Abstract
The long- and short-term effects of salt on biological nitrogen and phosphorus removal processes were studied in an aerobic granular sludge reactor. The microbial community structure was investigated by PCR-denaturing gradient gel electrophoresis (DGGE) on 16S rRNA and amoA genes. PCR products obtained from genomic DNA and from rRNA after reverse transcription were compared to determine the presence of bacteria as well as the metabolically active fraction of bacteria. Fluorescence in situ hybridization (FISH) was used to validate the PCR-based results and to quantify the dominant bacterial populations. The results demonstrated that ammonium removal efficiency was not affected by salt concentrations up to 33 g/liter NaCl. Conversely, a high accumulation of nitrite was observed above 22 g/liter NaCl, which coincided with the disappearance of Nitrospira sp. Phosphorus removal was severely affected by gradual salt increase. No P release or uptake was observed at steady-state operation at 33 g/liter NaCl, exactly when the polyphosphate-accumulating organisms (PAOs), “Candidatus Accumulibacter phosphatis” bacteria, were no longer detected by PCR-DGGE or FISH. Batch experiments confirmed that P removal still could occur at 30 g/liter NaCl, but the long exposure of the biomass to this salinity level was detrimental for PAOs, which were outcompeted by glycogen-accumulating organisms (GAOs) in the bioreactor. GAOs became the dominant microorganisms at increasing salt concentrations, especially at 33 g/liter NaCl. In the comparative analysis of the diversity (DNA-derived pattern) and the activity (cDNA-derived pattern) of the microbial population, the highly metabolically active microorganisms were observed to be those related to ammonia (Nitrosomonas sp.) and phosphate removal (“Candidatus Accumulibacter”).
INTRODUCTION
Numerous wastewaters, like those generated in seafood canning, pickling, and cheese-processing industries, can contain significant amounts of inorganic dissolved salts (14). The use of seawater for toilet flushing in ships, offshore installations, and regions with problems related to water supply also introduces salts into freshwater. Wastewaters with a high salt content require the activity of salt-tolerant microorganisms which may not be present in high numbers in microbial inocula from sewage treatment plants. Salt ions exert high osmotic pressure on the microorganisms, and most of the freshwater-based microbial populations are unable to survive at these high osmotic pressures and either die or become dormant under these conditions (40). Besides that, inorganic salts can affect the structure and settling properties of microbial flocs as well as the maximum solubility of oxygen and its transfer to the liquid phase, which can lead to oxygen limitation. Consequently, high salt concentrations have a negative influence on existing biological wastewater treatment plants, affecting organic matter, nitrogen, and phosphorus removal (17). So far, most studies on chemical oxygen demand (COD), N, and P removal from saline wastewaters have shown that inhibition takes places when salt concentrations exceed 1% (wt/vol) (50, 63, 68).
Currently, there is an increasing interest in the use of aerobic granular sludge (AGS) in wastewater treatment. The AGS process has numerous advantages compared to the activated sludge process, such as excellent settling properties, good biomass retention, strong compact structure, and the capability to withstand shocks of both organic and toxic compounds (6). Moreover, the granulation process takes place without reliance on surfaces for biofilm growth, which makes artificial carriers unnecessary. Developed in the last decade (29), an impressive number of studies have been performed to explore the potentials of the AGS technology in the treatment of high-strength organic wastewaters (10, 45, 61), toxic aromatic pollutants (32, 35, 73), heavy metals (38, 70, 71), and textile dyes (62). Additionally, AGS has been extensively applied for nutrient removal (19, 20, 43, 55). Only a few studies can be found in the literature related to the effect of salt on aerobic granules. Figueroa et al. (25) investigated the treatment of fish-caning effluents containing high salt concentrations (up to 30 g/liter NaCl) in an aerobic granular sludge sequencing batch reactor and observed that AGS biomass could withstand the saline conditions. Once the aerobic granules were formed, the presence of salt did not cause a detrimental effect on the operation of the reactor. Based on thermogravimetric analysis, Li and Wang (34) investigated the content of both inorganic and organic components in aerobic granule structures at low (1%, wt/vol) and high (5%, wt/vol) salinity levels. These authors observed that the surface of the granules was more regular and smoother, and granules were found to grow faster and larger at high salinity levels. Also, the porosity of aerobic granules was found to be reduced at salt concentrations as high as 5%.
However, none of the aforementioned studies performed a detailed study on the dynamics of the microbial community structure in the granules. A comprehensive evaluation of the microbiology and ecology of aerobic granules at increasing salt concentrations has not been conducted to date. The major aim of our study was to evaluate the effect of increasing salt concentrations on the main biological processes taking place in an aerobic granular sludge reactor and relate those observations to the dynamics of microbial populations. For this purpose, we used PCR-denaturing gradient gel electrophoresis (DGGE) on 16S rRNA and amoA genes. We compared PCR products obtained from genomic DNA and from rRNA after reverse transcription to identify bacteria present and metabolically active. Moreover, we used fluorescence in situ hybridization (FISH) with specific probes for the 16S rRNA of specific subpopulations to validate the PCR-based results and to quantify the dominant bacterial populations. This is one of the first molecular ecology-based characterizations of metabolically active microorganisms of a simultaneous nitrogen and phosphorus removal process.
MATERIALS AND METHODS
Reactor setup and operating conditions.
A sequencing batch reactor with a working volume of 2.7 liters, an internal diameter of 5.6 cm, and a total height of 90 cm was used in our experiments. The reactor was inoculated with aerobic granular biomass from a Nereda pilot reactor in the Epe wastewater treatment plant in The Netherlands. Air was introduced with a fine-bubble aerator located at the bottom of the reactor (4 liters/min). The dissolved-oxygen (DO) concentration and the pH were measured continuously. DO was kept at more than 90% air saturation, and the pH was controlled at 7.0 ± 0.2 by dosing with 1 M NaOH or 1 M HCl. The temperature was maintained at 20°C. To achieve nitrogen and phosphate removal, the reactor was operated in successive cycles of 3 h under alternating anaerobic and aerobic conditions comprising four phases: 60-min anaerobic feeding from the bottom of the reactor in a plug-flow regimen through the settled bed, 112-min aeration, 3-min settling, and 5-min effluent withdraw. Effluent was discharged from a port close to the middle of the reactor, and the volume exchange ratio was around 56%. The hydraulic retention time (HRT) was 5.3 h. The synthetic wastewater had the following composition: solution A, sodium acetate (NaAc) at 63 mM, MgSO4 · 7H2O at 3.6 mM, and KCl at 4.7 mM; solution B, NH4Cl at 35.4 mM, K2HPO4 at 4.2 mM, KH2PO4 at 2.1 mM, and 10 ml/liter trace element solution (64). One hundred fifty ml per cycle was dosed from both media together with 1,200 ml of tap water. The system was operated for 449 days and its operation was divided in four phases, which represented different saline conditions (Table 1). Sludge retention time (SRT) was maintained at approximately 30 days by periodically removing sludge from the reactor.
Table 1.
Operational phases according to salt concentration
| Phase | NaCl concn (g/liter) | Time of operation (days) |
|---|---|---|
| I | 0 | 15 |
| II | 11 | 317 |
| III | 22 | 71 |
| IV | 33 | 46 |
Batch experiments.
Several batch experiments were performed to access the short-term effect of salt on nitrogen and phosphorus removal. For this set of experiments, granules were taken from the reactor in the last day of operation of each operational phase (Table 1 shows the duration of each phase). Biomass was taken from the reactor immediately after the feeding phase. An incubation step with acetate under anaerobic conditions (established by sparging nitrogen into the solution) was performed to enhance the release of residual phosphate potentially present inside the cells and the accumulation of an extra polyhydroxyalkanoates by polyphosphate-accumulating organisms (PAOs). The granules then were placed in a sieve and washed with tap water. Equal amounts of granules (based on the wet weight) were introduced in different 250-ml flasks filled with a Tris-HCl buffer (pH 7.0) containing the same minerals of the synthetic media fed to the reactor (except acetate) and different NaCl concentrations (from 0 to 40 g/liter). The flasks were aerated through air diffusers, and the batch experiments lasted for 2 h. Samples were collected every 5 to 20 min, and only liquid sample was removed from each flask in the defined intervals of time, meaning that the amount of biomass per volume of liquid increased during the experiment. To avoid problems related to volume correction for the calculation of biomass concentration, the determination of biomass-specific rates did not rely on the concentration of biomass but on the total amount of biomass, which was constant during the batch tests. Similarly, instead of concentrations, the total amounts (in mg) of PO43−-P and NH4+-N present at certain times of the experiment were considered, which were obtained by summing the amount of PO43−-P and NH4+-N remaining in the flask and the amount removed by sampling. Biomass-specific rates then could be obtained by dividing the total amount of PO43−-P and NH4+-N at a specific time by the constant amount of volatile suspended solids (VSS).
