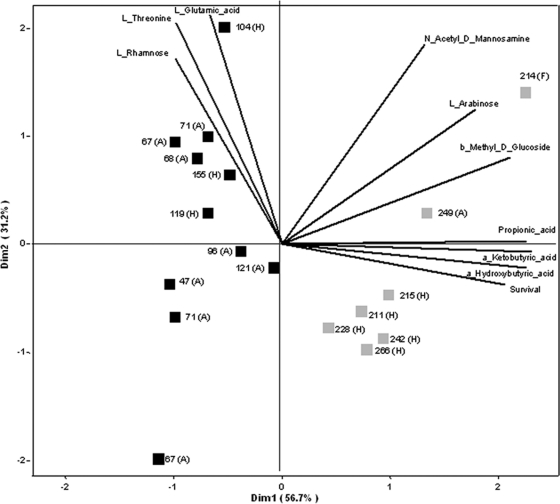
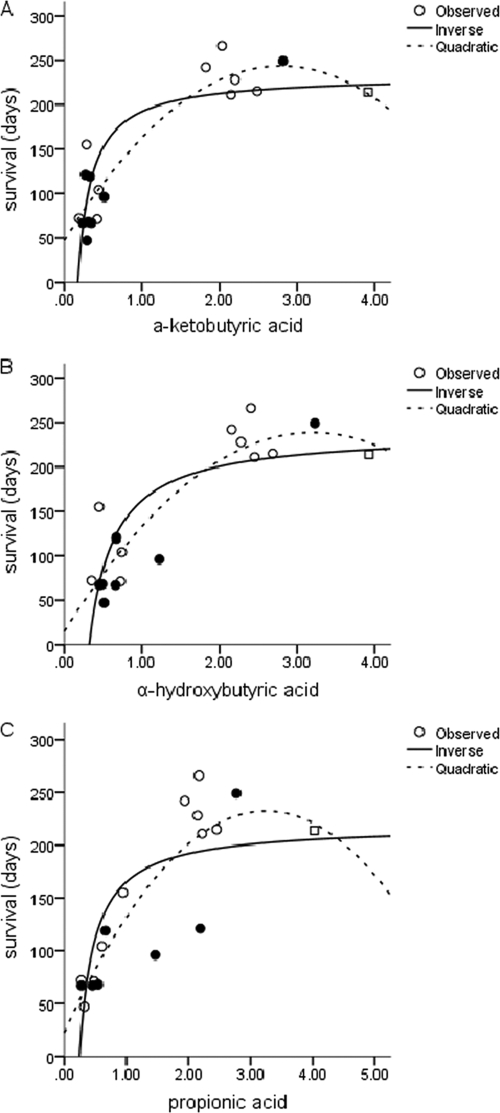
The cluster diagram based on the Biolog phenotypic microarray data revealed a distinct clustering in relation to survival time (Fig. 3). Seven strains (1 animal, 1 food, and 5 human isolates) formed a separate cluster, with all surviving more than 200 days. This means that the metabolic profiles of the isolates surviving more than 200 days were distinctly different from the metabolic profiles of those isolates surviving less than 200 days. Out of the 192 carbon sources, 8 were found to differ significantly with respect to oxidative activity between strains surviving longer than 200 days compared to strains surviving shorter than 200 days. The substrates l-rhamnose, l-glutamic acid, and l-threonine were oxidized at a significantly low rate by the strains surviving longer than 200 days compared to the strains surviving less than 200 days (fold differences of 0.54 [P = 0.032], 0.71 [P = 0.032] and 0.55 [P = 0.006], respectively). The substrates α-ketobutyric acid (P < 0.001), α-hydroxybutyric acid (P < 0.001), propionic acid (P < 0.001), β-methyl-d-glucoside (P = 0.003), and l-arabinose (P = 0.033) were oxidized at a significantly high rate by the strains surviving longer than 200 days compared to the strains surviving less than 200 days. Principal component analysis and factor analysis of the Biolog metabolic profiling data revealed the separation of two distinct clusters of E. coli O157 isolates (correlating to survival times) and showed that the oxidization rates of propionic acid, α-ketobutyric acid, and α-hydroxybutyric acid were indeed most responsible for the observed separation of the two (Fig. 3). Regression analysis showed that the relation between survival time and oxidative activity on these three fatty acids was best described by inverse or quadratic relations (Fig. 4). For α-ketobutyric, α-hydroxybutyric, and propionic acid, the quadratic relation explained 86%, 82%, and 78%, respectively, of the observed variation in survival time (Table 2).
