Abstract
Internal egg hatching in Caenorhabditis elegans, “worm bagging,” is induced by exposure to bacteria. This study demonstrates that the determination of worm bagging frequency allows for advanced insight into the degree of bacterial pathogenicity and is highly predictive of the survival of worm populations. Therefore, worm bagging frequency can be regarded as a reliable population-wide stress reporter.
TEXT
Caenorhabditis elegans worms normally lay eggs that hatch outside the parental body. However, eggs can also be retained and hatch inside the parental body, a process called “worm bagging” due to the creation of a bag of worms, or “worm bag” (WB) (22, 26). Internal hatching is modulated by genes (28) and is not restricted to the widely used laboratory strain N2 (19). Internal hatching is rare when worms are maintained under standard laboratory conditions. However, axenic condition (24), a transfer from solid to liquid medium (23), and adverse environmental conditions, such as starvation (9) and exposure to toxic compounds (8, 27) and bacteria (1, 20), can increase the frequency of worm bags. It was proposed that worm bagging may be an adaptive response, as the parental body can provide enough food under starvation conditions for larvae to reach the resistant dauer stage (10, 16). Retention of progeny could also provide physical protection and transport for small larvae, which could be released in more favorable environments.
C. elegans is a widely used model organism for host-pathogen interaction studies (2, 6, 13, 17, 25). The impact of bacteria on worms is evaluated through analysis of life history traits, such as survival. In survival assays, worm bags are generally disregarded or internal hatching is prevented by the use of genetically sterile mutants, by ablation of germ line cells despite the established role of the germ line in both life span and immune response (3), or by chemical sterilization, e.g., 5-fluorodeoxyuridine treatment (11), which can also affect the bacteria and significantly modify host-pathogen interactions.
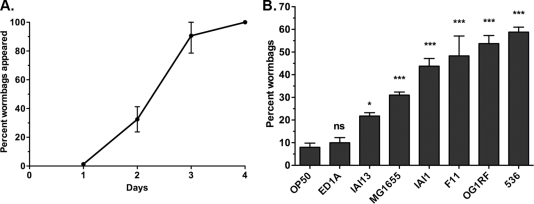
This study investigates (i) how bacteria modulate worm bag frequency, (ii) the correlation of worm bag frequency with life span at the population level, and (iii) the impact of internal hatching on nematode reproductive fitness. The C. elegans N2 (ancestral) strain was exposed to the following Gram-negative Escherichia coli commensal and pathogenic strains: control strain OP50 (7) (sequenced by R. C. May, N. J. Loman, A. S. Haines, M. J. Pallen, C. Boehnisch, C. W. Penn, C. H. Lee, and J. Kim [personal communication]), ED1A (18), IAI13 (21), MG1655 (5), IAI1 (21), F11 (18), and 536 (18). The virulence of these strains was previously established using the C. elegans sterile fer-15 strain (14). The Gram-positive Enterococcus faecalis strain OG1RF was also used (15). All assays were performed at 25°C on monoxenically seeded nematode growth medium agar plates with age-synchronized L4 worms developed on OP50 (4). The proportions of worm bags were determined over the 4-day reproductive period (Fig. 1A). The worm bag frequency varied from 8% on OP50 to 59% on the pathogenic 536 strain (Fig. 1B).
Fig. 1.
Timeline and frequency of bacterium-induced internal egg hatching. (A) Cumulative percentage of worm bags appearing over the course of the C. elegans reproductive period. Data are compiled, with all bacterial strains included, from all experiments of worm bag frequency determination (n = 55). Means ± standard deviations (SDs) are graphed. (B) Frequency of worm bags in C. elegans populations exposed to E. coli strains OP50 (n = 381), ED1A (n = 291), IAI13 (n = 245), MG1655 (n = 582), IAI1 (n = 378), F11 (n = 235), and 536 (n = 905) and to E. faecalis strain OG1RF (n = 313). Data are compiled from at least three independent experiments. Bagging frequencies were analyzed using a one-way analysis of variance (ANOVA), followed by Dunnett's multiple-comparison test against OP50. Means ± standard errors of the mean (SEM) are graphed. ns, not significant; *, P < 0.05; ***, P < 0.0001.