Analytical measurements.
Ammonium nitrogen (NH4+-N), nitrate nitrogen (NO3−-N), and nitrite nitrogen (NO2−-N) were measured through a flow injection analyzer (QuikChem 8500; Lachat Instruments, Inc.). Phosphate (PO43−-P) was quantified by Hach Lange cuvette tests (LCK 350). Biomass concentrations were determined according to standard methods (3).
Nucleic acid extraction.
Samples for genomic DNA and RNA extraction were collected from the reactor in different weeks of operation from phases II to IV. No sample was collected during phase I. In total, five or three samples of each experimental phase were taken into consideration for further DNA- and RNA-based analysis, respectively. Biomass was centrifuged, and the same amount of biomass was introduced in the DNA/RNA extraction kit tubes. DNA and RNA extraction were performed, respectively, by using the UltraClean Microbial DNA isolation kit and UltraClean Microbial RNA isolation (MO BIO, Carlsbad, CA) by following the manufacturer's instructions. The extracted DNA and RNA products were evaluated on 1% (wt/vol) agarose gel and stored at −20°C until further use. About 10 ng of the extracted DNA was used as the template for the PCR in which specific primers for either the bacterial 16S RNA gene or amoA gene (encoding the site of ammonia monooxygenase of ammonia-oxidizing bacteria) were used.
Reverse transcription of RNA and PCR amplification.
The reverse transcription of isolated RNA into cDNA was performed using the iScript cDNA synthesis kit (Bio-Rad, CA) according to the manufacturer's protocol. The reverse transcription procedure was carried out for two samples of each experimental phase (except phase I). One μl (80 to 100 ng) of the RNA template was used in this step. The set of primers used for 16S rRNA gene amplification was BAC341f (containing a 40-bp GC clamp) and BAC907rM (where M is A/C) (58). Both genomic DNA and cDNA (0.5 μl each) were used as templates for the amplification reactions. The PCR program for the 16S rRNA gene included a precooling phase of the thermocycler at 4°C for 1.5 min and initial denaturation at 95°C for 5 min, followed by 32 cycles of denaturing at 95°C for 30 s, annealing at 57°C for 40 s, and elongation at 72°C for 40 s. After the last cycle, a final elongation at 72°C for 30 min took place, and the amplification ended at 12°C. For the amoA gene amplification, the primers used were amoA-1F-GC and amoA-2R (31). The cycling regimen for the amplification of the amoA gene was 4 min at 94°C (initial denaturation) and then 35 cycles consisting of 30 s at 94°C (denaturation), 40 s at 60°C (annealing), and 40 s at 72°C (elongation), with a final extension for 30 min at 72°C. The quality of the PCR products of both genes (16S rRNA and amoA) was evaluated on a 1% (wt/vol) agarose gel.
DGGE.
Denaturing gradient gel electrophoresis (DGGE) was performed using the Bio-Rad DCode system (Bio-Rad, Richmond, CA). Electrophoresis was run in 1-mm-thick gels containing either 6% polyacrylamide (for 16S RNA PCR products) or 8% polyacrylamide (for amoA gene PCR products). The gels were prepared with denaturing gradients ranging from 20 to 70% for the 16S RNA fragments and from 10 to 50% for the amoA fragments (100% is defined as 7 M urea and 40% [vol/vol] deionized formamide), and they were submerged in 1× TAE buffer (40 mM Tris, 40 mM acetic acid, 1 mM EDTA, pH 7.4) for 16S RNA fragments or 0.5× TAE buffer for amoA fragments. Approximately 250 ng of the GC-clamped PCR products was added to individual lanes on the gel. The electrophoresis of 16S rRNA PCR products was run for 16 h at a constant voltage of 100 V and a temperature of 60°C. For the amoA gene PCR products, the electrophoresis lasted for 5 h at 200 V and a temperature of 55°C. After electrophoresis, the gels were stained for about 30 min with a 5 ml 1× TAE solution containing Sybr green nucleic acid stain (Molecular Probes, Eugene, OR) in the dark and visualized in a Safe Imager Blue-Light transilluminator (Invitrogen, Carlsbad, CA). Images were acquired with a GeneSnap system (Syngene, Cambridge, United Kingdom).
Band isolation, DNA sequencing, and phylogenetic analysis.
To obtain a substantially pure PCR product for DNA sequencing, individual bands from DGGE gels of both 16S RNA and amoA genes were carefully excised using sterile razor blades, placed in 1.5-ml microcentrifuge tubes containing 40 μl of 1× Tris-HCl buffer, and stored for 48 h at 4°C. A volume of 1 μl of the DNA eluted from the DGGE band was used for reamplification with non-GC-clamped primers by following the same PCR programs described above. DNA sequencing analysis was carried out by a commercial company (Macrogen, South Korea). The obtained 16S rRNA and amoA genes sequences were compared to those stored in GenBank using the basic local alignment search tool (BLAST) algorithm (http://www.ncbi.nlm.nih.gov/blast). Subsequently, they were imported into the ARB software (http://www.arb-home.de) and aligned by using the ARB automatic aligner. The alignment was further verified and corrected manually. A phylogenetic tree was generated by performing neighbor-joining algorithms.
FISH.
Granule samples collected from the reactor during phases II to IV for FISH analysis were crushed, washed with 1× phosphate-buffered saline (PBS), and immediately fixed with 4% (wt/vol) paraformaldehyde in PBS solution for 3 h at 4°C. After fixation, cells were centrifuged at 13,000 × g for 1 min, washed twice in 1× PBS, and resuspended in an ethanol-PBS solution (1:1) for storage at −20°C. In the hybridization step, fixed samples were spread on gelatin-coated microscope slides and were placed in the oven at 46°C for drying. The coverslips containing the dried cells then were dehydrated in three steps (3 min for each step) with 50, 80, and 96% (vol/vol) ethanol. After dehydration, 10 μl of a hybridization buffer solution containing 0.9 M NaCl, 0.02 M Tris-HCl, 35% (vol/vol) formamide for all probes and 0.02% (wt/vol) SDS and including fluorescently labeled oligonucleotide probes (0.5 pmol for Cy3/Cy5-labeled and 0.83 pmol for fluorescein-labeled probes) were added to the cells. The hybridization was carried out in a humid chamber for at least 1.5 h of incubation at 46°C. A subsequent washing step to remove unbound oligonucleotides was performed by immersing the gelatin-coated slides in a buffer containing 20 mM Tris-HCl (pH 8), 0.01% (wt/vol) sodium dodecyl sulfate, 0.08 mM NaCl, and 0.005 mM EDTA for 10 min at 48°C. The wells of the slides were rinsed with Milli-Q water, dried by compressed air, and embedded in 2 μl of Vectashield H-1000 mounting oil for fluorescence (Vector Laboratories, Burlingame, CA). Slides were observed with an epifluorescence microscope (Axioplan 2; Zeiss), and image acquisition was performed with a Leica D350F camera. The hybridization experiments were performed using different fluorochromes for each probe to validate the results. The images were exported in .jpg format from the Zeiss microscopy imaging software (AxioVision version 4.7). The quantification of probe-targeted bacterial cells labeled with different dyes (Cy3 and Fluos) and the determination of the fraction of positive signal from each probe relative to the signal visualized with general bacterial probes (Cy5) was performed with quantitative imaging software (Leica QWin). At least 10 recorded images from the samples representing an experimental run of the reactor were taken into account for the quantification analysis. The rRNA-targeted oligonucleotide probes, labeled with three different fluorescent dyes (Cy3, Fluos, and Cy5), are listed in Table 2. According to Oehmen et al. (47), the PAOmix combination (PAO462, PAO651, and PAO846) is the most sensitive and specific for “Candidatus Accumulibacter” from the currently available probes for these bacteria. To differentiate clades of PAOs (clade I and clade II, capable and not capable of reducing nitrate, respectively), specific probes developed by Flowers et al. (26) were used. GAO phenotype bacteria also were targeted by combinations of the probes GAOQ431 and GAOQ989 and designated GAOmix.
