Abstract
Brown rot basidiomycetes have an important ecological role in lignocellulose recycling and are notable for their rapid degradation of wood polymers via oxidative and hydrolytic mechanisms. However, most of these fungi apparently lack processive (exo-acting) cellulases, such as cellobiohydrolases, which are generally required for efficient cellulolysis. The recent sequencing of the Postia placenta genome now permits a proteomic approach to this longstanding conundrum. We grew P. placenta on solid aspen wood, extracted proteins from the biodegrading substrate, and analyzed tryptic digests by shotgun liquid chromatography-tandem mass spectrometry. Comparison of the data with the predicted P. placenta proteome revealed the presence of 34 likely glycoside hydrolases, but only four of these—two in glycoside hydrolase family 5, one in family 10, and one in family 12—have sequences that suggested possible activity on cellulose. We expressed these enzymes heterologously and determined that they all exhibited endoglucanase activity on phosphoric acid-swollen cellulose. They also slowly hydrolyzed filter paper, a more crystalline substrate, but the soluble/insoluble reducing sugar ratios they produced classify them as nonprocessive. Computer simulations indicated that these enzymes produced soluble/insoluble ratios on reduced phosphoric acid-swollen cellulose that were higher than expected for random hydrolysis, which suggests that they could possess limited exo activity, but they are at best 10-fold less processive than cellobiohydrolases. It appears likely that P. placenta employs a combination of oxidative mechanisms and endo-acting cellulases to degrade cellulose efficiently in the absence of a significant processive component.
INTRODUCTION
Brown rot basidiomycetes are the principal recyclers of lignocellulose in coniferous forests and also cause the most destructive type of decay in wooden structures (14, 57). Unlike the closely related white rot fungi (20), they remove little of the lignin that shields the cellulose and hemicelluloses in sound wood from enzymatic attack (10). Instead, brown rot fungi apparently initiate decay by producing small extracellular reactive oxygen species that depolymerize cellulose and hemicellulose (15, 17). In addition, they acidify their extracellular milieu, which may promote the hydrolysis of these structural polysaccharides (16). However, brown rot fungi also produce cellulases (3, 5, 10, 19, 25, 28, 37, 47, 50) and are so efficient at removing polysaccharides from wood that it is reasonable to conclude that these glycoside hydrolases (GHs) also have an important role. One objection is that the porosity of wood undergoing brown rot has generally been considered too low for enzymes to infiltrate (12), but recent observations that brown rot fungi partially depolymerize lignin suggest that their GHs may in fact have access to the underlying polysaccharides (29, 54, 55).
It has long been noted that few brown rot fungi produce detectable levels of highly processive cellulases (i.e., cellobiohydrolases), which release soluble cellobiose from insoluble cellulose by repeated exo attack and are key components of the synergistic cellulase systems most cellulolytic organisms employ (60). Instead, the cellulases detectable in cultures of most brown rot fungi consist only of endoglucanases, most of which are thought to introduce random endo scissions into cellulose (10). However, a random endocellulolytic mechanism is inherently inefficient, because it generates a high proportion of cello-oligomers with a degree of polymerization (DP) of ≥6 that have low water solubility and remain hydrogen bonded to the surrounding cellulose (60). This inefficiency seems at odds with the rapid removal of wood cellulose that characterizes brown rot fungi (10). Accordingly, we and others have suggested that some brown rot endoglucanases may exhibit moderate processivity (5, 56), as also observed with some GH family 9 (GH9) bacterial endoglucanases (46, 61).
The recent sequencing of the Postia placenta genome now provides a way to examine this question for one brown rot fungus (30, 48). The P. placenta genome encodes no currently known type of cellobiohydrolase and only a few GHs that are likely to exhibit endoglucanase activity. Moreover, these putative P. placenta endoglucanases belong to the GH5 and GH12 families, which are generally nonprocessive. They also lack any known type of cellulose-binding module (CBM), a structural feature that confers processivity on some GH9 endoglucanases (61). The genomic data thus do not favor our original hypothesis, suggesting instead that a nonprocessive cellulase system operating in conjunction with small cellulolytic oxidants may suffice for P. placenta to degrade cellulose efficiently. However, it remains unclear which cellulases P. placenta expresses on lignocellulose or whether they exhibit any processivity.