Worm bag frequency can be modulated by food availability (9). However, this does not seem to be the case in this study. The numbers of live cells (mean ± SD) in bacterial lawns were significantly different (unpaired t test, P < 0.0001) between the OP50 (3.83 × 109 ± 8.46 × 108 CFU/lawn) and ED1A (1.15 × 1010 ± 9.56 × 108 CFU/lawn) strains, but the strains induced similar bagging frequencies (Fig. 1B). In contrast, the ED1A and 536 (1.45 × 1010 ± 5.12 × 108 CFU/lawn) strains generated similar numbers of live cells but induced different bagging frequencies (Fig. 1B). Thus, the nature of each bacterial strain, not the quantity of cells, modulates worm bag frequency.
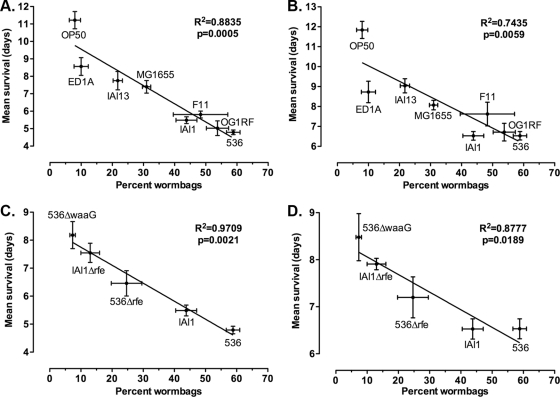
Next, the correlation between bagging frequency and life span was established. Life span was defined as the time from the end of L4 stage until death of the adult worm (4). When worm bags were included in the analysis, the bagging frequency correlated significantly with the mean life span (Fig. 2A) as well as with the median and 25% survival rates (see Fig. S1A and B in the supplemental material). This may be expected, because worm bags have lower average survival rates and their proportion in populations can be very high and increases with bacterial pathogenicity (Fig. 1B). However, the correlations between worm bag frequency and survival remained significant even when worm bags were excluded from the survival analyses (Fig. 2B; see also Fig. S1C and D in the supplemental material). Attenuated lipopolysaccharide-truncated E. coli mutants were constructed by gene replacement through homologous recombination using pKD3 template plasmid (12) and the GTGAATTTACTGACAGTGAGTACTGATCTCATCAGTATTTTGTGTAGGCTGGAGCTGCTTC and CATAGAGGAAGAATGCTAGCAAAAAGAGCACCAGCATGACCATATGAATATCCTCCTTAG primers for the rfe gene. The correlation between worm bag frequency and life span is also significant in worm populations exposed to attenuated strains, independently of whether worm bags are included or excluded (Fig. 2C and D). Therefore, worm bag frequency is highly predictive of the survival of worm populations exposed to different bacterial strains. In addition, worm bag frequency provides advanced insight into the population's life span because 90% of worm bags appear by the third day (Fig. 1A), while complete survival studies can last over 2 weeks.
Fig. 2.
Correlation of worm bag frequency with mean life span in C. elegans. Correlations between worm bag frequency and mean life span were calculated using linear regression to mean survival when worm bags were included in (A and C) or excluded from (B and D) the survival assays. Worms (including and excluding worm bags, respectively [n]) were exposed to E. coli strains OP50 (n = 381 and 344), ED1A (n = 291 and 261), IAI13 (n = 245 and 193), MG1655 (n = 582 and 402), IAI1 (n = 378 and 211), F11 (n = 235 and 120), and 536 (n = 905 and 368) and E. faecalis strain OG1RF (n = 313 and 144) (A and B) and to attenuated lipopolysaccharide-truncated E. coli mutants 536Δrfe (n = 210 and 167), 536ΔwaaG (n = 214 and 198), and IAI1Δrfe (n = 153 and 133) (C and D). Data are compiled from at least three independent experiments. Means ± SEM are graphed.