Table 2.
Oligonucleotide probes and their targeted microbial groups
| Probe | Sequence (5′–3′) | Target group | Mix | Reference |
|---|---|---|---|---|
| PAO 462 | CCGTCATCTACWCAGGGTATTAAC | PAO clustera | PAOmix | 11 |
| PAO 651 | CCCTCTGCCAAACTCCAG | PAO clustera | PAOmix | 11 |
| PAO 846 | GTTAGCTACGGCACTAAAAGG | PAO clustera | PAOmix | 11 |
| Acc-I-444 | CCCAAGCAATTTCTTCCCC | Clade IAb | PAOmix | 26 |
| Acc-II-444 | CCCGTGCAATTTCTTCCCC | Clade IIAc | PAOmix | 26 |
| GAO Q431 | TCCCCGCCTAAAGGGCTT | “Candidatus Competibacter phosphates” | GAOmix | 10 |
| GAO Q989 | TTCCCCGGATGTCAAGGC | “Candidatus Competibacter phosphatis” | GAOmix | 10 |
| Nso 190 | CGATCCCCTGCTTTTCTCC | Ammonia-oxidizers β-proteobacteria | 42 | |
| Ntspa 662 | GGA ATT CCG CGC TCC TCT | Genus Nitrospira | 16 | |
| Nit 1035 | CCT GTG CTC CAT GCT CCG | Nitrobacter spp. | 65 | |
| Neu 653 | CCC CTC TGC TGC ACT CTA | Most halophilic and halotolerant Nitrosomonas spp. | 66 | |
| Nso 1225 | CGC CAT TGT ATT ACG TGT GA | Nitrosomonas | 42 | |
| EUB 338 I | GCTGCCTCCCGTAGGAGT | Most bacteria | EUBmix | 1 |
| EUB 338 II | GCAGCCACCCGTAGGTGT | Planctomycetes | EUBmix | 15 |
| EUB 338 III | GCTGCCACCCGTAGGTGT | Verrucomicrobiales | EUBmix | 15 |
Nucleotide sequence accession numbers.
The 16S rRNA sequences determined in the course of this work were deposited in GenBank under the following accession numbers: JF276761 to JF276773.
RESULTS
Long-term and short-term effect of salt on nitrification/denitrification and phosphate removal.
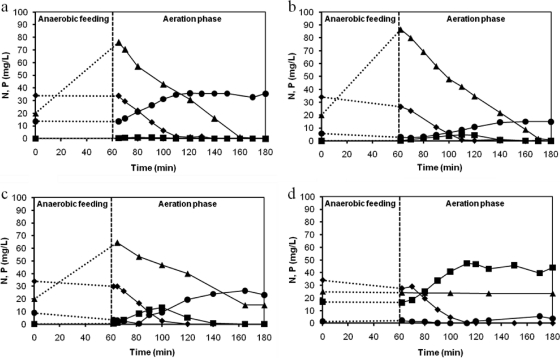
Figure 1 shows the ammonia, nitrate, and nitrite concentration profiles obtained in the cycle measurements performed at the end of phases I to IV, when a pseudo-steady-state condition was achieved. The first measured sample was taken 2 min after the aeration phase had started (for mixing purposes). As can be seen, the ammonium concentration after anaerobic feeding was lower than expected based on the influent concentration and the dilution in the reactor. This fact is due to ammonium adsorption, a phenomenon that was investigated in a separate study (4). Moreover, the nitrite and nitrate concentrations depicted at time zero were calculated based on their concentrations at the end of the cycle and the dilution in the reactor. Due to the plug-flow feeding regimen of the reactor, both nitrite and nitrate were pushed up during the feeding phase, avoiding the growth of ordinary heterotrophs which will use the influent COD to denitrify these nitrogen compounds. The ammonium removal efficiency was not affected by salt in the tested concentrations (up to 33 g/liter NaCl). Ammonia was depleted within 60 min of aeration at all salt concentrations. Nitrate was the main product of nitrification during phases I to III. During phase III, intermediate nitrite accumulated but subsequently was either oxidized to nitrate by nitrite-oxidizing bacteria (NOB) or denitrified to N2 by denitrifying polyphosphate-accumulating organisms (DPAOs) or glycogen-accumulating organisms (DGAOs). During operation at 33 g/liter NaCl (phase IV), a significant amount of nitrite (up to 20 mg/liter N) accumulated, which indicated that NOB were more sensitive than ammonia-oxidizing bacteria (AOB) to the highest salinity level tested. In contrast to nitrification, phosphate removal was severely affected by an increased salt concentration. Both phosphate release during anaerobic feeding and phosphate uptake in the subsequent aeration phase decreased with the increase in NaCl concentration from 11 to 22 g/liter. At 11 and 22 g/liter NaCl, a decrease in the P uptake rate can clearly be noticed during the course of an operational cycle, exactly during the period when high nitrite concentrations accumulated, as shown in Fig. 1b and c. Furthermore, when nitrite concentrations started to decrease, the P uptake increased again. The P release and uptake stopped completely when a pseudo-steady-state operation was reached at 33 g/liter NaCl. In this operational phase, nitrite accumulated and was not further nitrified by NOB. Furthermore, denitrification by denitrifying GAOs (PAOs were not present anymore) was almost negligible.
Fig. 1.
Ammonium (♦), nitrate (•), nitrite (■), and phosphate (▴) profiles acquired in the cycle measurements performed at the end of phase I (no salt) (a), phase II (11 g/liter NaCl) (b), phase III (22 g/liter NaCl) (c), and phase IV (33 g/liter NaCl) (d). The starting ammonium and phosphate concentrations depicted at time zero were calculated based on the influent concentration (60 mg/liter NH4+-N and 20 mg/liter PO43−-P) and the dilution in the reactor. Nitrite and nitrate concentrations at time zero were calculated based on their concentrations at the end of the cycle and the dilution in the reactor.
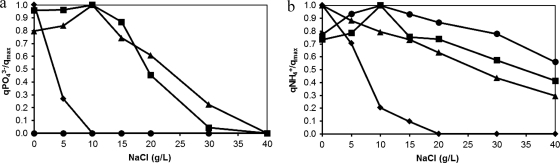
Batch experiments were conducted to evaluate the short-term effect of salt on the nitrification/denitrification and phosphate removal potential of the biomass at different salt concentrations (0, 5, 10, 15, 20, 30, and 40 g/liter NaCl). Specific ammonium and phosphate uptake rates obtained for each salt concentration tested were expressed as a fraction of the maximum value obtained (Fig. 2). In the test using biomass collected at the end of phase I not acclimated to salt (0 g/liter NaCl), maximum P uptake was obtained when no extra salt was present (Fig. 2a). Even at low salt concentrations (5 g/liter NaCl), the specific P uptake rate was only around 25% of the maximum rate. From 10 to 40 g/liter NaCl, no P uptake was observed. On the contrary, the additional release of phosphate was observed. For the biomass collected at the end of phase II and phase III (adapted to 11 and 22 g/liter NaCl, respectively), maximum P uptake was observed at 10 g/liter of salt. The specific PO43−-P uptake rate remained practically constant from 0 to 10 g/liter NaCl in the test with biomass acclimated to 11 g/liter. At higher salt concentrations (30 g/liter), the P uptake decreased to almost zero. For the biomass acclimated to 22 g/liter of NaCl, specific rates obtained at 0, 5, and 15 g/liter were roughly the same, and a significant decrease was observed from 15 to 40 g/liter NaCl. In the test with biomass collected at the end of phase IV (adapted to 33 g/liter), no phosphate uptake was observed at all salt concentrations and minor P release was observed.
Fig. 2.