To address these questions, we have performed shotgun liquid chromatography-tandem mass spectrometry (LC/MS-MS) analyses of the proteins that P. placenta secretes while biodegrading wood and have identified the likely cellulases in this data set. In addition, we have expressed these GHs heterologously and report here their activities on cellulosic substrates. Finally, to assess the randomness of cellulose hydrolysis by the P. placenta cellulases, we have compared their abilities to saccharify cellulose with predictions generated by computer modeling.
MATERIALS AND METHODS
Reagents and organisms.
All chemicals were analytical grade and, unless otherwise indicated, purchased from Sigma-Aldrich (Milwaukee, WI). Filter paper was Whatman number 1. Phosphoric acid-swollen cellulose (PASC) was prepared from microcrystalline cellulose (Avicel) as described previously (42), and PASC with reduced end groups (rPASC) was prepared from PASC by treatment with NaBH4 (40). Viscometric analyses (44) showed that treatment with phosphoric acid and NaBH4 had no discernible effect on the DP of the cellulose in these preparations.
P. placenta MAD-698-R (ATCC 44394) and Gloeophyllum trabeum MAD-617 (ATCC 11539) were maintained on potato dextrose agar slants and grown on malt extract agar at 28°C for 7 days to prepare inocula.
Proteomic analysis of extracts from colonized wood.
Petri plates (150-mm diameter; containing 1.5% malt extract, 0.2% yeast extract, and 1.5% agar) were inoculated with several agar plugs each of pregrown P. placenta inoculum, and were incubated at 22°C for several days until covered with confluent mycelial lawns. For proteomic experiments, aspen sapwood chips (each approximately 80 mg [dry weight], with dimensions of approximately 20 by 6 by 3 mm) were first autoclaved twice, with an intervening gap of 4 to 5 days, using a wet cycle of 60 min. A 36-g portion of sterilized chips was then placed in each pregrown petri plate culture, along with five to seven sterilized, marked chips whose individual dry weights had been determined beforehand. The plates were incubated at 22°C, and triplicates were harvested at 1, 2, 4, 6, and 8 weeks. The preweighed aspen chips from each plate were then removed, dried at 50°C under vacuum for 72 h, and reweighed to determine representative dry weight losses.
The remaining wood chips in each set of three were then pooled and stirred in several volumes of 0.5 M NaCl for 4 h at room temperature to extract proteins. The supernatant fraction was decanted, the chips were stirred in additional 0.5 M NaCl overnight at 4°C, and this supernatant fraction was also decanted. The pooled supernatant fractions were filtered through several layers of cheesecloth and centrifuged at 8,000 × g at 4°C to remove debris. The resulting filtrate was concentrated using a 10-kDa-cutoff polyethersulfone membrane in an ultrafiltration unit (Millipore, Bedford, MA) and then dialyzed against 20 mM sodium acetate buffer (pH 5.0). The dialyzed concentrate was centrifuged again at 10,000 × g for 10 min at 4°C to remove particulates, after which the supernatant fraction was lyophilized. At the conclusion of the 8-week experiment, each of the lyophilized samples was resuspended in 200 liters of 7 M guanidine HCl, mixed by vortexing, and incubated at 37°C for 20 min. Freshly prepared ammonium bicarbonate (20 μl of a 1 M solution) and Tris(2-carboxyethyl)phosphine (1 μl; Thermo Fisher Scientific, Rockford, IL) were then added to each sample.
For protein quantification, a 5-μl portion of each sample was removed, combined with 5 μl of Laemmli sample buffer (Bio-Rad, Hercules, CA), and loaded onto a 4 to 20% gradient Tris-HCl Criterion Stain Free gel (Bio-Rad). Following electrophoresis in sodium dodecyl sulfate (SDS) running buffer (0.1% SDS, 25 mM Tris, 192 mM glycine), gels were imaged using the Criterion Stain Free imaging system (Bio-Rad). Images were saved in TIF file format and analyzed using the gel analysis tool in the ImageJ program (http://rsb.info.nih.gov/ij/index.html). Sample protein content was estimated by comparing sample peak areas with those of Precision Plus protein standards (Bio-Rad) that were run on the same gel. Protein recoveries from the colonized wood were variable but typically within the range 10 to 100 μg per petri plate.
