Abstract
RNase R is a highly processive, hydrolytic 3′-5′ exoribonuclease belonging to the RNB/RNR superfamily which plays significant roles in RNA metabolism in bacteria. The enzyme was observed to be essential for growth of the psychrophilic Antarctic bacterium Pseudomonas syringae Lz4W at a low temperature. We present results here pertaining to the biochemical properties of RNase R and the RNase R-encoding gene (rnr) locus from this bacterium. By cloning and expressing a His6-tagged form of the P. syringae RNase R (RNase RPs), we show that the enzyme is active at 0 to 4°C but exhibits optimum activity at ∼25°C. The enzyme is heat labile in nature, losing activity upon incubation at 37°C and above, a hallmark of many psychrophilic enzymes. The enzyme requires divalent cations (Mg2+ and Mn2+) for activity, and the activity is higher in 50 to 150 mM KCl when it largely remains as a monomer. On synthetic substrates, RNase RPs exhibited maximum activity on poly(A) and poly(U) in preference over poly(G) and poly(C). The enzyme also degraded structured malE-malF RNA substrates. Analysis of the cleavage products shows that the enzyme, apart from releasing 5′-nucleotide monophosphates by the processive exoribonuclease activity, produces four-nucleotide end products, as opposed to two-nucleotide products, of RNA chain by Escherichia coli RNase R. Interestingly, three ribonucleotides (ATP, GTP, and CTP) inhibited the activity of RNase RPs in vitro. The ability of the nonhydrolyzable ATP-γS to inhibit RNase RPs activity suggests that nucleotide hydrolysis is not required for inhibition. This is the first report on the biochemical property of a psychrophilic RNase R from any bacterium.
INTRODUCTION
RNA processing and degradation play a crucial role in the regulation of gene expression. Although a number of proteins, such as endoribonucleases, exoribonucleases, helicases, and RNA-binding proteins, work in a concerted manner to bring about processing and/or degradation of target RNAs, exoribonucleases play a major role in these processes (2, 11). Bacterial exoribonucleases generally show the 3′-5′ polarity of degradation, with one notable exception (20). RNase R, which is a member of RNB/RNR superfamily of exoribonucleases, exhibits (3′→5′) polar activity with a hydrolytic mode of action and plays an important role in bacterial physiology (11). It was first discovered as a product of vacB, an essential gene for virulence in Shigella flexneri, and later rediscovered in Escherichia coli as RNase R for its action on rRNA (10). This highly processive enzyme has been implicated in quality control of rRNA, mRNA degradation and tmRNA processing. Subsequently, RNase R was shown to be upregulated in response to multiple stress conditions such as cold shock, stationary phase, and starvation in E. coli (1, 5, 7). Although most of these studies were performed in E. coli, the gene (rnr) encoding RNase R has been observed in all sequenced bacterial genomes including in Mycoplasma that has the smallest genome of all free-living organisms. Recently, studies have been initiated on the RNase R from many other bacteria (6, 12–14, 19, 24, 33). In our studies with the psychrotrophic Pseudomonas syringae we observed that RNase R is a component of the novel degradosome, a multisubunit RNA degrading complex that also comprises the endoribonuclease RNase E and RhlE helicase (25). More importantly, we discovered that rnr knockout mutants of the psychrotroph were severely cold-sensitive. These mutants were also defective in the processing of 3′-ends of 16S and 5S rRNAs. The study implicated a new role for RNase R in rRNA processing, not yet shown in any other bacteria (26). Recruitment of RNase R to the degradosome complex, its need during growth at low temperature, and its novel role in rRNA processing prompted us to undertake biochemical characterization of this enzyme from P. syringae. To achieve this goal, we characterized the rnr gene locus and the RNase R enzyme from P. syringae Lz4W. This is the first report on the characterization of this exoribonuclease from any psychrotrophic organism. We present here the general properties of the RNase RPs enzyme with respect to substrate specificity, temperature optima, metal ion requirement, and activity on structured RNA substrates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The psychrophilic P. syringae strain Lz4W (32) was grown in Antarctic bacterial medium (ABM) composed of 5 g of peptone and 2.5 g of yeast extract liter−1 or on ABM-agar (1.5%), as described earlier (30). E. coli cells were grown in Luria-Bertani medium (31). When required, growth media were supplemented with antibiotics: ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; and tetracycline, 20 μg ml−1. For growth analysis, bacterial cells from overnight cultures were inoculated into fresh medium at a dilution of 1:100, and the turbidity of the cultures (i.e., the optical density at 600 nm [OD600]) was measured at various time intervals.
Enzymes, reagents, and general molecular biology techniques.
All chemicals were reagent grade. Nucleotides and the ATP analogue, ATP-γS, were purchased from Roche. Restriction enzymes, T4 polynucleotide kinase, and other DNA modifying enzymes were bought from New England Biolabs (NEB), unless otherwise mentioned. Oligonucleotides were bought from a commercial source (BioServe Biotechnology, India). Protein markers were from Amersham Biosciences. RNA markers were from Ambion.
General molecular biology techniques, including the isolation of genomic DNA, restriction analysis, PCR, ligation, transformation, RNA isolation, and reverse transcription-PCR (RT-PCR) analysis, were performed as described previously (31). Plasmids were isolated by using a plasmid isolation kit (Qiagen). DNA sequencing reactions were carried out using double-stranded plasmid DNA as templates and an ABI Prism dye terminator cycle sequencing method (Perkin-Elmer) and analyzed on an automated DNA sequencer (ABI model 3700; Applied Biosystems).
Cloning, nucleotide sequencing, and RT-PCR analysis of rnr locus from P. syringae.