Fig. 3.
Principal component analysis (PCA) ordination diagram of Biolog data for the different E. coli O157 strains used in this study. The length and direction of the lines represent the contribution of the oxidative activity of specific substrates to the first two dimensions (i.e., the explained variance). Only substrates showing a significant different oxidative activity between both groups of short and long survivors are shown. Black squares represent strains surviving less than 200 days, and gray squares more than 200 days. The numbers represent the days of survival; the letters indicate whether the strain is from animal (A), human (H), or food (F) origin.
Fig. 4.
Best-fit relation of survival times of E. coli O157 strains in manure-amended soil (closed circles, animal isolates; open circles, human isolates; open square, food isolate) and normalized oxidative activity on α-ketobutyric acid (A), α-hydroxybutyric acid (B), and propionic acid (C).
Table 2.
Biolog PM-measured parameter estimates and goodness-of-fit values for inverse and quadratic relations between TTDL and normalized oxidative activity on various substrates
| Carbon source | Relation |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Inversea |
Quadraticb |
||||||||
| Constant | a | P | R2 | Constant | a | b | P | R2 | |
| α-Ketobutyric acid | 232.70 | −40.66 | <0.001 | 0.72 | 47.25 | 140.51 | −25.15 | <0.001 | 0.86 |
| α-Hydroxybutyric acid | 238.92 | −76.88 | <0.001 | 0.67 | 15.04 | 139.58 | −21.79 | <0.001 | 0.82 |
| Propionic acid | 218.75 | −53.61 | <0.001 | 0.65 | 22.51 | 129.07 | −19.85 | <0.001 | 0.78 |
Survival time (TTDL in days) = constant − (a/substrate), where “substrate” is the observed normalized oxidative activity.
Survival time (TTDL in days) = constant + (b · substrate) + (b · substrate)2, where “substrate” is the observed normalized oxidative activity.
E. coli O157 efficiently colonizes the gut of ruminants and humans, and the colon is believed to be the primary site of such colonization (30). The gastrointestinal tract is a complex microenvironment containing a variety of short-chain fatty acids (SCFAs), which are major anaerobic microbial fermentation products of complex carbohydrates. The three principal organic acids present in the intestine are acetate (conjugate base of acetic acid), propionate (conjugate base of propionic acid), and butyrate (conjugate base of butyric acid) (2). The concentrations of propionic acid, α-ketobutyric acid, and α-hydroxybutyric acid in the Biolog system were, respectively, 27.0, 19.8, and 38.4 mM (at a pH close to neutral), which are in the range of the total SCFA concentrations in the distal ileum (5), the cattle colon (16), and liquid dairy manure (38). SCFAs often inhibit the growth of pathogens like E. coli O157, but this depends on the type of acid, pH, and temperature (59). E. coli O157 is increasingly capable to grow in the presence of organic acids at elevated pH and temperature (59). Although a direct mechanistic link has not been made in the present study, the strong relation between carbon metabolism in vivo and survival in vitro suggests that the increased capacity to oxidize certain carbon compounds is a strong advantage in surviving a manure-amended soil environment.