Meanwhile, in preparation for proteomic analysis, the samples in guanidine HCl-ammonium bicarbonate-Tris(2-carboxyethyl)phosphine were incubated at 60°C for 45 min and cooled to room temperature. Iodoacetamide (20 μl of a 200 mM solution) was added to alkylate cysteine residues, and the samples were incubated in the dark at room temperature for 30 min. The samples were then diluted with 50 mM ammonium bicarbonate to reduce the guanidine HCl concentration to 900 mM. Trypsin (mass spectrometry grade; Promega, Madison, WI) was rehydrated according to the manufacturer's instructions, and 6 μg was added to each sample. After incubation overnight at 37°C, the resulting peptides were isolated on a reversed-phase C18 solid-phase extraction cartridge (Supelco, St. Louis, MO) and eluted with 1.8 ml of 80% aqueous acetonitrile. The recovered peptides were lyophilized and stored at −20°C.
For LC/MS-MS analysis, the lyophilized peptides were resuspended in 0.1% formic acid at a concentration of 0.2 to 0.9 μg original undigested protein per μl. A 1-μl portion of each sample was then injected onto a Jupiter C18 resin reversed-phase column (5-μm particle size, 38 cm long, 150-μm inner diameter; Phenomenex, Torrance, CA) connected to an Agilent (Santa Clara, CA) 1100 high-performance liquid chromatograph. The peptides were eluted at 2 μl/min with solutions of 0.1% aqueous formic acid (solvent A) and 0.1% formic acid in acetonitrile and water, 9:1 (solvent B), using the following conditions: 0 to 15 min, isocratic at 100% solvent A; 15 to 20 min, linear gradient to 20% solvent B; 20 to 75 min, linear gradient to 50% solvent B; 75 to 80 min, linear gradient to 95% solvent B; 80 to 85 min, isocratic at 95% solvent B. The column was reequilibrated in 100% solvent A for 40 min between sample injections. Eluted peptides were introduced into an LTQ mass spectrometer (Thermo Fisher, Waltham, MA) by electrospray ionization. Spectra were collected in a data-dependent mode, with the five most intense ions in each survey scan selected for collisional-induced dissociation in subsequent scans. Raw data sets were analyzed by the SEQUEST program (9) using a protein database created from the P. placenta genome sequence (http://genome.jgi-psf.org/Pospl1/Pospl1.download.html) (30). Tryptic peptides with established scoring criteria (51) are analyzed in this work.
Heterologous expression of P. placenta and G. trabeum GHs.
For isolation of total RNA (P. placenta and G. trabeum) or genomic DNA (G. trabeum), the fungi were grown from blended agar plate inocula in a medium containing 20 g malt extract, 5 g glucose, 5 g peptone, and 10 g ball-milled aspen wood per liter. The cultures, containing 250 ml in 2-liter Erlenmeyer flasks, were shaken at 150 rpm for 5 days at 29°C, after which mycelia were harvested by centrifugation at 3,000 × g for 5 min at 4°C. For total RNA isolation, approximately 100 mg of P. placenta mycelium was ground with a mortar and a pestle in liquid nitrogen and extracted using an RNeasy minikit (Qiagen, Valencia, CA) as described in the manufacturer's instructions. The RNA was then treated with RNase-free DNase (Qiagen, Valencia, CA) and quantified with a Nanodrop NT1000 instrument (Thermo Fisher Scientific, Waltham, MA). First-strand cDNA was synthesized using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI) according to the manufacturer's instructions with 2 μg of RNA and 0.5 μg of oligo(dT) in 25-μl reactions. Genomic DNA was isolated from G. trabeum mycelium as described earlier (4, 53).
For expression of the P. placenta GHs PpCel5A, PpCel5B, PpCel12A, and PpXyn10A, the published genome sequence (30) was used to design forward and reverse PCR primers for amplification of cDNAs encoding the predicted mature proteins, using the first-strand cDNA as the template (Table 1). For expression of the G. trabeum endoglucanase GtCel5A, two degenerate PCR primers, designated here DGt1 and DGt2 (Table 1), were designed on the basis of partial peptide sequences published for this enzyme (5) and were used to amplify a 131-bp fragment from genomic DNA. Based on the sequence of this fragment, upstream primers (RGsp1 for primary PCR and RGsp2 for nested PCR) and downstream primers (LGsp1 for primary PCR and LGsp2 for nested PCR) were designed to extend the gene sequence using a universal GenomeWalker kit (ClonTech, Mountain View, CA). Based on the sequence of the resulting 1,415-bp fragment, forward and reverse primers were designed to amplify the full-length GtCel5A cDNA (Table 1). The predicted coding region contains 1,077 bp and encodes 358 amino acids. The gene includes seven introns and a 21-bp secretion signal as predicted by SignalP 3.0 software (http://www.cbs.dtu.dk/services/SignalP/).