The amplification and cloning of the RNase R encoding gene (rnr) of P. syringae has been reported earlier (25). We amplified the rnr upstream region by PCR, using a set of forward [Prnr_Fw, 5′-CTACCAATTTC(C/A)CCA(C/T)CTGGGC-3′] and reverse (Prnr_RP, 5′-GGGATCGAGGGTTTGCCAATCGGCCAT-3′) primers, and cloned it into pMOSBlue plasmid (Amersham) to generate pMOSrnr. The rnr downstream region was amplified by using the forward primer (RR_FP8, 5′-GCCGAGCTGCGCAAAAGTCGTGAATTG-3′) located within the rnr gene and the reverse primer (S6_RP3, 5′-CGTTGTARCGGAAGTTGTCTTCCAGCTC-3′) corresponding to a conserved region of rpsF gene that encodes S6 ribosomal protein. All PCR amplified products were cloned in the EcoRV site of pMOSBlue and sequenced. The-overlapping nucleotide sequences were aligned to get the complete sequence of the rnr locus of P. syringae (GenBank accession no. HQ122447).
The primers Rnr_RTFP (5′-CTAAAGACGCAGGCAAGCCTTCCAAGC-3′) and Trm_RTRP (5′-TTACATCAGCCACAACACCCTGGTGC-3′) were used for RT-PCR analysis of cotranscripts from the rnr-trmH operon.
Overexpression and purification of P. syringae RNase R (RNase RPs) protein.
For biochemical characterization of RNase RPs protein, the rnr gene of P. syringae was subcloned from pMOSrnr into the pET28a expression vector (Novagen). Briefly, the rnr gene was excised out of pMOSrnr and ligated between the NdeI and SacI sites of pET28a. The resultant pETrnr-His was then analyzed by nucleotide sequencing to confirm the rnr ORF is in-frame with the vector-encoded six-histidine (His6) tag at the N-terminal end of the recombinant RNase R.
RNase RPs was expressed by IPTG (isopropyl-β-d-thiogalactopyranoside) induction of E. coli BL21(DE3) harboring the pETrnr-His plasmid. To increase the yield of RNase RPs in soluble cytoplasmic fractions, cells grown at 37°C (OD600 of ∼0.6) were shifted to 15°C in which they were kept shaking in the presence of IPTG (0.2 mM) overnight (∼16 h). His-tagged RNase RPs was then isolated from the cell lysates under native conditions using Ni-NTA-agarose (Qiagen) chromatography according to the supplier's instructions. All buffers contained protease inhibitors (Complete EDTA-free cocktail; Roche). Final purification of the protein for biochemical analysis was made by gel filtration on a Superose-12 column. Purified proteins were kept at −20°C in buffer containing 25 mM Tris-Cl (pH 7.5), 150 mM KCl, and 10% glycerol.
RNA substrates and exoribonuclease activity assays.
Poly(A) and other synthetic RNA substrates were labeled at their 5′ ends using [γ-32P]ATP and T4 polynucleotide kinase. The unincorporated nucleotides were removed by gel filtration on Sephadex G50 columns. For preparing the 32P-labeled malE-malF RNA substrate, the plasmid containing the malE-malF intergenic region under the control of T7 promoter (17) was linearized by digestion with HindIII and used as a template for transcription by T7 RNA polymerase in the presence of [α-32P]UTP using an in vitro transcription kit (MAXIscript; Ambion). The unincorporated radioactive nucleotides were removed by using NucAway spin columns (Ambion), and the transcripts (318 nucleotides [nt]) were checked by 8 M urea-polyacrylamide gel electrophoresis (PAGE) as described previously (25).
RNase R assays were typically carried out in 20-μl reaction mixtures containing 25 mM Tris-HCl (pH 7.0), 100 mM KCl, 0.25 mM MgCl2, 5 mM dithiothreitol, 10 pmol of 32P-labeled substrate, and the indicated amount of enzyme. Unless otherwise mentioned, the reactions were performed for 5 min at 22°C and stopped by adding 10 mM EDTA, 0.2% sodium dodecyl sulfate (SDS) and 1 mg of proteinase K ml−1. Two volumes of formamide dye mix that contained 5 mM EDTA, 10% SDS, 0.025% bromophenol blue, and 95% formamide were then added, and the mixture was then boiled for 5 min and snap-frozen. RNA degradation products were resolved on denaturing gels (8 M urea–8% polyacrylamide). For resolving smaller products, sequencing gels (8 M urea–20% polyacrylamide) was used. The cleavage products were visualized by using a phosphorimager (Fuji) and quantified by using ImageGauge software (Fuji). To compare the relative activities, we made sure that the substrates were not completely digested. The data were obtained from the ratio of signal intensities of products arising due to degradation and the total input of substrates (combined signals from degraded and undegraded smears of heterogeneous substrates) and then normalized for protein amounts and reaction time. The data points are presented as mean of values derived from at least two independent experiments. Since the synthetic substrates used in assays were heterogeneous, their concentrations were calculated by using their average sizes: 150 nt for poly(A), poly(G), and poly(C) and 100 nt for poly(U).
Other methods.
Proteins were quantified by the dye binding method of Bradford (4), using bovine serum albumin as a standard. SDS-PAGE, transfer of proteins onto Hybond C membrane, and Western analyses of proteins with RNase R-specific polyclonal antibodies or anti-His tag antibodies (Santa Cruz Biotechnology) were carried out as described earlier (25). Analysis of the molecular mass of RNase R by gel filtration under different salt conditions was performed on a Superdex 200 column (Amersham Biosciences). The protein molecular markers for gel-filtration analysis were aldolase, catalase, ferritin, and thyroglobulin which had the molecular masses of 158, 232, 440, and 669 kDa, respectively.
RESULTS
rnr gene locus of Antarctic P. syringae.