Very little research has been performed to investigate the metabolic capacities of E. coli O157 in relation to their isolation origin and environmental persistence. Recently, it was revealed that all E. coli O157 isolates derived from a single large spinach-associated outbreak displayed a phenotype that was characterized by the inability to metabolize N-acetyl-d-galactosamine (48). It was suggested that this phenotype could be exploited as a biomarker for a selective advantage to E. coli O157 with respect to survival in the plant environment. The results of the current study also suggest that metabolic markers could be used for the recognition of environmental fitness. These results indicate that studying difference in phenotypes might give valuable additional information on the ecology and risk assessment of different strains and isolates within the same serotype. Recently, several polymorphisms associated with altered physiology were identified in bovine-associated LSPA lineage II isolates (20). These include reduced cellulose production (which may result in reduced transmissibility to humans and reduced pathogenicity) and reduced functioning of the ethanolamine utilization (eut) operon (probably resulting in impaired pathogenicity). This highlights the importance of linking genetic information with physiological and phenotypic characteristics in order to better understand the role of mutations in the ecology of pathogens like E. coli O157.
Synopsis and conclusions.
The current results indicate that some E. coli O157 strains have metabolic capacities that allow increased survival in a soil environment. Especially, the ability to grow on propionic acid, α-ketobutyric acid, and α-hydroxybutyric acid was highly correlated with survival time. The increased environmental survival might result in a higher probability of transmission to humans. Indeed, human isolates survived significantly longer than animal isolates, and the majority of isolates that were characterized by the long-surviving metabolic profile were human isolates. However, it should be noted that not all human isolates showed extended survival; moreover, not all showed an increased capacity to oxidize the three specific fatty acids. It might be that these strains were acquired by humans by more direct ways of transmission like contact with animals/feces or consumption of undercooked meat. In turn, one animal isolate showed survival exceeding 200 days and showed a metabolic profile similar to that of the long-surviving human isolates and the food isolate. This animal isolate could be a potential risk strain for humans given its similarity to the majority of the human isolates with respect to survival and metabolic characteristics.
Although the genetic diversity of E. coli O157 strains has been described extensively (9, 44, 66), phenotypic diversity has been little explored. The absence of a relation between virulence gene profile and manure-amended survival time, in spite of the presence of a strong correlation between metabolic profile and survival time, highlights the importance of considering phenotypic diversity among E. coli O157 strains along with studying genetic diversity. Phenotypic characteristics, which are the result of the interaction between genotype and the environment, can be highly valuable markers for different aspects of pathogen ecology (survival, transmission, and virulence) and may help our understanding on various aspects of pathogen behavior. In addition, phenotypic characteristics could provide defined targets for intervention strategies.
Supplementary Material
ACKNOWLEDGMENTS
Part of this work was performed at the RIKILT Institute for Food Safety, Wageningen University and Research Centre, Wageningen, The Netherlands. We thank Michael Ziman (Biolog, Hayward, CA) for helpful assistance on the interpretation of the phenotypic microarray results, Fimme van Wal and Albert de Boer of the Central Veterinary Institute (Wageningen University and Research Centre, Lelystad, The Netherlands) for conducting the LSPA assays, and Kim van der Zwaluw (Centre for Infectious Disease Control, The Netherlands) for the strains and the PFGE profiles.
This work was financed by the Dutch Ministry of Economic Affairs, Agriculture and Innovation.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Allerberger F., Sessitsch A. 2009. Incidence and microbiology of salad-borne disease. CAB Rev. 4:1–13 [Google Scholar]