Table 1.
Primers and vectors used in this study
| Name | Direction | Restriction siteb | Sequencea |
|---|---|---|---|
| DGt1 | Forward | NA | GAYWSNGAYAAYWSNGGNAC |
| DGt2 | Reverse | NA | CTYTGNCCNCCNCCNTTRTG |
| LGsp1 | Forward | NA | CTGGAACGAGCAACACCTGCGTCACCAAC |
| LGsp2 | Forward | NA | ACCTGCGTCACCAACAACACCGTGGAGG |
| RGsp1 | Reverse | NA | TTACCGCCACCCGTCTCAGTGTTGATCGC |
| RGsp2 | Reverse | NA | GCGAGACGGTTGTTAGCACAGAGCCAAA |
| GtCel5A | Forward | EcoRI | GAATTCCAAAATGTTCAAGGCACT |
| GtCel5A | Reverse | NotI | GCGGCCGCTGCGTTGGCAATCGGA |
| PpCel5A | Forward | XhoI | TTTTCTCGAGAAAAGACAACTGATTCGTCTGGGTGGTGTGA |
| PpCel5A | Reverse | NotI | TTTTGCGGCCGCTCAAGGTAAGTTCGGCCTCACAGCA |
| PpCel5B | Forward | EcoRI | TTTGAATTCCAATTCTCTACTCGTCTGGGTGGT |
| PpCel5B | Reverse | NotI | TTTTGCGGCCGCTCACGGCAGGTAGGGGCG |
| PpCel12A | Forward | XhoI | TTTTCTCGAGAAAAGACAAACTACCCTTACTGGCCAGTAT |
| PpCel12A | Reverse | NotI | TTTTGCGGCCGCTTAGGTCTCGACGGAGACGCTGAAGCTCTC |
| PpXyn10A | Forward | XhoI | TTTTCTCGAGAAAAGAAATGGTCTACCGCAGGCCGCTT |
| PpXyn10A | Reverse | NotI | TTTTGCGGCCGCTCAGATAGATTGACCCTGCAAGGCGA |
Restriction sites are shown in bold, and Kex2 cleavage sites are underlined.
NA, not applicable.
The P. placenta cDNAs encoding the mature peptides for PpCel5A, PpCel5B, PpCel12A, and PpXyn10A, as well as the full-length cDNA encoding GtCel5A (secretion signal included), were amplified in 20-μl PCRs using Phusion Hot Start DNA polymerase (Finnzymes, Espoo, Finland) as specified in the manufacturer's protocols. Amplicons were visualized on agarose gels, purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA), digested with restriction enzymes, ligated into expression vector pPICZB or PICZαA (Invitrogen, Carlsbad, CA), and transformed into Escherichia coli DH5α competent cells. Recombinant GHs were transformed into Pichia pastoris strain GS115 or KM71H by electroporation and expressed using an EasySelect Pichia expression kit (Invitrogen, Carlsbad, CA) as described in the manufacturer's protocols. After small-scale screenings for 4-nitrophenylcellobiosidase (4-NPCase) activity, which was assayed as described below, one positive transformant of each gene was grown in buffered complex glycerol medium, and the resulting cell pellets were then resuspended in buffered complex medium containing 0.5% or 1.0% methanol for expression according to the manufacturer's instructions. Four cultures (200 ml each, in 2-liter Erlenmeyer flasks) were grown at 29°C and shaken at 250 rpm. Optimal incubation times, which were approximately 3 days, were determined by monitoring 4-NPCase activity as described below, using 40 μl of harvested culture filtrate.
Enzyme purifications.