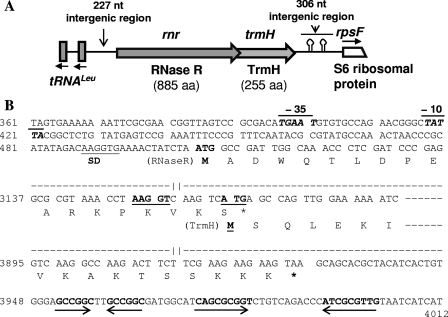
Bioinformatic analysis of genome sequences of different bacteria available in the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov) suggested that the rnr (encoding exoribonuclease R) locus is highly conserved among different Pseudomonas species, in which the trmH gene (encoding a putative tRNA/rRNA methyl transferase) constitutes the second gene of an rnr-trmH operon. Downstream of the dicistronic operon had another highly conserved gene (rpsF) encoding the S6 ribosomal protein in these bacteria. For the characterization of rnr locus from the Antarctic P. syringae Lz4W, we used the cloned rnr gene in pMOSBlue plasmid (named as pMOSrnr) as described in Materials and Methods. The nucleotide sequence analysis of the cloned DNA confirmed that it encodes RNase R homologue (RNase RPs) of the organism. We then characterized the entire rnr locus of P. syringae Lz4W by cloning and sequencing of the downstream and upstream regions of the rnr gene (Fig. 1). The downstream region was amplified by using a set of forward primer (RR_FP8) located within the rnr and the reverse primer (S6_RP3) corresponding to a conserved region within the rpsF gene that encodes S6 ribosomal protein. The amplified DNA was cloned and sequenced. As expected, this amplified DNA segment contained the trmH gene, apart from the partial sequences of rnr and rpsF genes. Analysis of the sequence also showed that the reading frames of RNase R and TrmH (encoding proteins of 885 and 255 amino acids, respectively) of P. syringae overlaps with each other by 4 nt, as in the case of other Pseudomonas species. The trmH gene was, however, separated from the downstream rpsF gene by 306 nt, which contained two stem-loop structures (Fig. 1B) that may function as a transcriptional terminator. RT-PCR analyses of cellular RNAs indicated that rnr and trmH genes are cotranscribed (see Fig. S1 in the supplemental material).
Fig. 1.
Gene organization in the rnr locus of P. syringae Lz4W. (A) Schematic representation of different genes in the 4,468-bp sequenced region (accession no. HQ122447), shown as block arrows on a line. The directions of transcription have been indicated by the arrows. The putative hairpin structures that occur downstream of the rnr-trmH operon and may act as transcription terminator signals have been indicated. Two leucine-tRNA genes transcribed divergently from rnr are shown as vertical blocks. (B) Nucleotide sequence of the relevant regions of the rnr-trmH operon. Only the sequence of the promoter region of the rnr gene, the overlapping region of RNase R and TrmH reading frames, and the downstream region containing the inverted repeats forming putative hairpin structures are shown. The numbers at left of the sequence correspond to the nucleotide numbers in accession no. HQ122447. The sequences not shown here have been indicated by dashed lines with gaps. Marked on the sequences are the “−10” and “−35” regions of the putative rnr promoter shown as a line at the top, putative ribosome binding Shine-Dalgarno (SD) sequences by a line below, and the inverted repeats by opposing arrows below the sequences. Amino acid residues encoded by relevant regions of the reading frames are given in single-letter code. Translational start codons (ATG) are shown in boldface, while the stop codons are indicated by an asterisk (*).
We also cloned and sequenced the upstream region of rnr using the primers set (Prnr_Fw and Prnr_RP) as described in Materials and Methods. The nucleotide sequence analysis indicated that the upstream region contains not only a putative rnr promoter but also divergently transcribing genes for two tRNALeu (Fig. 1). The putative rnr promoter consisting of “−10” (TATTA) and “−35” (TGAAT) boxes separated by 16 nt was located 80 nt upstream of the “ATG” start codon of RNase R reading frame. A six-nucleotide sequence (AAGGTG) showing imperfect complementarity to the 16S rRNA 3′-end (3′UUCCUC-5′) of P. syringae (24) was observed 11 nt upstream of the “ATG” codon, which is likely to act as the Shine-Dalgarno (SD) sequence for translation initiation. On the other hand, a 5-nt putative SD sequence (AAGGT) which is perfectly complementary to the 3′ end of 16S rRNA sequence was located 6 nt upstream of the “ATG” start codon of trmH reading frame of the bicistronic operon.
The deduced amino acid sequence of RNase RPs was 71 to 89% identical (the lowest with P. aeruginosa and highest with P. fluorescence) to the homologues of different Pseudomonas species and only ca. 54% identical (70% similar) to E. coli RNase R (RNase REc). CLUSTAL W analysis of the homologues from different gammaproteobacteria indicated that RNase R of different Pseudomonas species form a distinct cluster which is separated from the clusters representing E. coli/Salmonella group and Vibrio and Photobacterium group (see Fig. S2 in the supplemental material). Similar clustering of the homologues were also observed with TrmH; the deduced amino acid sequences of TrmH of P. syringae Lz4W displayed 80 to 90% identity (representing P. aeruginosa and P. fluorescence, respectively), with the homologue from E. coli showing 56% identity.
Cloning, expression, and purification of RNase RPs.
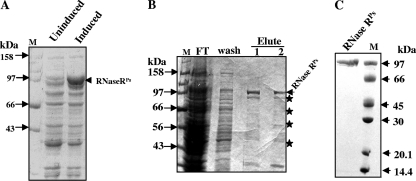
For biochemical characterization of RNase RPs, E. coli BL21 harboring pETrnr-His was induced by IPTG at a low temperature as described in Materials and Methods. The cells produced His-tagged RNase R protein, which migrated near the 97-kDa marker band in SDS-PAGE (Fig. 2A). This was consistent with the calculated molecular mass of the RNase RPs protein (∼98.7 kDa). The overexpressed protein was purified using Ni-NTA chromatography under native conditions as described in Materials and Methods. A few minor proteins were coeluted with RNase RPs from the Ni-NTA column (Fig. 2B). When these eluted fractions were subjected to Western analysis using polyclonal anti-His antibody, the proteins cross-reacted with the antibody (data not shown), suggesting that the coeluted proteins probably represent the degraded RNase R fragments that have retained the His tag. The intact RNase RPs protein was separated and purified from the degraded protein fragments by glycerol gradient centrifugation, or by gel-filtration on Superose-12 as shown in Fig. 2C. The purified protein fractions that produced a single band on SDS-PAGE were used for subsequent biochemical analysis. The yield of purified RNase RPs was ∼10 mg per liter of culture.