Internal hatching can result in the early death of individuals, reducing population survival significantly when the worm bag frequency reaches 20% (Fig. 2A to D; see also Fig. S1A to D in the supplemental material). However, it is not necessarily lethal to worms. Some worm bags expulse internally hatched larvae and live as long as some non-worm bag individuals. Because pathogenic bacteria induce high frequencies of worm bags, to disregard or prevent worm bagging when investigating host-pathogen interactions can lead to important underestimations of the bacterial pathogenicity. Exclusion of worm bags from survival assays resulted in overestimations of worm population survival by up to 27%.
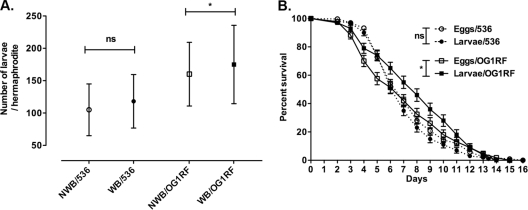
Internal hatching decreases survival, but it also has the potential to reduce reproductive output, i.e., fitness, because retained larvae could damage gonads. Offspring production and survival were quantified on E. coli 536 and E. faecalis OG1RF, the two strains inducing worm bag frequencies more than 50%. Individual nematodes were maintained in 12-well plates and transferred every 24 h into new plate wells. The plates were further incubated for 24 h to allow the eggs to hatch. All internally hatched larvae were released or freed themselves by the end of the reproductive period. Reproductive output was measured as the total number of larvae produced per individual worm (4). Worm bags had as much or more progeny than non-worm bag individuals (Fig. 3A). The impact of internal hatching versus external hatching on the offspring's survival was also evaluated. Internally hatched worms survived as well as or better than externally hatched worms (Fig. 3B). Thus, internal hatching does not negatively impact brood size or offspring survival.
Fig. 3.
Impact of internal hatching on C. elegans fitness. (A) Average brood size per non-worm bag (NWB) individual (open symbols) or worm bag (WB) individual (filled symbols) exposed to E. coli 536 (circles; NWB, n = 127; WB, n = 171) and E. faecalis OG1RF (squares; NWB, n = 61; WB, n = 54). Average brood sizes are as expected at 25°C. Data are compiled from at least two independent experiments. Significance of results was determined by an unpaired t test. ns, not significant; *, P < 0.05. Means ± SDs are graphed. (B) Survival of the internally hatched (larvae; filled symbols) and externally hatched (eggs; open symbols) offspring exposed to E. coli 536 (dashed lines; eggs, n = 328; larvae, n = 189) and E. faecalis OG1RF (solid lines; eggs, n = 127; larvae, n = 122). Data were compiled from at least two independent experiments. Survival assays were analyzed using the Gehan-Breslow-Wilcoxon test by comparing conditions by pairs and allowing for unequal hazard ratios. Means ± SEM are graphed. ns, not significant; *, P < 0.05.
Because worm bagging is induced by exposure to pathogens and because its frequency correlates with the survival of non-worm bag individuals, it cannot be neglected as an irrelevant by-product and can be regarded as a population-wide stress reporter. This study shows that worm bag frequency can be used as a proxy for reliable survival determination and allows for advanced insight into the degree of bacterial pathogenicity.
Supplementary Material
Acknowledgments
We acknowledge J. J. Ewbank (Centre d'Immunologie de Marseille-Luminy, Inserm/CNRS/Université de la Méditerranée, Marseille, France) for providing C. elegans strain N2, U. Dobrindt (Institut für Molekulare Infektionsbiologie, Bayerische Julius-Maximilians-Universität Würzburg, Würzburg, Germany) for providing E. coli bacterial mutant strain 536ΔwaaG, and P. Courvalin (Unité des Agents Antibactériens, Institut Pasteur, Paris, France) for providing bacterial strain E. faecalis OG1RF.