- 2. Aluwong T., Kobo P. I., Abdullahi A. 2010. Volatile fatty acids production in ruminants and the role of monocarboxylate transporters: a review. J. Biotechnol. 9:6229–6232 [Google Scholar]
- 3. Aminul Islam M., Heuvelink A. E., Talukder K. A., de Boer E. 2006. Immunoconcentration of Shiga toxin-producing Escherichia coli O157 from animal faeces and raw meats by using Dynabeads anti-E. coli O157 and the VIDAS system. Int. J. Food Microbiol. 109:151–156 [DOI] [PubMed] [Google Scholar]
- 4. Anjum M. F., et al. 2007. Pathotyping Escherichia coli by using miniaturized DNA microarrays. Appl. Environ. Microbiol. 73:5692–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Argenzio R. A., Southworth M., Stevens C. E. 1974. Sites of organic acid production and absorption in the equine gastrointestinal tract. Am. J. Physiol. 226:1043–1050 [DOI] [PubMed] [Google Scholar]
- 6. Avery S. M., Buncic S. 2003. Escherichia coli O157 diversity with respect to survival during drying on concrete. J. Food Prot. 66:780–786 [DOI] [PubMed] [Google Scholar]
- 7. Baker D. R., et al. 2007. Differences in virulence among Escherichia coli O157:H7 strains isolated from humans during disease outbreaks and from healthy cattle. Appl. Environ. Microbiol. 73:7338–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berger C. N., et al. 2010. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 12:2385–2397 [DOI] [PubMed] [Google Scholar]
- 9. Besser T. E., et al. 2007. Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl. Environ. Microbiol. 73:671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beutin L., Krause G., Zimmermann S., Kaulfuss S., Gleier K. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bono J. L., et al. 2007. Association of Escherichia coli O157:H7 tir polymorphisms with human infection. BMC Infect. Dis. 7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brandl M. T. 2006. Fitness of human enteric pathogens on plants and implications for food safety. Annu. Rev. Phytopathol. 44:367–392 [DOI] [PubMed] [Google Scholar]
- 13. Chapman P. A., Siddons C. A., Zadik P. M., Jewes L. 1991. A improved selective medium for the isolation of Escherichia coli O157. J. Med. Microbiol. 35:107–110 [DOI] [PubMed] [Google Scholar]
- 14. Chapman P. A., et al. 1993. Cattle as a possible source of verotoxin-producing Escherichia coli O157 contaminations in man. Epidemiol. Infect. 111:439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desmarchelier P. M., et al. 1998. A PCR specific for Escherichia coli O157 based on the rfb locus encoding O157 lipopolysaccharide. J. Clin. Microbiol. 36:1801–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diez-Gonzalez F., Callaway T. R., Kizoulis M. G., Russell J. B. 1998. Grain feeding and the dissemination of acid-resistant Escherichia coli from cattle. Science 281:1666–1668 [DOI] [PubMed] [Google Scholar]
- 17. Dowd S. E., et al. 2010. Microarray analysis and draft genomes of two Escherichia coli O157:H7 lineage II cattle isolates FRIK966 and FRIK2000 investigating lack of shiga toxin expression. Foodborne Pathog. Dis. 7:763–773 [DOI] [PubMed] [Google Scholar]
- 18. Dowd S. E., Williams J. B. 2008. Comparison of shiga-like toxin II expression between two genetically diverse lineages of Escherichia coli O157:H7. J. Food Prot. 71:1673–1678 [DOI] [PubMed] [Google Scholar]
- 19. Doyle M. P., Erickson M. C. 2008. Summer meeting 2007 - the problems with fresh produce: an overview. J. Appl. Microbiol. 105:317–330 [DOI] [PubMed] [Google Scholar]
- 20. Eppinger M., Mammel M. K., LeClerc J. E., Ravel J., Cebula T. A. 2011. Genome signatures of Escherichia coli O157:H7 isolates from the bovine host reservoir. Appl. Environ. Microbiol. 77:2916–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ethelberg S., et al. 2004. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg. Infect. Dis. 10:842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fabich A. J., et al. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76:1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Franz E., et al. 2008. Manure-amended soil characteristics affecting the survival of E. coli O157:H7 in 36 Dutch soils. Environ. Microbiol. 10:313–327 [DOI] [PubMed] [Google Scholar]
- 24. Franz E., van Bruggen A. H. 2008. Ecology of E. coli O157:H7 and Salmonella enterica in the primary vegetable production chain. Crit. Rev. Microbiol. 34:143–161 [DOI] [PubMed] [Google Scholar]