Fig. 2.
Overexpression and purification of His-RNase RPs. (A) SDS-PAGE analysis of cell lysates from uninduced and IPTG-induced cultures of E. coli BL21 harboring pETrnr-His. (B) SDS-PAGE profile of the eluted protein fractions of His-tagged RNase RPs from Ni-NTA column. Starred protein bands and RNase R in the eluted fractions cross-reacted with anti-His antibodies. (C) Coomassie blue-stained gel of purified RNase R. In this experiment, Ni-NTA-fractionated RNase RPs was further purified by gel filtration on Superose-12. Lane M contains protein markers. The molecular masses of the marker proteins have been labeled.
Substrate preference and end product analysis of RNase RPs activity.
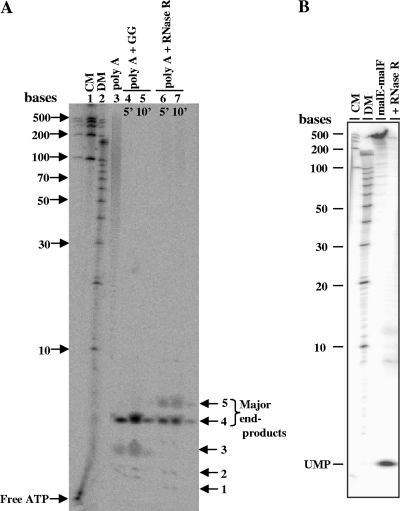
The exoribonuclease activity of purified His-RNase RPs was examined on 32P-end-labeled synthetic RNA substrates such as poly(A), poly(G), poly(U), and poly(C) to check any nucleotide preference. Maximum activity was observed on poly(A) as seen with other enzymes of RNB family (9, 34). A quantitative estimation of degradation activity, as described in Materials and Methods, showed following specific activities (pmol min−1 ng of protein−1): poly(A), 1.9; poly(U), 1.7; poly(C), 0.29; and poly(G), 0.05. Thus, poly(A) and poly(U) were better than poly(G) and poly(C) as substrates for degradation. We then analyzed the products of RNase RPs activity on 5′[32P]-end-labeled poly(A) (Fig. 3A) and internally 32P-labeled malE-malF RNA (Fig. 3B). The cleavage product of malE-malF was nucleotide monophosphates (Fig. 3B), as expected for exonucleolytic degradation. However, as depicted in Fig. 3A, the predominant limit product (also called end product) was a tetramer produced from 5′-32P-end-labeled poly(A) degradation. A certain amount of pentamer was also observed. This is interesting because the limit product produced by E. coli RNase R (RNase REc) activity is a dinucleotide (9, 21). We also noticed that the activity of P. syringae degradosome complex (glycerol gradient purified) on poly(A) also produced tetramer as major product, with only a trace amount of trimer (Fig. 3A, lanes 4 and 5). This suggests that degradation of RNA chains mediated by degradosome is also incomplete, implying a requirement for oligoribonuclease (16).
Fig. 3.
Analysis of degradation products of RNase RPs. (A) 5′-end 32P-labeled poly(A) was used as a substrate, and the degradation products were analyzed on 20% sequencing gel (8 M urea-PAGE). Lanes 1 and 2 contain RNA century markers (CM) and decade markers (DM), respectively. Lane 3 is a control (without enzyme). Lanes 4 and 5 represent the activity of the degradosomal complex (marked as GG for glycerol gradient fraction) on poly(A). Lanes 6 and 7 depict the activity of purified RNase RPs on poly(A). Limit products (end products) of degradation have been labeled with arrows. (B) 32P-internally labeled malE-malF RNA (318 nt) was used as a substrate for degradation. Samples in each lane are labeled at the top of the panel. The sizes of the molecular markers and the cleavage product of the nucleotide monophosphate (UMP) are marked.
Temperature optima and thermal stability of RNase RPs.
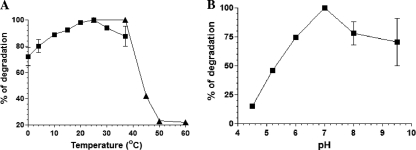
We examined the effect of temperature on the activity of purified RNase RPs on poly(A) substrate. The activity assays were carried out at different temperatures. As shown in Fig. 4A, the enzyme was active at lower temperatures (0 and 4°C) but showed maximum activity at 22°C. A lesser activity at 37°C and above, compared to 22°C, hinted that the enzyme might not be heat stable. To test this, RNase RPs was preincubated at different temperatures (22, 37, 45, 50, and 60°C) for 5 min, followed by assay for activity at 22°C. No activity was observed when the enzyme was preincubated at the temperatures above 37°C, suggesting that the RNase RPs is extremely heat labile (Fig. 4A). Thus, the RNase RPs appears to be a psychrophilic enzyme with an optimum activity at ∼22°C. This is interesting because most other enzymes from this psychrotrophic bacterium displayed increased activity with increase in temperature and had maximum activity at 37°C (29).
Fig. 4.
Temperature optima and heat stability (A) and pH-dependent activity (B) of RNase RPs. (A) 5′-end-32P-labeled poly(A) substrate was used for degradation in the presence of 10 ng of RNase RPs in each reaction at the indicated temperatures. Products were resolved on 8 M urea–8% PAGE, visualized by using a phosphorimager, and quantified as described in Materials and Methods. Heat stability was tested by preincubating the enzyme at the indicated temperatures (22, 37, 45, 50, and 60°C) for 5 min, and then degradation assays were performed at 22°C for quantification. Quantified values for the temperature-dependent activity (■) and heat stability (▴) of RNase RPs are plotted on the same graph. (B) pH-dependent activity of RNase RPs on poly(A). Assays were performed at 25°C. Error bars shown on graphs indicate the deviation in the values.
pH optimum and metal ion requirement for RNase RPs activity.