Short-term effect of salt on (a) phosphate- and (b) ammonium-specific uptake rates. Biomass was adapted to 0 g/liter NaCl (♦), 11 g/liter NaCl (■), 22 g/liter NaCl (▴), and 33 g/liter NaCl (•).
The specific ammonium uptake rate was substantially reduced at salt concentrations exceeding 10 g/liter. This is especially the case in the test with non-salt-adapted biomass, in which no ammonium uptake was observed at salt concentrations exceeding 20 g/liter NaCl (Fig. 2b). Maximum specific ammonium uptake rates were observed at an NaCl concentration of 10 g/liter for the tests using biomass acclimated to 11 and 33 g/liter NaCl. For the test where biomass adapted to 22 g/liter was used, maximum ammonium uptake rate was obtained at 0 g/liter NaCl.
Nitrate and nitrite measurements also were performed for all batch experiments. Nitrate was the main nitrification product at all salt concentrations for the tests using biomass adapted to 0 and 11 g/liter of salt, while nitrite concentrations always were lower than 2 mg/liter N. For the test using granules acclimated to 22 and 33 g/liter, nitrite was the main product of nitrification, reaching concentrations of up to 9 mg/liter N (for biomass adapted to 22 g/liter NaCl) and 20 mg/liter N (for biomass adapted to 33 g/liter NaCl).
According to the nitrogen balance performed taking into account the nitrogen compounds measured (NH4+-N, NO3−-N, and NO2−-N), the highest denitrification activity was observed in the experiment using biomass acclimated to 11 g/liter NaCl, when nitrogen removal ranged between 70 and 85% for all salt concentrations tested. In the experiment using granules adapted to 22 and 33 g/liter NaCl, the maximum N removal obtained was around 45 and 20%, respectively.
Microbial community analysis by DGGE of 16S rRNA gene fragments.
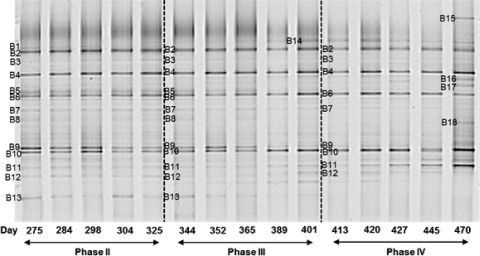
The microbial community of the aerobic granular sludge reactor was investigated by DGGE of the 16S rRNA gene fragments. Figure 3 shows the DGGE banding patterns representing the general bacterial community of the experimental phases II to IV. DNA extraction of the biomass started to be performed after 275 days of operation, when the reactor had already been run at 11 g/liter NaCl for a long time. No biomass was collected during phase I when no NaCl was added to the culture medium fed to the reactor.
Fig. 3.
Bacterial DGGE results from phases II (11 g/liter NaCl), III (22 g/liter NaCl), and IV (33 g/liter NaCl). Five representative samples from phases II to IV were taken into account. The operational days can be seen just below gel lanes. Most of the bands indicated by a number were sequenced successfully and used for phylogenetic analysis. Some of them (particularly bands 1, 2, 3, and 16) were not taken into account due to the unsatisfactory sequencing results.
The DGGE profile indicates a greatly diverse bacterial population, with some bands present in all of the experimental phases, albeit at different intensities. Most of the dominant bands appeared across the whole experimental period, while minor variations could be observed. The DNA-derived pattern of the samples taken from the reactor during all three phases resulted in 18 bands. Some bands were excised from different lanes, and sequencing results showed that their nucleotide sequences were the same. Most of the bands enumerated were sequenced successfully and used for phylogenetic analysis. Conversely, some of them (particularly bands B1, B2, B3, and B16) gave ambiguous sequences and were not included in the analysis. Some bands, such as B1, B2, and B3, were even excised from three different gels loaded with the same samples and sequenced, but none of the sequence results showed a pure sequence and therefore had to be neglected.
The banding pattern of phase II resulted in approximately 13 bands (named B1 to B13). These bands remained until the end of this experimental phase, meaning that no variation in the microbial composition occurred within this period. As can be seen from the phylogenetic tree (Fig. 4), some interesting bacteria were present which probably are directly linked to the main biological conversions taking place in the reactor. Band B9 was closely related to “Candidatus Accumulibacter,” which is commonly considered the main microorganism involved in the enhanced biological phosphorus removal (EBPR) process. Band B8, which showed high sequence similarity to Nitrosomonas sp., and band B13, closely affiliated with Nitrospira sp., are representatives of the AOB and NOB populations, respectively. Bands B10 and B11 clustered well with the Denitromonas group. Band B5, B6, and B7 were closely related to uncultured bacteria identified in biological phosphorus removal systems belonging to the Bacteroidetes phylum. Actually, bands B5 and B6 showed similar mobility in the DGGE gel and gave identical sequences. Therefore, only one was used for the phylogenetic analysis. Band B4 belonged to the Cytophagaceae family, and band B12 was closely related to Dokdonella soli (gammaproteobacteria).
Fig. 4.
Phylogenetic analysis of the bacterial 16S rRNA sequences excised from the DGGE gel. Sequences determined in this study are printed in boldface. The bar indicates 10% sequence difference. The sequence of Nitrosopumilus maritimus (Archaea) was used as an outgroup but was pruned from the tree.
In phase III, when the salt concentration was increased to 22 g/liter, bands B5, B7, and B9 tended to disappear, showing weaker intensities compared to those in phase II. A new band (B14), representing an uncultured bacterium belonging to the family Flavobacteriaceae and detected in a saline environment (shoreline), started to appear in the beginning of phase III, probably due to the affinity for the increased salt concentration.
When the salt concentration was increased from 22 to 33 g/liter (transition of phase III to phase IV), bands B8 and B13 completely disappeared. As observed in phase III, the intensity of band B9 continued decreasing, being present just in the beginning of phase IV. The intensity of band B11 was even increased during long-term operation at 33 g/liter. Curiously, the last sample shown in the gel, collected from the reactor during the end of phase IV (day 470 of operation), showed new bands (B15, B16, B17, and B18). As mentioned before, band 16 did not show satisfactory sequence results. Band B17 is closely related to Denitromonas sp., similarly to band B11. Band B18 (Denitromonas group) also was closely related to an uncultured bacterium isolated from a denitrifying culture, and band B15 represented an uncultured bacterium belonging to the Cryomorpaceae family, which was isolated from surface seawater.
It should be noted that in two previous DGGE gels where just samples from the phases II and III were loaded, we observed other bands that did not appear in the gel shown in Fig. 3. All of these extra bands retrieved in preliminary DGGE analysis represented the GAO population and appeared in the down part of the DGGE gel, meaning that these bacteria had a lower GC content than the others. Therefore, it is possible that the bands representing the GAO population have run out of the gel depicted in Fig. 3. However, even though they were not retrieved in the DGGE gel displayed in Fig. 3, the bands retrieved in previous DGGE analyses representing the GAO population were taken into account in the phylogenetic analysis.
Comparative DGGE analysis of DNA and RNA.
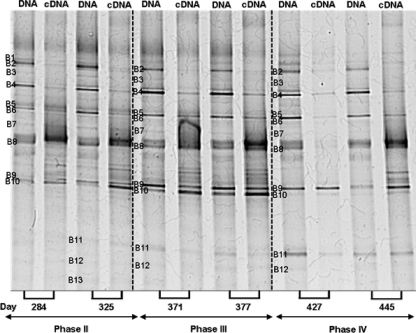
Since the community structure remained rather stable over time, and taking into account that some bacteria could be present in the granule structure even when they are not active anymore due to the long solid retention time, an analysis to compare the diversity and the activity of the microbial community was performed. The diversity is related to the presence of bacterial populations that are above the detection limit of DGGE and can be inferred by the number of DNA-derived bands in a DGGE gel. Conversely, the activity is assumed to be related to the RNA-derived bands, which reflect the predominant active populations (18). Therefore, an RNA analysis was performed to verify which bacterial groups are more metabolically active in the aerobic granules. DGGE profiles obtained with both PCR-amplified bacterial 16S rRNA gene and reverse-transcribed PCR amplified 16 rRNA (cDNA) were compared (Fig. 5).
Fig. 5.