- 25. Fremaux B., Prigent-Combaret C., Vernozy-Rozand C. 2008. Long-term survival of Shiga toxin-producing Escherichia coli in cattle effluents and environment: an updated review. Vet. Microbiol. 132:1–18 [DOI] [PubMed] [Google Scholar]
- 26. Friedrich A. W., et al. 2002. Escherichia coli harboring shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84 [DOI] [PubMed] [Google Scholar]
- 27. Gagliardi J. V., Karns J. S. 2002. Persistence of Escherichia coli O157:H7 in soil and on plant roots. Environ. Microbiol. 4:89–96 [DOI] [PubMed] [Google Scholar]
- 28. Gannon V. P. J., et al. 1997. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J. Clin. Microbiol. 35:656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geeraerd A. H., Valdramidis V. P., Van Impe J. F. 2005. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int. J. Food Microbiol. 102:95–105 [DOI] [PubMed] [Google Scholar]
- 30. Herold S., Paton J. C., Srimanote P., Paton A. W. 2009. Differential effects of short-chain fatty acids and iron on expression of iha in Shiga-toxigenic Escherichia coli. Microbiology 155:3554–3563 [DOI] [PubMed] [Google Scholar]
- 31. Hiramatsu R., et al. 2002. Characterization of Shiga toxin-producing Escherichia coli O26 strains and establishment of selective isolation media for these strains. J. Clin. Microbiol. 40:922–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hussein H. S., Sakuma T. 2005. Invited review: prevalence of Shiga toxin-producing Escherichia coli in dairy cattle and their products. J. Dairy Sci. 88:450–465 [DOI] [PubMed] [Google Scholar]
- 33. Islam M., Doyle M. P., Phatak S. C., Millner P., Jiang X. 2004. Persistence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J. Food Prot. 67:1365–1370 [DOI] [PubMed] [Google Scholar]
- 34. Jamieson R. C., Gordon R. J., Sharples K. E., Stratton G. W., Madani A. 2002. Movement and persistence of fecal bacteria in agricultural soils and subsurface drainage water: a review. Can. Biosyst. Eng. 44:1.1–1.9 [Google Scholar]
- 35. Kim J., Nietfeldt J., Benson A. K. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. U. S. A. 96:13288–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kovarova-Kovar K., Egli T. 1998. Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol. Mol. Biol. Rev. 62:646–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kulasekara B. R., et al. 2009. Analysis of the genome of the Escherichia coli O157:h7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 77:3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lamptey J., Moo-Young M., Robinson C. W. 1986. Use of manure liquor as nutrient source for ethanol fermentation. Biotechnol. Lett. 8:525–528 [Google Scholar]
- 39. Lejeune J. T., Abedon S. T., Takemura K., Christie N. P., Sreevatsan S. 2004. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg. Infect. Dis. 10:1482–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lim J. Y., et al. 2010. Phenotypic diversity of Escherichia coli O157:H7 strains associated with the plasmid O157. J. Microbiol. 48:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lowe R. M. S., et al. 2009. Escherichia coli O157:H7 strain origin, lineage, and Shiga toxin 2 expression affect colonization of cattle. Appl. Environ. Microbiol. 75:5074–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lowe R. M. S., et al. 2010. Factors influencing the persistence of Escherichia coli O157:H7 lineages in feces from cattle fed grain versus grass hay diets. Can. J. Microbiol. 56:667–675 [DOI] [PubMed] [Google Scholar]
- 43. Lynch M. F., Tauxe R. V., Hedberg C. W. 2009. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect. 137:307–315 [DOI] [PubMed] [Google Scholar]
- 44. Manning S. D., et al. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martinez J. L. 2006. Role of non-clinical environments in the selection of virulence and antibiotic resistance determinants in pathogenic bacteria. J. Biol. Sci. 6:1–8 [Google Scholar]
- 46. Meyer-Broseta S., Bastian S. N., Arné P. D., Cerf O., Sanaa M. 2001. Review of epidemiological surveys on the prevalence of contamination of healthy cattle with Escherichia coli serogroup 0157:H7. Int. J. Hyg. Environ. Health 203:347–361 [DOI] [PubMed] [Google Scholar]
- 47. Mubiru D. N., Coyne M. S., Grove J. H. 2000. Mortality of Escherichia coli O157:H7 in two soils with different physical and chemical properties. J. Environ. Qual. 29:1821–1825 [Google Scholar]