The optimum pH at which RNase RPs is active was tested by degradation of poly(A) in the presence of 100 mM KCl, 0.25 mM MgCl2, and 25 mM Tris-HCl (for pH values between 6 and 9.5) or sodium acetate buffer (for pH 4.5 and 5.2). The highest activity was observed at pH 6.5 to 7.5, suggesting a broad pH optima for the enzyme's activity (Fig. 4B). RNase REc (9) also displays high activity over a broad pH range (7.5 to 9.5) and both differ from Mycoplasma RNase R, which displayed activity peak at pH 8.5 (19).
The effect of various divalent cations on the activity of RNase RPs was assessed using poly(A) as a substrate. The cations (Mg2+, Mn2+, Ca2+, and Zn2+) in the reaction buffer containing 25 mM Tris-Cl (pH 7.5) and 100 mM KCl were tested by varying their amounts. RNase RPs catalyzed the degradation of poly(A) in the presence of Mg2+, Mn2+, and Ca2+ ions (see Fig. S3 in the supplemental material). However, it was noted that the enzyme also degraded poly(A) in the absence of exogenously added divalent cations (lane 2 [see Fig. S3A, B, and C in the supplemental material]). This suggested that the enzyme-bound metal ions or that trace amount of cations present in the reaction buffer might be responsible for such degradation. To confirm that this is the case, when EDTA was used in the buffer to remove any trace amount of divalent cations, the enzyme lost poly(A) degradation activity (lane 3 in panels A and B and lane 7 in C of Fig. S3 in the supplemental material). Subsequently, by varying the amounts of EDTA, we established that 1 mM EDTA in the reaction mixture is just sufficient to abolish the intrinsic initial degradation activity. We then used this condition to assess divalent cation requirements for stimulation of activity. As expected, enzyme activity increased with the addition of increased concentration of Mg2+ and Mn2+ to the reaction mixture (see Fig. S3A and B in the supplemental material). Interestingly, at lower concentration (0.1 mM) Mn2+ was better than Mg2+ in stimulating the activity, displaying 0.76 and 0.42 pmol of poly(A) degradation min−1, respectively. However, Mn2+ at a higher concentration (>0.5 mM) was inhibitory, but Mg2+ (1 to 5 mM) addition increased the activity further (∼0.82 pmol min−1). On the other hand, Ca2+ stimulated the poly(A) degradation maximally at 0.2 mM (0.74 pmol min−1) but inhibited it at 0.7 mM (see Fig. S3C in the supplemental material). Poly(A) was not degraded at all when Zn2+ (50 μM to 0.7 mM) was tested under similar conditions (data not shown), suggesting that Zn2+ does not stimulate the catalytic activity of RNase RPs. This is in contrast to the Mycoplasma RNase R that was reported to be highly active in the presence of Zn2+ compared to Mg2+ at its optimal concentration (0.05 to 0.25 mM) (19). RNase REc, on the other hand, was reported to be more active at 0.1 to 0.5 mM Mg2+ (highest in 0.25 mM) and in 10 μM Zn2+ (9), suggesting subtle alterations in the metal ion requirements of the enzymes from different species.
Among the monovalent cations (K+, Li+, Na+, and Rb+) that were tested in the assays, maximum RNase RPs activity was observed with K+. Sodium and rubidium ions were also effective, almost similar to potassium ion, but lithium was completely inhibitory for the activity (data not shown). When the KCl concentration in the assay buffer was varied, the highest activity was observed between 50 and 150 mM KCl (see Fig. S4A and B in the supplemental material). The activity dropped drastically (∼5-fold) at 300 mM concentration and above. The effect was more pronounced when malE-malF RNA was used as the substrate (see Fig. S4B in the supplemental material). This was surprising since E. coli RNase R required 300 mM KCl for optimum activity (9).
RNase RPs oligomerization and reduction of activity.
Since the P. syringae RNase R displayed higher activity in the presence of 50 to 150 mM KCl but poor activity at 300 mM and above, we examined the oligomerization state of the RNase RPs in the presence of 50 to 500 mM KCl by gel-filtration analysis. As indicated in the gel elution profile (see Fig. S4C in the supplemental material), the enzyme largely remained as monomer in the buffer containing 50 mM KCl but entered a higher oligomerization state in the presence of 300 and 500 mM KCl. Hence, RNase RPs oligomerization might have an inhibitory effect on the activity. The exact oligomerization state was not determined accurately here due to the broad nature of the peaks for the high-molecular-weight regions of the eluted fractions. However, the elution profiles (indicated by the elution time) of the RNase R oligomers (14 min) and monomers (22.7 min) compared to the marker proteins, e.g., thyroglobulin (669 kDa, 10.2 min), ferritin (440 kDa, 11.4 min), catalase (232 kDa, 13.5 min), and aldolase (158 kDa, 14.8 min), suggested that RNase RPs largely forms dimers in 300 to 500 mM KCl.
RNase R activity on structured RNA substrates.
REP (for repetitive extragenic palindrome) sequences comprised of highly stable stem-loop structures in RNA play an important role in protecting transcripts against degradation and also in translation. RNase R plays important role in their metabolism in cells (8). We examined the activity of RNase RPs on the intergenic malE-malF RNA with a REP sequence (18, 23, 28). As shown in Fig. 5A, the malE-malF RNA was completely degraded in a time-dependent manner by purified RNase RPs at 25°C, without generating any cleavage intermediates. At the 5- and 10-min time points when malE-malF was only partially digested, the remaining RNAs retained their original length, confirming the processive hydrolysis by RNase RPs on the structured substrate.
Fig. 5.