All purification steps were undertaken at 4°C unless otherwise specified. The harvested cultures were centrifuged (10 min, 11,000 × g) to remove yeast cells and then concentrated and dialyzed against sodium phosphate buffer (50 mM, pH 6.0) by repeated ultrafiltration through a polyethersulfone 10-kDa-cutoff membrane (Millipore, Bedford, MA). Portions (2 ml) of crude enzyme preparation were applied to a DEAE-Sepharose CL-6B anion exchange column (30 by 3 cm; Pharmacia) that had been preequilibrated with the same buffer. The column was eluted at 1 ml/min and ambient temperature with a linear gradient of 0 to 0.4 M NaCl in the above-described buffer over 60 min. Fractions containing 4-NPCase activity were pooled, concentrated by ultrafiltration as described above, and subjected to gel permeation chromatography on a Superdex 75 10/30 column (30 by 1 cm; Amersham) in 50 mM sodium phosphate buffer (pH 6.0) containing 0.2 M NaCl, at ambient temperature and a flow rate of 0.5 ml/min. Active fractions were pooled, concentrated as described above, dialyzed against sodium citrate buffer (20 mM, pH 5.0), and stored at −80°C until use. The size and purity of the purified recombinant enzymes were analyzed by SDS polyacrylamide gel electrophoresis (SDS-PAGE) on 4 to 15% gradient gels (Ready Gels; Bio-Rad, Hercules, CA). Deglycosylation of the enzymes was performed with an enzymatic kit (E-DEGLY; Sigma, St. Louis, MI) under denaturing conditions according to the supplier's protocol.
Enzyme activity assays.
All GH assays were done in triplicate. Apparent numbers of reducing ends were determined using a glucose standard curve for cellulase measurements and a xylose standard curve for xylanase measurements. Activities were recorded as micromoles of reducing ends produced per minute, and 95% confidence intervals for the mean activities were calculated using Student's t formalism. Protein concentrations were determined by the Coomassie blue dye-binding method using a kit (Bio-Rad) and bovine serum albumin as the standard (1).
In addition to the heterologously expressed GHs from P. placenta and G. trabeum, the Cel5A core enzyme from Hypocrea jecorina (Trichoderma reesei) was included in these assays for comparison. This endoglucanase, originally designated EGIII (42) and more recently EGII (31), contains a CBM in its native form, but for this work the core enzyme without a CBM was produced as described earlier (43).
4-NPCase activity was determined as previously described with minor modifications (5). The reaction mixtures (50 μl) contained approximately 0.1 nmol enzyme in 50 mM sodium citrate buffer (pH 4.5) and were incubated for 20 min followed by the addition of 1 ml of 5% (wt/vol) Na2CO3 (39).
Carboxymethylcellulose (CMC) hydrolysis was assayed in 300-μl mixtures containing 0.05 nmol enzyme in 2- to 10-min reactions, using 1% (wt/vol) CMC in 50 mM sodium citrate buffer (pH 4.5) at 50°C. Xylanase activity was assayed similarly, except that 1% (wt/vol) birchwood xylan and 50 mM sodium citrate buffer (pH 5.0) were employed. Reducing sugars were determined as glucose or xylose equivalents by the dinitrosalicylic acid method as described previously (13), except that the boiling time was extended to 30 min for complete color development. It should be noted that our CMCase and xylanase results are approximate due to the nonlinear response of the dinitrosalicylic acid method to dissolved reducing sugars.
Avicelase activity was determined in 300-μl reaction mixtures that contained 0.4 nmol (9.6 to 15.2 μg) enzyme, 1% (wt/vol) substrate, 100 mM sodium citrate buffer (pH 4.5), 1 mM CaCl2, 0.02% (wt/vol) bovine serum albumin, and 0.02% (wt/vol) NaN3. The capped mixtures were rotary shaken at 200 rpm and 37°C for 18 h. To separate insoluble and soluble fractions, reactants were filtered using 0.45-μm Spin-X microcentrifuge filters (Costar, New York, NY). Soluble reducing sugars were determined as glucose equivalents by the p-hydroxybenzoic acid hydrazide (PAHBAH) method as described previously (34), except that the boiling time was extended to 15 min for full color development. Enzymatic hydrolyses of PASC and rPASC were conducted as described for Avicel, except that the reaction time was 2 h and the enzyme amount was 0.2 nmol. Filter paper hydrolyses were conducted using disks (3.3 mg) of the paper under the same conditions as described for Avicelase measurements, except that the time was 16 h and the temperature was 50°C.