- 48. Mukherjee A., Mammel M. K., LeClerc J. E., Cebula T. A. 2008. Altered utilization of N-acetyl-d-galactosamine by Escherichia coli O157:H7 from the 2006 spinach outbreak. J. Bacteriol. 190:1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murinda S. E., Batson S. D., Nguyen L. T., Gillespie B. E., Oliver S. P. 2004. Phenotypic and genetic markers for serotype-specific detection of Shiga toxin-producing Escherichia coli O26 strains from North America. Foodborne Pathog. Dis. 1:125–135 [DOI] [PubMed] [Google Scholar]
- 50. Ogden I. D., Fenlon D. R., Vinten A. J. A., Lewis D. 2001. The fate of Escherichia coli O157 in soil and its potential to contaminate drinking water. Int. J. Food Microbiol. 66:111–117 [DOI] [PubMed] [Google Scholar]
- 51. Olukoya D. K. 1986. Nutritional variation in Escherichia coli. J. Gen. Microbiol. 132:3231–3234 [DOI] [PubMed] [Google Scholar]
- 52. O'Reilly K. M., et al. 2010. Associations between the presence of virulence determinants and the epidemiology and ecology of zoonotic Escherichia coli. Appl. Environ. Microbiol. 76:8110–8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Peleg M. 2003. Microbial survival curves: interpretation, mathematical modeling, and utilization. Comments Theor. Biol. 8:357–387 [Google Scholar]
- 54. Perelle S., Dilasser F., Grout J. L., Fach P. 2004. Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world's most frequent clinical cases. Mol. Cell. Probes 18:185–192 [DOI] [PubMed] [Google Scholar]
- 55. Ritchie J. M., Wagner P. L., Acheson D. W. K., Waldor M. K. 2003. Comparison of Shiga toxin production by hemolytic-uremic syndrome-associated and bovine-associated Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 69:1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saridakis C. E., Johnson R. P., Benson A., Ziebell K., Gyles C. L. 2004. Influence of animal origin and lineage on survival of Escherichia coli O157:H7 strains in strong and weak acid challenges. J. Food Prot. 67:1591–1596 [DOI] [PubMed] [Google Scholar]
- 57. Semenov A. V., Franz E., van Bruggen A. H. C. 2010. COLIWAVE a simulation model for survival of E. coli O157:H7 in dairy manure and manure-amended soil. Ecol. Model. 221:599–609 [Google Scholar]
- 58. Semenov A. V., Franz E., Van Overbeek L., Termorshuizen A. J., Van Bruggen A. H. C. 2008. Estimating the stability of Escherichia coli O157:H7 survival in manure-amended soils with different management histories. Environ. Microbiol. 10:1450–1459 [DOI] [PubMed] [Google Scholar]
- 59. Shin R., Suzuki M., Morishita Y. 2002. Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. J. Med. Microbiol. 51:201–206 [DOI] [PubMed] [Google Scholar]
- 60. Sivapalasingam S., Friedman C. R., Cohen L., Tauxe R. V. 2004. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67:2342–2353 [DOI] [PubMed] [Google Scholar]
- 61. Steinberg K. M., Levin B. R. 2007. Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc. R. Soc. B Biol. Sci. 274:1921–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tuttle J., et al. 1999. Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 122:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vanaja S. K., Springman A. C., Besser T. E., Whittam T. S., Manning S. D. 2010. Differential expression of virulence and stress fitness genes between Escherichia coli O157:H7 strains with clinical or bovine-biased genotypes. Appl. Environ. Microbiol. 76:60–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. van Boekel M. A. J. S. 2002. On the use of the Weibull model to describe thermal inactivation of microbial vegetative cells. Int. J. Food Microbiol. 74:139–159 [DOI] [PubMed] [Google Scholar]
- 65. Vidovic S., Block H. C., Korber D. R. 2007. Effect of soil composition, temperature, indigenous microflora, and environmental conditions on the survival of Escherichia coli O157:H7. Can. J. Microbiol. 53:822–829 [DOI] [PubMed] [Google Scholar]
- 66. Yang Z., et al. 2004. Identification of common subpopulations of non-sorbitol-fermenting, β-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Y., et al. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang Y., et al. 2010. Lineage and host source are both correlated with levels of Shiga toxin 2 production by Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 76:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.