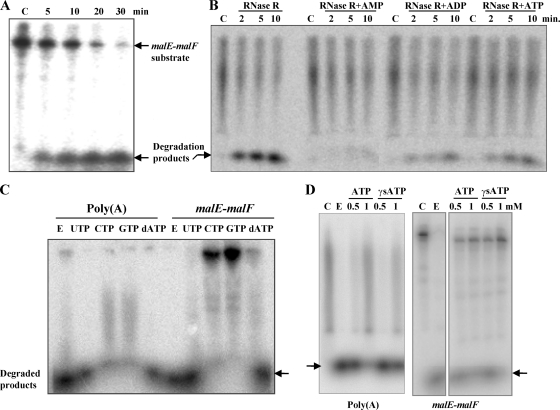
Nucleotide inhibition of RNase RPs activity on malE-malF and poly(A) substrates. 32P-labeled substrates were used for degradation at 25°C. (A) Degradation activity of RNase RPs on intergenic malE-malF RNA substrate, which contains a REP sequence with stem-loop structures (17, 21). (B) Effects of ATP, ADP, and AMP on RNase R-mediated degradation of 5′-end-labeled poly(A). (C and D) The effects of the nucleotides UTP, CTP, GTP, and dATP and inhibition by nonhydrolyzable ATP-γS of RNase RPs on poly(A) and malE-malF RNA degradation are shown in panels C and D, respectively. A total of 10 ng of RNase R and 5 mM nucleotides were used in the reaction mixtures in panel C. Lanes C represent control samples (without the enzyme), while lanes E represent the second control with enzyme but without any added nucleotides.
Since temperature affects the stability of RNA secondary structures, we assessed the temperature-dependent degradation of malE-malF RNA by the psychrophilic RNase RPs (see Fig. S5 in the supplemental material). The degradation of malE-malF reached maximum at ∼25°C, but the activity at 0 to 4°C was lower (∼25% of maximal activity) on this structured substrate compared to the activity (∼70% of maximal activity) on single-stranded poly(A). The activity curve remained more or less unchanged between 25 to 37°C, probably due to slow inactivation of the enzyme at these temperatures.
Nucleotide inhibition of RNase RPs activity in vitro.
Intriguingly, poly(A) degradation by the degradosome from P. syringae (27) is somewhat inhibited in the presence of ATP. We wondered whether this was due to the inhibition of the RNase R exonuclease activity by ATP. To examine this, we incubated 32P-labeled poly(A) substrate with purified RNase R in the presence of 5 mM ATP used in the degradosomal activity assay. Quiet remarkably, ATP inhibited the activity of His-RNase R (Fig. 5B). The effect of ADP and AMP on RNase R activity was also checked in the same experiment. The extent of inhibition observed was as follows: AMP > ADP > ATP, showing ca. 75, 52, and 43% inhibition, respectively, in that order, compared to control (i.e., without added nucleotides) activity. These results, although consistent with product inhibition of poly(A) cleavage reactions, were found later to be novel. We observed that other ribonucleotides (e.g., GTP and CTP) could also inhibit the degradation by RNase RPs (Fig. 5C). The inhibition was not restricted to poly(A) only but was observed with other RNA, such as malE-malF. Interestingly, UTP and dATP were inefficient in inhibiting the cleavage reactions (Fig. 5C). This ruled out the possibility of a general mechanism by which metal ion chelating by nucleotides would lead to enzyme inhibition. Quantification of inhibitory activity based on phosphorimages suggested that ATP, GTP, and CTP at 5 mM could inhibit ∼85% of the poly(A) cleavage in which 10 pmol of substrate and 5 nM protein were incubated at 25°C for 10 min. The specific activity of the enzyme in this condition was 9.25 nmol of poly(A) min−1 nmol of protein−1. When ATP was tested at lower concentration (0.5 and 1.0 mM), the activity was inhibited by ∼20 and ∼65%, respectively. Since AMP was better than ATP in the inhibition, we also checked if the inhibition was mediated by ATP-hydrolysis, by performing assays in the presence of ATP-γS. The nonhydrolyzable ATP-γS also inhibited the RNase R activity (Fig. 5D), suggesting that the inhibition of RNase R activity does not require ATP hydrolysis.
DISCUSSION
Study of the rnr locus in P. syringae Lz4W suggests that the gene organization is the same as seen in other Pseudomonas genomes (www.pseudomonas.com). Two genes of the rnr-trmH operon, which encode the exoribonuclease R and a putative tRNA/rRNA methylase enzyme, respectively, are highly conserved. The occurrence of overlapping reading frames between the two genes suggests that they are transcriptionally and translationally coupled and possibly functionally linked. However, while the inactivation of rnr caused cold sensitivity in a P. syringae mutant (26), we did not observe any discernible phenotype by inactivation of trmH (P. Mittal and M. K. Ray, unpublished data). Since Mycoplasma RNase R is sensitive to ribose-methylated RNA substrate (19), it is possible that TrmH-mediated methylation of RNA substrates can act as a regulator of the physiological activities of RNase RPs. At present, any functional significance of the coupling of trmH with rnr in the Pseudomonas genomes remains to be established.
RNase R is widespread in eubacteria, but it has been extensively characterized from only mesophilic E. coli and pathogenic Mycoplasma and to some extent from Salmonella enterica serovar Typhimurium and Streptococcus pneumonia (12). Our study for the first time provides an insight into the characteristics of an RNase R homologue from a cold-adapted bacterium. The biochemical properties of the RNase R from Antarctic P. syringae strain Lz4W show that there are a number of common properties in the exoribonuclease from these organisms, despite a few significant differences, as mentioned below. The major similarity, as expected, lies in the ability of both RNase RPs and RNase REc enzymes to degrade secondary structured RNAs, without the help of RNA helicases. Both enzymes prefer divalent cations Mg2+ and Mn2+ for this activity. In both cases, the enzymes were highly processive degrading RNA chains from 3′→5′ direction and produced nucleotide 5′-monophosphates as the major hydrolytic products of both single-stranded and double-stranded structured RNA substrates. However, the enzymes differed from each other in their thermal stability and temperature-dependent activities. The enzymes also differed in the lengths of the limit products of RNA substrates, tetranucleotides (with a small amount of pentanucleotides) in the case of RNase RPs, and dinucleotides (with smaller amounts of trinucleotide) in the case of RNase REc (9). Interestingly, M. genitalium RNase R (RNase RMg) was shown to produce mostly trinucleotides (and only a small amount of dinucleotides) as the end products (19). Although the molecular basis of the production of different lengths of end products of RNA chain by different RNase R homologues is not known, the proposed catalytic pocket residues in the RNB domain of the enzyme might play an important role, as shown for RNase II and RNase R from E. coli (3, 12, 21, 22). Structural models of RNase REc (3, 34) based on the RNase II crystal structures (15, 35) have placed the catalytic site deep in a putative funnel-like structure of RNB domain into which the 3′ ends of the RNA chains are threaded for cleavage. Therefore, it is possible that the strength of anchoring of RNA 3′ ends in the active-site domain of RNase R of different organisms may vary, leading to the release of different end products which fail to remain bound to the enzyme. It is important to note that Pseudomonas species contain only one member of RNB/RNR exoribonuclease family (based on sequenced genomes), which has greater identity to RNase R. The possibility that RNase RPs maintains certain features of RNase II, such as the generation of 4 nt as a limit product, possibly due to sequence conservation in the RNB site (22), was not obvious from a sequence alignment of RNase RPs, RNase REc, and RNase IIEc (see Fig. S6 in the supplemental material). At present, any implication of the variation of limit product size or their role in adaptation of the RNB/RNR enzymes to environment is an open question.