DGGE showing the DNA- and RNA (cDNA)-derived bands for each sample. Two representative samples from phases II to IV were taken into account. Bands with the same number retrieved from different lanes gave identical sequence results.
DGGE fingerprints of the PCR-amplified and reverse-transcribed PCR-amplified 16S rRNA genes were similar. Most of the bands in the DGGE profile from DNA-derived PCR products also were detected in the profiles from RNA-derived PCR products (i.e., cDNA), although their intensities differ in most cases. In this analysis, two representative samples from phases II to IV were taken into account. The respective operational days of each sample are indicated below the gel lanes. Bands were numbered from B1 to B13 in the same way as in the previous DGGE gel (Fig. 3). In accordance with findings for that gel, sequencing results showed that the microorganisms found in each band were the same. The only exceptions were bands B9 and B10, which switched position according to the sequence determined with the previous DGGE gel. Moreover, the same band excised from different lanes showed identical sequence results.
The intensity of several bands (B1, B2, B3, B4, and B6) decreased from the DNA to the cDNA profile. On the contrary, the intensity of band B8 (closely related to Nitrosomonas sp.) increased from the DNA to cDNA banding pattern. This result suggests that Nitrosomonas sp. showed high metabolic activity. The intensity of the band referred to “Candidatus Accumulibacter” (retrieved in band B10 but not in band B9, as in the previous DGGE gel) also was higher in the cDNA than the DNA banding pattern, particularly for phases II and III. During phase IV, band B10 was almost imperceptible. Bands B11 to B13 did not show expression results in the DNA/cDNA DGGE gel.
Ammonia-oxidizing bacterial community structure.
To get further insight into the AOB community in the aerobic granular sludge reactor exposed to increased salt levels, PCR-DGGE analysis using amoA-specific primers was performed. The DGGE banding pattern showed only one dominant band in all of the experimental phases (results not shown). The sequence results indicated that this band showed high sequence similarity (more than 99%) with Nitrosomonas sp., which is in accordance with the DGGE analysis using 16S rRNA-specific primers. All minor bands excised from the gel gave the same sequence result.
FISH analysis.
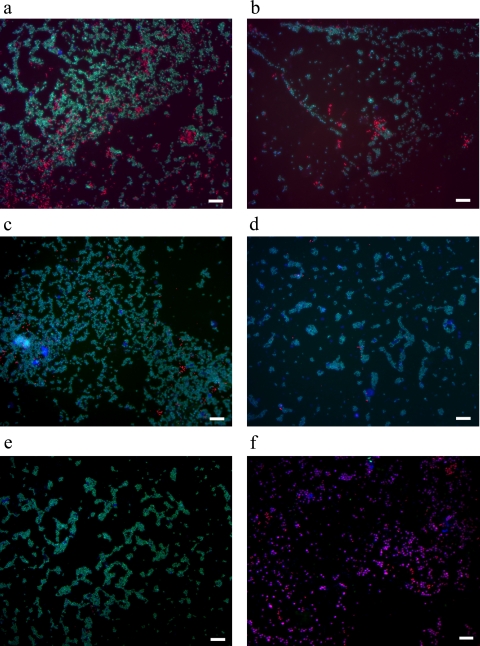
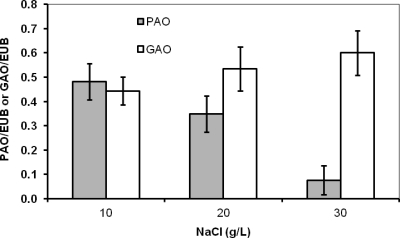
Several specific oligonucleotide probes (Table 2) were used for FISH analysis to complement the PCR-DGGE results and to quantify the different populations among phases II to IV. Results obtained with FISH analysis indicated that almost all bacteria present in the reactor in all operational phases belonged to either the PAO or GAO group (Fig. 6). From phases II to IV, the fraction of PAO in the whole bacterial community decreased significantly (Fig. 6a to e). The quantification analysis (Fig. 7) of the probe-targeted PAO cells (Cy3; in red) and GAO cells (Fluos; in green) and the determination of the fraction of positive signal from each probe relative to the signal visualized with general bacteria probes (Cy5; in blue) revealed that the proportions of PAO and GAO in the whole microbial community in phase II were quite similar. In phase III, the percentage of PAOs decreased while that of the GAOs increased. One week after phase IV was started, the fraction of GAO became even more dominant (Fig. 6c). Just a few cells of PAOs were present after the second week (Fig. 6d), and no positive signal was found for these organisms at the end of the third week of phase IV (Fig. 6e). Nitrifying bacterial cells (AOB and NOB) were detected in substantially lower numbers than in the PAO-plus-GAO population (Fig. 6f). FISH analysis also showed that almost all PAOs belonged to clade I (PAOI), which is capable of using nitrate as an electron acceptor for anoxic P uptake.
Fig. 6.
Fluorescent in situ analysis of the PAO/GAO populations among all bacteria present in the reactor at the end of phase II (a), end of phase III (b), first week of phase IV (c), second week of phase IV (d), and third week of phase IV (e) using combinations of specific probes for PAO (PAO462, PAO462, and PAO846; shown in red), GAO (GAOQ431 and GAOQ989; shown in green), and general bacteria EUB338 (EUB338I, EUB338II, and EUB338III; shown in blue). (f) Fractions of populations using probe combinations of PAO+GAO (using PAOmix/GAOmix probes; shown in red) and AOB+NOB (using combination of probes Nso1225/Nso190/Neu653/Ntspa662/Nit1035; shown in green) within the whole bacterial community (EUB338 mix) in the granules during phase II. The scale bar indicates 20 μm.
Fig. 7.
Proportion of PAOs and GAOs in the whole microbial community in phases II, III, and IV. For the quantification analysis, data were averaged from at least 10 pictures as described in Materials and Methods.
DISCUSSION
Effect of increasing NaCl concentrations on PAO-GAO competition.
Our results demonstrated that phosphate removal was severely affected by increasing salt concentrations, particularly at 22 and 33 g/liter NaCl. Both phosphate release during anaerobic feeding and phosphate uptake in the aeration phase were observed to decrease gradually during phases II to IV. P release has been reported to significantly drop when salinity was increased, especially above 5 g/liter NaCl (13). Phosphorus removal was completely inhibited when the chloride concentration was higher than 2.5 g/liter Cl− (∼4 g/liter NaCl) in an experiment performed by Hong et al. (30) using an anaerobic/anoxic/oxic reactor. The effect of salt on phosphorus removal also was reported by Uygur and Kargi (63), who described a decrease in P removal from 84 to 22% when the NaCl concentration was increased from 0 to 6% (wt/vol).
Our results from the reactor cycle measurements suggested that the considerable amount of nitrite accumulated during the operational cycle at salt concentrations equal to or greater than 11 g/liter NaCl contributed to the reduction of phosphorous removal capability. As reported in the literature, nitrite can negatively affect the P uptake activity of PAOs under both anoxic and aerobic conditions. Concentrations of up to 2 mg/liter NO2−-N are already inhibitory for PAOs according to Saito et al. (56), who reported the complete inhibition of aerobic P uptake at nitrite concentrations above 6 mg/liter N. A severe inhibition of both anoxic and aerobic P uptake at 6 to 8 mg/liter NO2− also was reported by Meinhold et al. (41). In our work, the effect of salt was detrimental to NOB, which was reflected in the accumulation of nitrite. As a cascade effect, phosphate uptake was reduced when nitrite concentrations were above 4 mg/liter N. When nitrite concentrations started to decrease the P uptake rate increased, which suggested a reversible inhibitory effect. Therefore, the gradual deterioration of phosphate removal likely was caused by a combined effect of both salt and nitrite. However, further research is necessary to confirm the detrimental effect of nitrite on the activity of PAOs. Pijuan et al. (53) reported that free nitrous acid (whose concentration depends on nitrite concentration and pH) had a stronger effect on PAOs than on GAOs, giving a competitive advantage to GAOs in EBPR systems. This observation is also in line with our results, which showed the enrichment of GAOs and the disappearance of PAOs as the salt concentration was increased and nitrite accumulated and started to be left over at the end of the cycle.