Distributions of reducing ends in the soluble and insoluble fractions after cellulase treatments were conducted with filter paper and with rPASC as the substrates. We used rPASC rather than PASC in these assays, because the relatively low DP of unreduced PASC causes it already to have a high level of insoluble reducing ends, which makes it difficult to quantify newly produced insoluble reducing ends accurately. Reactions with both filter paper and rPASC were terminated by filtering them through 0.45-μm-pore-size Spin-X microcentrifuge filters, adding additional sodium citrate buffer to the filter cup (100 mM, pH 4.5), and centrifuging again. The two filtrates were then combined to assess the number of soluble reducing ends by the PAHBAH method. To assess the number of insoluble reducing ends, the entire microcentrifuge filter cup containing the rinsed residual cellulose was immersed in a tube of PAHBAH reagent and boiled just as the tube containing the soluble fraction was. The apparent extents of saccharification in soluble/insoluble reducing end determinations were 2.0 to 14.1% for rPASC assays and 0.4 to 2.0% for filter paper assays.
To aid computer modeling of cellulose hydrolysis by the heterologously expressed GHs (see below), the sugars they produced from rPASC at low extents of saccharification were identified and quantified by high-performance ion chromatography with pulsed amperometric detection as described earlier (23), except that the column was eluted isocratically with a mixture of H2O-70 mM sodium acetate-100 mM NaOH, 83:7:10. Elution times for authentic standards were as follows: glucose, 2.3 min; cellobiose, 3.4 min; cellotriose, 5.1 min; cellotetraose, 8.4 min; cellopentaose, 14.3 min.
Modeling of soluble/insoluble ratio trends.
Computer simulations of cellulose hydrolysis were implemented in Visual Basic for Microsoft Excel (the computer code and spreadsheet are available from the authors). In the computer model, rPASC was represented by 100,000 cellulose chains with a DP of 200, and filter paper was represented by 20,000 cellulose chains with a DP of 1,850 (59). Since binding of cellulases is sensitive to the direction of the cellulose chains, the method preserves this information. A Monte Carlo approach (18) was taken to determine the evolution of the molecular weight distribution as bonds were hydrolyzed. The calculation sequence can be summarized as follows: (i) select a bond at random; (ii) if the bond is already cut, select a different bond at random; (iii) if the next bond toward the reducing end is cut, select a different bond at random; (iv) if within four residues of a chain end, cleave with the specified end probability, and if not, cleave with the specified middle probability; (v) if a bond was cleaved in step iv, move two bonds toward the reducing end; (vi) if the new bond is intact, cleave with the specified probability of procession. If a cleavage results, return to step vi. If no cleavage results, return to step i. Step iii was required to match the observation that most cellulases do not efficiently hydrolyze cellobiose. The probabilities in this calculation sequence were set to various values, and the resulting populations of model cello-oligomers were stored in the computer. The model molecular weight distributions generated at various levels of bond cleavage were then analyzed to calculate soluble/insoluble reducing end ratios and extents of apparent saccharification.
Nucleotide sequence accession numbers.
The cDNA sequences encoding PpCel5A, PpCel5B, PpCel12A, PpXyn10A, and GtCel5A have been deposited in the National Center for Biotechnology Information (NCBI) database and assigned the accession numbers HM052797, HM052798, HM052799, HM052800, and HM052796, respectively.
RESULTS AND DISCUSSION
Proteomic analysis of GHs expressed on wood.
We grew P. placenta on solid aspen wood, one of its natural substrates (11), and harvested specimens at intervals to determine dry weight losses. The percent weight losses and standard deviations, obtained using five to seven replicates for each harvest, were as follows: week 1, −1 ± 1; week 2, 3 ± 2; week 4, 28 ± 8; week 6, 44 ± 6; week 8, 46 ± 12. This extent of decay establishes that our cultures were cellulolytic, because wood degraded to this extent by P. placenta is more than 50% deficient in glucan (6, 21). Since this culture system elicited a complete decay system, we adopted it to assess which cellulolytic enzymes the fungus employs to facilitate lignocellulose biodegradation.