However, as pointed out above, the important difference between the psychrophilic RNase RPs and the mesophilic RNase REc appears to lie in the temperature optimum of activity. The P. syringae enzyme displayed highest activity at ∼22°C, while the E. coli enzyme showed maximum activity at 37°C (9). The enzymes also differed in their thermal stability, with the RNase RPs displaying the heat-labile nature of psychrophilic protein. Thus, the P. syringae RNase R appears to have become a “cold-loving” enzyme during the course of evolution, probably from a mesophilic ancestor, as has been suggested earlier based on many other enzymes which display maximal activity at 37°C (29). However, the maximum activity displayed by RNase RPs at ∼22°C is interesting since it points out a different facet of evolution of the multicomponent protein complexes. While the entire RNA degradosome complex consisting of RNase E, RNase R, and Rhl E, as well as the isolated RNase E enzyme of P. syringae display highest activity at 37°C (27), RNase R has adapted to function optimally at a lower temperature. This suggests that the psychrophilic nature of RNase RPs is very important in low-temperature physiology when the enzyme works alone outside the complex, as evidenced from the lack of cold sensitivity in RNase E C-terminal-deleted strain of P. syringae in which degradosomal proteins fail to associate (P. Mittal and M. K. Ray, unpublished observation). However, it may also reflect that it has evolved faster than RNase E for cold adaptation and/or that RNase R has been recruited to the degradosome complex at a later stage of evolution in the Antarctic P. syringae.
The ability of purified RNase RPs to degrade the structured malE-malF substrate is consistent with the general property of RNase R from other organisms to degrade rRNAs and the structured RNAs containing REP sequences (7, 34). This, however, raises an interesting question as to why the degradosomal complex of P. syringae (25) would need the RNA helicase RhlE at all in the complex when only RNase E-RNase R together would have been sufficient to degrade RNAs with secondary structures. The protein complex without helicase would have been highly economical to the cold-adapted organism, since this would have reduced the ATP (energy) requirements of cells. The answer to this is not quite obvious. However, one possible explanation would be that the degradation by RNase R alone is probably influenced by the different properties of RNA chains, e.g., the nucleotide composition, as seen in case of E. coli (34), or as shown in the present study that RNase RPs fails to degrade poly(G) and poly(C) homopolymers. These homopolymers are not only poor substrates but also have poor affinity toward RNase R of E. coli (34). Therefore, in the metabolism of G- and C-rich transcripts produced from the (G+C) rich genomes of Pseudomonas, the degradosome-associated RNA helicase might aid in enhancing the binding affinity of the complex to RNA substrates and also possibly help modulate the structures so that the binding sites for endoribonuclease are exposed for cleavage and then degradation by exoribonuclease. It is also plausible that in vivo the mRNAs remain associated with proteins to form ribonucleoprotein particles, remodeling of which would require the helicase activity to make RNA accessible to RNases.
It is most surprising that RNase RPs is inhibited by ATP, GTP, and CTP. This has not been reported earlier, and its significance could not be ascertained at present. In fact, the inhibition by ATP is counterintuitive as the helicase component in the degradosome complex requires ATP for duplex RNA unwinding activity. The opposite modes of activity imparted by ATP on two proteins in the same complex might be a novel way of regulating the overall activity of protein complexes in cells. As for the mechanism of inhibition, such as whether the inhibitory nucleotides bind to the product (nucleotide monophosphates) exit site in the catalytic pocket of RNase RPs and thereby prevents further degradation of the substrates, needs to be examined in the future. Interestingly, we have noted that one of the crystal structures of RNase II (pdb 2ix0) in the Protein Data Bank (www.pdb.org) shows a CTP bound at the interface between CSD2 and RNB domain of the enzyme. The CTP bound at this position would block the entry of RNA chain into the active site, which could also be a possible mechanism for inhibition in the case of RNase RPs, if the inhibitory nucleotides bind to the analogous site of the enzyme. However, it is important for us to know why RNase RPs is essential for the growth of P. syringae only at low temperatures. We are currently investigating the structure-function relationship of the enzyme in cleaving different RNA substrates and their physiological implications during the growth of P. syringae at low temperatures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gabriele Klug, E. Evguenieva-Hackenberg, and D. P. Kasbekar for reading the manuscript.