From our analysis of the microbial community structure, we conclude that the decrease in phosphate removal observed during phase III (22 g/liter NaCl) is directly linked to the gradual disappearance of the microorganism closely related to “Candidatus Accumulibacter” (band B9 of the 16S rRNA DGGE gel). This trend continued in the beginning of phase IV (33 g/liter NaCl), when band B9 was hardly detected and P release and uptake still were occurring, although to a minimal extent. A complete absence of those particular microorganisms during the course of phase IV also coincided with the complete deterioration of P release and removal. Batch experiments confirmed that the long exposure of the biomass to 33 g/liter NaCl was detrimental for “Candidatus Accumulibacter” and consequently for phosphate removal. In the batch test using biomass adapted to 22 g/liter, P removal still was observed at 30 g/liter NaCl. This suggests that “Candidatus Accumulibacter” still is capable of P removal at 30 g/liter NaCl, but it is outcompeted by GAOs in the bioreactor. It should be pointed out that even though granulation was stable during the whole experiment, the microbial population structure was changing faster than can be expected if no growth occurs (based on SRT). During phase IV (33 g/liter NaCl), PAOs completely disappeared in 3 weeks according to FISH analysis despite an SRT of 30 days.
In parallel to the disappearance of the PAOs, we observed that GAOs became more dominant at increasing salt concentrations, as was demonstrated using FISH analysis. GAOs can perform carbon transformations similar to those of PAOs, but no release/uptake of phosphorus is involved in their metabolism, and they are considered competitors for PAOs (33). GAOs were shown to dominate laboratory-scale cultures fed with acetate (12, 48, 72) and usually are associated with the deterioration of EBPR systems (46). Some operational factors, such as a long SRT (27), excessive aeration (7), high temperatures (39), and a low phosphorus/carbon ratio in the feed (36, 37) are described in literature and favor growth of GAOs over PAOs. In our study, the dominance of GAOs at the expense of decreasing numbers of PAOs was directly related to the increase in salt concentration. From our experiments, we observed that PAOs could adapt quite well to 11 g/liter NaCl and reasonably well to 22 g/liter NaCl, but they could not tolerate 33 g/liter NaCl, losing the competition with GAOs. So far, no report has shown that high salinity levels favor GAOs over PAOs, and our study is the first to show the complete disappearance of PAOs and the dominance of GAOs under such conditions.
Effect of increasing NaCl concentrations on nitrogen conversions.
We have shown that the ammonia removal rates were not affected by increases in salt concentrations up to 33 g/liter NaCl. The only work described in the literature that investigated the effect of salt on nutrient removal using aerobic granules was done by Figueroa et al. (25), who observed a slight decrease in ammonia removal efficiency when salt content was higher than 10 g/liter Cl− (around 16 g/liter NaCl). In our study, the relatively long period of operation (more than 300 days) at moderate salt concentrations, particularly 11 g/liter NaCl, seemed to improve the salt tolerance of the ammonia-oxidizing bacteria, suggesting that this acclimation procedure can be used to successfully achieve good performance of ammonium removal at high salinity levels.
In spite of the fact that ammonia removal was not affected when the salt concentration was increased, we observed that the only AOB encountered by DGGE in the system (belonging to the Nitrosomonas group) disappeared during the course of phase III. This indicates that either other ammonium-oxidizing bacteria not detected in DGGE analysis were metabolically active in the system (i.e., oxidizing ammonium to nitrite) or the population of Nitrosomonas decreased in such a way that it was not detected anymore by the DGGE analysis of amplified 16S rRNA fragments. To investigate in more detail the AOB population, PCR-DGGE was done using amoA gene-based primers as molecular markers. As opposed to the DGGE profile derived from 16S rRNA genes, the analysis using amoA-specific primers showed that Nitrosomonas sp. was present during the full experimental period. This observation is in line with the operational performance of the bioreactor. Since the same sequencing result was derived from the bands excised from 16S rRNA and amoA DGGE analysis, it can be inferred that the composition of the AOB population did not change but that the organisms adapted to increased salt concentrations.
As demonstrated by FISH analysis, nitrifying bacteria represented just a small fraction (between 1 and 2%) of the total microbial community dominated by PAOs and GAOs. Given the small fraction of AOB in the system, it is possible that during amplification through PCR using 16S rRNA gene-targeted group-specific primers some AOB could not be amplified. This result emphasizes the importance of using specific primers for functional genes to detect slow-growing microorganisms in a complex mixture of microorganisms. It should be noted that the amoA gene analysis does not reflect a quantitative evaluation but a qualitative one. Therefore, it is based simply on the presence or absence of a particular bacterial group, and it does not give information on their quantity. Even if a specific microorganism is present in a substantially smaller amount, as the nitrifying bacteria in our case, functional gene primers can detect the target bacteria. Moussa et al. (44) also verified that Nitrosomonas europaea was the only detectable ammonia-oxidizing species at high salt concentrations (30 and 40 g/liter NaCl), although they observed other species of AOB at lower salt concentrations (up to 16 g/liter NaCl).
A perfect match between the analytical data and the microbial diversity also was obtained when considering the nitration step. The change from nitrate to nitrite as the main nitrification product was accompanied by the complete disappearance of the only NOB detected in DGGE analysis, which was closely related to Nitrospira sp. This is particularly clear during operation at 33 g/liter NaCl (phase IV), when high nitrite concentrations were observed. Batch experiments using biomass adapted to 33 g/liter NaCl also showed that practically all of the ammonium was oxidized to nitrite at all NaCl concentrations tested. This confirmed that the NOB were strongly inhibited at 33 g/liter NaCl. Similarly, Chen et al. (9) observed that nitrite oxidizers disappeared when salt concentrations were increased from 10 to 18.2 g/liter Cl− (16 to 30 g/liter NaCl). Moussa et al. (44) found that Nitrospira sp. was the dominant nitrite oxidizer at salt concentrations of up to 10 g/liter Cl− (16.5 g/liter NaCl). Several works described in the literature mentioned that Nitrospira was the dominant NOB (8, 49, 60), although most of them are not related to operation at high-salinity conditions.
From our experiment, we can conclude that the AOB in the granular sludge system investigated were much less susceptible to osmotic stress than the NOB, which is in agreement with many former observations (13, 22, 23, 57, 59).
Additional microbial community structure changes at increasing salt concentrations.
Apparently, microorganisms belonging to the Denitromonas group have higher affinity for high salt concentrations, since the intensity of some bands, such as B10 and B11, increased as salinity levels were increased. Moreover, other bands, like B17 and B18, representing Denitromonas species, also appeared during the long-term operation at the highest salinity level tested, when a highly turbid effluent was observed. In our previous work regarding the effect of salt on the microbial diversity of nitrifying sludge (5), we also observed the appearance of Denitromonas sp. at high salt concentrations (20 g/liter NaCl). Denitromonas is a genus of Betaproteobacteria, which are found in considerable numbers in denitrifying reactors (24). These organisms are scarcely mentioned in the literature, and the only research which reported the presence of Denitromonas in very high salinity conditions (6% NaCl) was performed by Xiao et al. (69).
Given that actively growing cells contain increased levels of rRNA, as has been described in previous research (2, 21, 33, 52, 54), while dormant cells showing low metabolic activity are associated with low rRNA content (67), the band intensity of the cDNA profile in DGGE analysis can be related to the ribosome content and provides information about the activity of the microbial population. From the comparative analysis of the diversity (DNA-derived pattern) and the activity (cDNA-derived pattern) of the microbial population, we observed that the highly metabolically active microorganisms were those related to ammonia (Nitrosomonas sp.) and phosphate removal (“Candidatus Accumulibacter”), a result supported by the increased intensity of their corresponding bands from the DNA to cDNA banding patterns. For “Candidatus Accumulibacter” in particular, the high metabolic activity was detected only up to 22 g/liter NaCl, again confirming the detrimental effect of their exposition at 33 g/liter NaCl. All other bands corresponded to microorganisms with low metabolic activity, most of them belonging to the Bacteroidetes group, which represents heterotrophic bacteria probably growing at the expense of other active bacteria (i.e., PAOs, GAOs, and nitrifiers), and they were not crucial for the biological processes occurring within the granules. The relevance of this finding lies in the fact that although the microbial population within the granules is considerably diverse, just the microorganisms directly related to the main biological conversions, such as PAOs and AOB, have shown to have high metabolic activity.