This work was supported in part by a fellowship from the Nestlé Foundation to M.L. and by ANR grant ANR-06-BLAN-0406/AGING to I.M.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Aballay A., Yorgey P., Ausubel F. M. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539–1542 [DOI] [PubMed] [Google Scholar]
- 2. Alegado R. A., Campbell M. C., Chen W. C., Slutz S. S., Tan M. W. 2003. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell. Microbiol. 5:435–444 [DOI] [PubMed] [Google Scholar]
- 3. Alper S., et al. 2010. The Caenorhabditis elegans germ line regulates distinct signaling pathways to control lifespan and innate immunity. J. Biol. Chem. 285:1822–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baeriswyl S., et al. 2010. Modulation of aging profiles in isogenic populations of Caenorhabditis elegans by bacteria causing different extrinsic mortality rates. Biogerontology 11:53–65 [DOI] [PubMed] [Google Scholar]
- 5. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474 [DOI] [PubMed] [Google Scholar]
- 6. Breger J., et al. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calafato S., Swain S., Hughes S., Kille P., Sturzenbaum S. R. 2008. Knock down of Caenorhabditis elegans cutc-1 exacerbates the sensitivity toward high levels of copper. Toxicol. Sci. 106:384–391 [DOI] [PubMed] [Google Scholar]
- 9. Chen J., Caswell-Chen E. P. 2004. Facultative vivipary is a life-history trait in Caenorhabditis elegans. J. Nematol. 36:107–113 [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J., Caswell-Chen E. P. 2003. Why Caenorhabditis elegans adults sacrifice their bodies to progeny. Nematology 5:641–645 [Google Scholar]
- 11. Couillault C., Ewbank J. J. 2002. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect. Immun. 70:4705–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dhakal B. K., et al. 2006. Caenorhabditis elegans as a simple model host for Vibrio vulnificus infection. Biochem. Biophys. Res. Commun. 346:751–757 [DOI] [PubMed] [Google Scholar]
- 14. Diard M., et al. 2007. Caenorhabditis elegans as a simple model to study phenotypic and genetic virulence determinants of extraintestinal pathogenic Escherichia coli. Microbes Infect. 9:214–223 [DOI] [PubMed] [Google Scholar]
- 15. Dunny G. M., Brown B. L., Clewell D. B. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. U. S. A. 75:3479–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fielenbach N., Antebi A. 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22:2149–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garsin D. A., et al. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U. S. A. 98:10892–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson J. R., et al. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141–1150 [DOI] [PubMed] [Google Scholar]
- 19. McCulloch D., Gems D. 2003. Evolution of male longevity bias in nematodes. Aging Cell 2:165–173 [DOI] [PubMed] [Google Scholar]
- 20. O'Quinn A. L., Wiegand E. M., Jeddeloh J. A. 2001. Burkholderia pseudomallei kills the nematode Caenorhabditis elegans using an endotoxin-mediated paralysis. Cell. Microbiol. 3:381–393 [DOI] [PubMed] [Google Scholar]
- 21. Picard B., et al. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R. (ed.). 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Plainview, NY: [PubMed] [Google Scholar]
- 23. Shook D. R., Johnson T. E. 1999. Quantitative trait loci affecting survival and fertility-related traits in Caenorhabditis elegans show genotype-environment interactions, pleiotropy and epistasis. Genetics 153:1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Szewczyk N. J., et al. 2006. Delayed development and lifespan extension as features of metabolic lifestyle alteration in C. elegans under dietary restriction. J. Exp. Biol. 209:4129–4139 [DOI] [PubMed] [Google Scholar]
- 25. Tenor J. L., McCormick B. A., Ausubel F. M., Aballay A. 2004. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr. Biol. 14:1018–1024 [DOI] [PubMed] [Google Scholar]
- 26. Trent C., Tsuing N., Horvitz H. R. 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104:619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Raamsdonk J. M., Hekimi S. 2009. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 5:e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Waggoner L. E., Zhou G. T., Schafer R. W., Schafer W. R. 1998. Control of alternative behavioral states by serotonin in Caenorhabditis elegans. Neuron 21:203–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.