This study was supported in part by the Council of Scientific and Industrial Research (CSIR; India) and in part by the Volkswagen Foundation (Germany) to M.K.R. Research fellowships awarded by the CSIR to P.I.R. and P.M. and by the University Grant Commission (UGC; India) to S.S. for carrying out the work are also acknowledged.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Andrade J. M., Cairrao F., Arraiano C. M. 2006. RNase R affects gene expression in stationary phase: regulation of ompA. Mol. Microbiol. 60:219–228 [DOI] [PubMed] [Google Scholar]
- 2. Arraiano C. M., et al. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34:883–923 [DOI] [PubMed] [Google Scholar]
- 3. Barbas A., et al. 2008. New insights into the mechanism of RNA degradation by ribonuclease II: identification of the residue responsible for setting the RNase II end product. J. Biol. Chem. 283:13070–13076 [DOI] [PubMed] [Google Scholar]
- 4. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 5. Cairrao F., Cruz A., Mori H., Arraiano C. M. 2003. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 50:1349–1360 [DOI] [PubMed] [Google Scholar]
- 6. Charpentier X., Faucher S. P., Kalachikov S., Shuman H. A. 2008. Loss of RNase R induces competence development in Legionella pneumophila. J. Bacteriol. 190:8126–8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C., Deutscher M. P. 2005. Elevation of RNase R in response to multiple stress conditions. J. Biol. Chem. 280:34393–34396 [DOI] [PubMed] [Google Scholar]
- 8. Cheng Z. F., Deutscher M. P. 2005. An important role for RNase R in mRNA decay. Mol. Cell 17:313–318 [DOI] [PubMed] [Google Scholar]
- 9. Cheng Z. F., Deutscher M. P. 2002. Purification and characterization of the Escherichia coli exoribonuclease RNase R: comparison with RNase II. J. Biol. Chem. 277:21624–21629 [DOI] [PubMed] [Google Scholar]
- 10. Cheng Z. F., Zuo Y., Li Z., Rudd K. E., Deutscher M. P. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 273:14077–14080 [DOI] [PubMed] [Google Scholar]
- 11. Deutscher M. P., Li Z. 2001. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic. Acids Res. Mol. Biol. 66:67–105 [DOI] [PubMed] [Google Scholar]
- 12. Domingues S., et al. 2009. Biochemical characterization of the RNase II family of exoribonucleases from the human pathogens Salmonella typhimurium and Streptococcus pneumoniae. Biochemistry 48:11848–11857 [DOI] [PubMed] [Google Scholar]
- 13. Erova T. E., et al. 2008. Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J. Bacteriol. 190:3467–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fonseca P., Moreno R., Rojo F. 2008. Genomic analysis of the role of RNase R in the turnover of Pseudomonas putida mRNAs. J. Bacteriol. 190:6258–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frazao C., et al. 2006. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 443:110–114 [DOI] [PubMed] [Google Scholar]
- 16. Ghosh S., Deutscher M. P. 1999. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl. Acad. Sci. U. S. A. 96:4372–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jager S., et al. 2001. An mRNA degrading complex in Rhodobacter capsulatus. Nucleic. Acids Res. 29:4581–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khemici V., Carpousis A. J. 2004. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 51:777–790 [DOI] [PubMed] [Google Scholar]
- 19. Lalonde M. S., et al. 2007. Exoribonuclease R in Mycoplasma genitalium can carry out both RNA processing and degradative functions and is sensitive to RNA ribose methylation. RNA 13:1957–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mathy N., et al. 2007. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129:681–692 [DOI] [PubMed] [Google Scholar]
- 21. Matos R. G., Barbas A., Arraiano C. M. 2009. RNase R mutants elucidate the catalysis of structured RNA: RNA-binding domains select the RNAs targeted for degradation. Biochem. J. 423:291–301 [DOI] [PubMed] [Google Scholar]
- 22. Matos R. G., Barbas A., Gómez-Puertas P., Arraiano C. M. 2011. Swapping the domains of exoribonucleases RNase II and RNase R: conferring upon RNase II the ability to degrade dsRNA. Proteins 79:1853–1867 [DOI] [PubMed] [Google Scholar]
- 23. McLaren R. S., Newbury S. F., Dance G. S., Causton H. C., Higgins C. F. 1991. mRNA degradation by processive 3′-5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J. Mol. Biol. 221:81–95 [PubMed] [Google Scholar]
- 24. Miyoshi A., et al. 2007. The role of the vacB gene in the pathogenesis of Brucella abortus. Microbes Infect. 9:375–381 [DOI] [PubMed] [Google Scholar]
- 25. Purusharth R. I., et al. 2005. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J. Biol. Chem. 280:14572–14578 [DOI] [PubMed] [Google Scholar]
- 26. Purusharth R. I., Madhuri B., Ray M. K. 2007. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3′ end processing of 16 and 5 S rRNA. J. Biol. Chem. 282:16267–16277 [DOI] [PubMed] [Google Scholar]
- 27. Purusharth R. I. 2006. Characterization of the RNA degrading machinery (degradosome) from the Antarctic psychrotrophic bacterium Pseudomonas syringae. Ph.D. thesis Jawaharlal Nehru University, New Delhi, India [Google Scholar]
- 28. Py B., Higgins C. F., Krisch H. M., Carpousis A. J. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169–172 [DOI] [PubMed] [Google Scholar]
- 29. Ray M., et al. 1998. Adaptation to low temperature and regulation of gene expression in Antarctic psychrotrophic bacteria. J. Biosci. 23:423–435 [Google Scholar]
- 30. Regha K., Satapathy A. K., Ray M. K. 2005. RecD plays an essential function during growth at low temperature in the antarctic bacterium Pseudomonas syringae Lz4W. Genetics 170:1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sambrook J., Fritsch E., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Shivaji S., et al. 1989. Isolation and identification of Pseudomonas spp. from Schirmacher Oasis, Antarctica. Appl. Environ. Microbiol. 55:767–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsao M. Y., Lin T. L., Hsieh P. F., Wang J. T. 2009. The 3′-to-5′ exoribonuclease (encoded by HP1248) of Helicobacter pylori regulates motility and apoptosis-inducing genes. J. Bacteriol. 191:2691–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vincent H. A., Deutscher M. P. 2006. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 281:29769–29775 [DOI] [PubMed] [Google Scholar]
- 35. Zuo Y., et al. 2006. Structural basis for processivity and single-strand specificity of RNase II. Mol. Cell 24:149–156 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.