In conclusion, our results demonstrate that Nitrosomonas sp. and Nitrospira were the only AOB and NOB, respectively, found in the system. AOB could tolerate NaCl concentrations of up to 33 g/liter (stable and complete ammonia removal), while NOB were severely affected (high nitrite accumulation). The increase in salt concentration had a strong effect on phosphorus removal. This observation could be directly linked to the gradual disappearance of “Candidatus Accumulibacter phosphatis.” P release and uptake stopped completely at steady-state operation at 33 g/liter NaCl. At high salinity, PAOs were outcompeted by GAOs in the bioreactor. Our findings related to process performance and molecular diversity analysis could help further the optimization of the simultaneous biological nitrogen and phosphorus removal processes in high-salinity environments.
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Amann R. I., et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aoi Y., Masaki Y., Tsuneda S., Hirata A. 2004. Quantitative analysis of amoA mRNA expression as a new biomarker of ammonia oxidation activities in a complex microbial community. Lett. Appl. Microbiol. 39:477–482 [DOI] [PubMed] [Google Scholar]
- 3. APHA 1998. Standard methods for examination of water and wastewater. American Public Health Association, New York, NY [Google Scholar]
- 4. Bassin J. P., Pronk M., Kraan R., Kleerebezem R., van Loosdrecht M. C. M. 2011. Ammonium adsorption in aerobic granular sludge, activated sludge and anammox granules. Water Res. 45:5257–5265 [DOI] [PubMed] [Google Scholar]
- 5. Bassin J. P., et al. 2011. Effect of different salt adaptation strategies on the microbial diversity, activity, and settling of nitrifying sludge in sequencing batch reactors. Appl. Microbiol. Biotechnol. doi:10.1007/s00253-011-3428-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beun J. J., et al. 1999. Aerobic granulation in a sequencing batch reactor. Water Res. 33:2283–2290 [DOI] [PubMed] [Google Scholar]
- 7. Brdjanovic D., et al. 1998. Impact of excessive aeration on biological phosphorus removal from wastewater. Water Res. 32:200–208 [Google Scholar]
- 8. Burrell P. C., Keller J., Blackall L. L. 1998. Microbiology of a nitrite-oxidizing bioreactor. Appl. Environ. Microbiol. 64:1878–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen G. H., Okabe S., Watanabe Y. 2003. Dynamic response of nitrifying activated sludge batch culture to increased chloride concentration. Water Res. 37:3125–3135 [DOI] [PubMed] [Google Scholar]
- 10. Chen Y., Jiang W. J., Liang D. T., Tay J. H. 2008. Biodegradation and kinetics of aerobic granules under high organic loading rates in sequencing batch reactor. Appl. Microbiol. Biotechnol. 79:301–308 [DOI] [PubMed] [Google Scholar]
- 11. Crocetti G., et al. 2000. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 66:1175–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crocetti G. R., Banfield J. F., Keller J., Bond P. L., Blackall L. L. 2002. Glycogen-accumulating organisms in laboratory-scale and full-scale wastewater treatment processes. Microbiology 148:3353–3364 [DOI] [PubMed] [Google Scholar]
- 13. Cui Y., Peng Y., Ye L. 2009. Effects of salt on microbial populations and treatment performance in purifying saline sewage using the MUCT process. Clean 37:649–656 [Google Scholar]
- 14. Dahl C., Sund C., Kristensen G. H., Vredenbregt L. 1997. Combined biological nitrification and denitrification of high-salinity wastewater. Water Sci. Technol. 36:345–352 [Google Scholar]
- 15. Daims H., Brühl A., Amann R., Schleifer K.-H., Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 16. Daims H., Nielsen J. L., Nielsen P. H., Schleifer K.-H., Wagner M. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dalmacija B., Karlovic E., Tamas Z., Misjovic D. 1996. Purification of high salinity wastewater by activated sludge process. Water Res. 30:295–298 [Google Scholar]
- 18. Dar S. A., Yao L., Dongen U. V., Kuenen J. G., Muyzer G. 2007. Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16S rRNA and dsrB genes as molecular markers. Appl. Environ. Microbiol. 73:594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Kreuk M. K., Heijnen J. J., van Loosdrecht M. C. M. 2005. Simultaneous COD, nitrogen and phosphate removal by aerobic granular sludge. Biotechnol. Bioeng. 90:761–769 [DOI] [PubMed] [Google Scholar]
- 20. De Kreuk M. K., van Loosdrecht M. C. M. 2004. Selection of slow growing organisms as a means for improving aerobic granular sludge stability. Water Sci. Technol. 49:9–17 [PubMed] [Google Scholar]
- 21. DeLong E. F., Wickham G. S., Pace N. R. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360–1363 [DOI] [PubMed] [Google Scholar]
- 22. Dincer A. R., Kargi F. 1999. Salt inhibition in nitrification and denitrification of saline wastewater. Environ. Technol. 20:1147–1153 [Google Scholar]
- 23. Dinçer A. R., Kargi F. 2001. Salt inhibition kinetics in nitrification of synthetic saline wastewater. Enzyme Microb. Technol. 28:661–665 [DOI] [PubMed] [Google Scholar]
- 24. Etchebehere C., Cabezas A., Dabert P., Muxi L. 2003. Evolution of the bacterial community during granules formation in denitrifying reactors followed by molecular, culture-independent techniques. Water Sci. Technol. 48:75–79 [PubMed] [Google Scholar]
- 25. Figueroa M., Mosquera-Corral A., Campos J. L., Méndez R. 2008. Treatment of saline wastewater in SBR aerobic granular reactors. Water Sci. Technol. 58:479–485 [DOI] [PubMed] [Google Scholar]
- 26. Flowers J. J., He S., Yilmaz S., Noguera D. R., McMahon K. D. 2009. Denitrification capabilities of two biological phosphorus removal sludges dominated by different “Candidatus Accumulibacter” clades. Environ. Microbiol. 1:583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fukase T., Shibata M., Miyaji Y. 1984. The role of an anaerobic stage on biological phosphorus removal. Water Sci. Technol. 17:69–80 [Google Scholar]
- 28. He S., Gall D. L., McMahon K. D. 2007. “Candidatus Accumulibacter” population structure in enhanced biological phosphorus removal sludges by polyphosphate kinase genes. Appl. Environ. Microbiol. 73:5865–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heijnen J. J., van Loosdrecht M. C. M. August 1998. Method for acquiring grain-shaped growth of a microorganism in a reactor. European and U.S. patent WO 98/37027
- 30. Hong C. C., Chan S.-K., Shim H. 2007. Effect of chloride on biological nutrient removal from wastewater. J. Appl. Sci. Environ. Sanit. 2:85–92 [Google Scholar]
- 31. Hornek R., et al. 2006. Primers containing universal bases reduce multiple amoA gene specific DGGE band patterns when analysing the diversity of beta-ammonia oxidizers in the environment. J. Microbiol. Methods 66:147–155 [DOI] [PubMed] [Google Scholar]
- 32. Jiang H. L., Tay J. H., Tay S. T. L. 2002. Aggregation of immobilized activated sludge cells into aerobically grown microbial granules for the aerobic biodegradation of phenol. Lett. Appl. Microbiol. 35:439–445 [DOI] [PubMed] [Google Scholar]
- 33. Lee S., Kemp P. 1994. Single-cell RNA content of natural marine planktonic bacteria measured by hybridization with multiple 16S rRNA-targeted fluorescent probes. Limnol. Oceanogr. 39:869–879 [Google Scholar]
- 34. Li Z. H., Wang X. C. 2008. Effects of salinity on the morphological characteristics of aerobic granules. Water Sci. Technol. 58:2421–2426 [DOI] [PubMed] [Google Scholar]
- 35. Liu Q. S., Liu Y., Show K. Y., Tay J. H. 2009. Toxicity effect of phenol on aerobic granules. Environ. Technol. 30:69–74 [DOI] [PubMed] [Google Scholar]
- 36. Liu W. T., Nakamura K., Matsuo T., Mino T. 1997. Internal energy-based competition between polyphosphate- and glycogen-accumulating bacteria in biological phosphorus removal reactor-effect of P/C feeding ratio. Water Res. 31:1430–1438 [Google Scholar]
- 37. Liu W. T., Mino T., Nakamura K., Matsuo T. 1996. Glycogen accumulating population and its anaerobic substrate uptake in anaerobic-aerobic activated sludge without biological phosphorus removal. Water Res. 30:75–82 [Google Scholar]
- 38. Liu Y., Yang S. F., Tan S. F., Lin Y. M., Tay J. H. 2002. Aerobic granules: a novel zinc biosorbent. Lett. Appl. Microbiol. 35:548–551 [DOI] [PubMed] [Google Scholar]
- 39. Lopez-Vazquez C. M., Song Y. I., Hooijmans C. M., Brdjanovic D., Moussa M. S., Gijzen H. J., van Loosdrecht M. C. M. 2007. Short-term temperature effects on the anaerobic metabolism of glycogen accumulating organisms. Biotechnol. Bioeng. 97:483–495 [DOI] [PubMed] [Google Scholar]
- 40. Madigan T. M., Martinko J. M., Parker J. 2002. Brock biology of microorganisms, 9th ed Prentice Hall, Upper Saddle River, NJ [Google Scholar]
- 41. Meinhold B., Arnold E., Isaacs S. 1999. Effect of nitrite on anoxic phosphate uptake in biological phosphorus removal activated sludge. Water Res. 33:1871–1883 [Google Scholar]
- 42. Mobarry B. K., Wagner M., Urbain V., Rittmann B. E., Stahl D. A. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mosquera-Corral A., De Kreuk M. K., Heijnen J. J., Van Loosdrecht M. C. M. 2005. Effects of oxygen concentration on N-removal in an aerobic granular sludge reactor. Water Res. 39:2676–2686 [DOI] [PubMed] [Google Scholar]
- 44. Moussa M. S., et al. 2006. Long term effects of salt on activity, population structure and floc characteristics in enriched bacterial cultures of nitrifiers. Water Res. 40:1377–1388 [DOI] [PubMed] [Google Scholar]
- 45. Moy B. Y. P., Tay J. H., Toh S. K., Liu Y., Tay S. T. L. 2002. High organic loading influences the physical characteristics of aerobic sludge granules. Lett. Appl. Microbiol. 34:407–412 [DOI] [PubMed] [Google Scholar]
- 46. Oehmen A., Saunders A. M., Vives M. T., Yuan Z., Keller J. 2006. Competition between polyphosphate and glycogen accumulating organisms in enhanced biological phosphorus removal systems with acetate and propionate as carbon sources. J. Biotechnol. 123:22–32 [DOI] [PubMed] [Google Scholar]
- 47. Oehmen A., et al. 2007. Advances in enhanced biological phosphorus removal: from micro to macro scale (review). Water Res. 41:2271–2300 [DOI] [PubMed] [Google Scholar]
- 48. Oehmen A., Yuan Z., Blackall L. L., Keller J. 2004. Short-term effects of carbon source on the competition of polyphosphate accumulating organisms and glycogen accumulating organisms. Water Sci. Technol. 50:139–144 [PubMed] [Google Scholar]
- 49. Okabe S., Satoh H., Watanabe Y. 1999. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65:3182–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Panswad T., Anan C. 1999. Impact of high chloride wastewater on an anaerobic/anoxic/aerobic process with and without inoculation of chloride acclimated seeds. Water Res. 33:1165–1172 [Google Scholar]
- 51. Peterson S. B., Warnecke F., Madejska J., McMahon K. D., Hugenholtz P. 2008. Environmental distribution and population biology of Candidatus accumulibacter, a primary agent of biological phosphorus removal. Environ. Microbiol. 10:2692–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pichard S. L., Paul J. H. 1993. Gene expression per gene dose, a specific measure of gene expression in aquatic microorganisms. Appl. Environ. Microbiol. 59:451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pijuan M., Ye L., Yuan Z. 2011. Could nitrite/free nitrous acid favour GAOs over PAOs in enhanced biological phosphorus removal systems? Water Sci. Technol. 63:345–351 [DOI] [PubMed] [Google Scholar]
- 54. Poulsen L. K., Ballard G., Stahl D. A. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qin L., Liu Y. 2006. Aerobic granulation for organic carbon and nitrogen removal in alternating aerobic-anaerobic sequencing batch reactor. Chemosphere 63:926–933 [DOI] [PubMed] [Google Scholar]
- 56. Saito T., Brdjanovic D., van Loosdrecht M. C. M. 2004. Effect of nitrite on phosphate uptake by phosphate accumulating organisms. Water Res. 38:3760–3768 [DOI] [PubMed] [Google Scholar]
- 57. Sánchez O., Aspé E., Martí M. C., Roeckel M. 2004. The effect of sodium chloride on the two-step kinetics of the nitrifying process. Water Environ. Res. 76:73–80 [DOI] [PubMed] [Google Scholar]
- 58. Schäfer H., Muyzer G. 2001. Denaturing gradient gel electrophoresis in marine microbial ecology. Methods Microbiol. 30:425–468 [Google Scholar]
- 59. Schenk H., Hegemann W. 1995. Nitrification inhibition by high salt concentrations in the aerobic biological treatment of tannery wastewater. GWF-Wasser/Abwasser 136:465–470 [Google Scholar]
- 60. Schramm A., De Beer D., Wagner M., Amann R. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwarzenbeck N., Borges J. M., Wilderer P. A. 2005. Treatment of dairy effluents in an aerobic granular sludge sequencing batch reactor. Appl. Microbiol. Biotechnol. 66:711–718 [DOI] [PubMed] [Google Scholar]
- 62. Sun X. F., et al. 2008. Biosorption of malachite green from aqueous solutions onto aerobic granules: kinetic and equilibrium studies. Bioresources Technol. 99:3475–3483 [DOI] [PubMed] [Google Scholar]
- 63. Uygur A., Kargi F. 2004. Salt inhibition on biological nutrient removal from saline wastewater in a sequencing batch reactor. Enzyme Microb. Technol. 34:313–318 [Google Scholar]
- 64. Vishniac W., Santer M. 1957. The thiobacilli. Bacteriol. Rev. 21:195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wagner M., Rath G., Koops H.-P., Flood J., Amann R. 1996. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. 34:237–244 [Google Scholar]
- 66. Wagner M., Rath G., Amann R., Koops H.-P., Schleifer K.-H. 1995. In-situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251–264 [Google Scholar]
- 67. Witzig R., et al. 2002. Microbiological aspects of a bioreactor with submerged membranes for aerobic treatment of municipal wastewater. Water Res. 36:394–402 [DOI] [PubMed] [Google Scholar]
- 68. Wu G., Guan Y., Zhan X. 2008. Effect of salinity on the activity, settling and microbial community of activated sludge in sequencing batch reactors treating synthetic saline wastewater. Water Sci. Technol. 58:351–358 [DOI] [PubMed] [Google Scholar]
- 69. Xiao Y., Roberts D. J., Zuo G., Badruzzaman M., Lehman G. S. 2010. Characterization of microbial populations in pilot-scale fluidized-bed reactors treating perchlorate- and nitrate-laden brine. Water Res. 44:4029–4036 [DOI] [PubMed] [Google Scholar]
- 70. Xu H., Liu Y. 2008. Mechanisms of Cd2+, Cu2+ and Ni2+ biosorption by aerobic granules. Sep. Purif. Technol. 58:400–411 [Google Scholar]
- 71. Yao L., Ye Z. F., Wang Z. Y., Ni J. R. 2008. Characteristics of Pb2+ biosorption with aerobic granular biomass. Chin. Sci. Bull. 53:948–953 [Google Scholar]
- 72. Zeng R. J., van Loosdrecht M. C. M., Yuan Z. G., Keller J. 2003. Metabolic model for glycogen-accumulating organisms in anaerobic/aerobic activated sludge systems. Biotechnol. Bioeng. 81:92–105 [DOI] [PubMed] [Google Scholar]
- 73. Zhu L., et al. 2008. A comparative study on the formation and characterization of aerobic 4-chloroaniline-degrading granules in SBR and SABR. Appl. Microbiol. Biotechnol. 79:867–874 [DOI] [PubMed] [Google Scholar]