At intervals of 1, 2, 4, 6, and 8 weeks, we harvested samples of the colonized wood and extracted them with concentrated NaCl to release extracellular proteins. After dialysis, lyophilization, and treatment of these samples with trypsin, we analyzed the resulting peptide mixtures by LC/MS-MS. SEQUEST analysis of the pooled data for all harvests showed the presence of 4,021 detectable peptides (see Table S1 in the supplemental material) attributable to 231 proteins in the predicted P. placenta proteome. Since P. placenta MAD-698-R is a dikaryon, most of the genes encoding these proteins exist as allelic pairs. Although the amino acid sequences of the allelic proteins differ slightly in some cases, the LC/MS-MS peptide data are most often consistent with assignment to either variant, and therefore we have considered all allelic pairs jointly without attempting to distinguish between them.
After allelic pairs have been combined, the data show that 34 of the proteins detected are likely GHs that fall into 20 of the GH families specified in the Carbohydrate-Active Enzymes (CAZy) database (http://www.cazy.org) (2). Most of these GHs are putative glycanases or glycosidases with potential roles in the biodegradation of wood polysaccharides (Table 2). Since a better understanding of cellulolytic mechanisms during brown rot was our principal goal, we focused on those enzymes in the data set that were likely to exhibit cellulase activity. These proteins were in the minority, consisting of just two GH5s (protein models 115648/108962 and 103675/117690) and one GH12 (model 121191/112685). In addition, the data show the presence of one GH10, a putative xylanase with no allelic variant (model 134783). Since some GH10 enzymes have appreciable cellulase activity (5, 49), we included it in the study.
Table 2.
Glycoside hydrolases detected by LC/MS-MS in tryptic digests of proteins extracted from P. placenta-colonized aspen wood
| Protein IDd | GH family no.a | Peptide count at week: |
Scoreb | Presence of signal peptidec | Putative function | ||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 4 | 6 | 8 | |||||
| jgi|Pospl1|57564/jgi|Pospl1|56576 | 2 | 0 | 2 | 0 | 0 | 2 | 1553 | + | β-Mannosidase |
| jgi|Pospl1|107557/n | 3 | 5 | 1 | 0 | 9 | 0 | 1733 | + | β-Glucosidase |
| jgi|Pospl1|128500/jgi|Pospl1|128225 | 3 | 0 | 1 | 1 | 0 | 0 | 1709 | − | β-Glucosidase |
| jgi|Pospl1|134890/jgi|Pospl1|51213 | 3 | 7 | 2 | 0 | 2 | 0 | 2272 | + | Exo-1,4-β-xylosidase |
| jgi|Pospl1|46915/jgi|Pospl1|95677 | 3 | 1 | 0 | 0 | 2 | 0 | 828 | + | β-Glucosidase |
| jgi|Pospl1|103675/jgi|Pospl1|117690 | 5 | 32 | 71 | 30 | 52 | 170 | 817 | + | Endo-1,4-β-d-glucanase |
| jgi|Pospl1|115648/jgi|Pospl1|108962 | 5 | 13 | 8 | 7 | 14 | 15 | 773 | + | Endo-1,4-β-d-glucanase |
| jgi|Pospl1|121831/jgi|Pospl1|134772 | 5 | 9 | 5 | 5 | 13 | 22 | 681 | − | Endo-1,4-β-mannanase |
| jgi|Pospl1|134783/n | 10 | 24 | 25 | 21 | 22 | 26 | 1017 | + | Endo-1,4-β-xylanase |
| jgi|Pospl1|121191/jgi|Pospl1|112685 | 12 | 11 | 1 | 8 | 5 | 9 | 920 | + | Endo-1,4-β-d-glucanase |
| jgi|Pospl1|113112/jgi|Pospl1|117345 | 15 | 30 | 8 | 14 | 27 | 20 | 1477 | + | Glucoamylase |
| jgi|Pospl1|112941/jgi|Pospl1|61809 | 16 | 1 | 4 | 0 | 3 | 1 | 640 | + | Glycosidase |
| jgi|Pospl1|113926/jgi|Pospl1|125346 | 16 | 2 | 0 | 0 | 0 | 0 | 683 | + | Glycosidase |
| jgi|Pospl1|114058/jgi|Pospl1|118230 | 18 | 1 | 0 | 0 | 1 | 0 | 344 | − | Chitinase |
| jgi|Pospl1|120960/jgi|Pospl1|119525 | 18 | 1 | 0 | 0 | 3 | 0 | 851 | + | Glycosidase |
| jgi|Pospl1|128923/jgi|Pospl1|101399 | 18 | 3 | 4 | 2 | 1 | 4 | 718 | + | Chitinase |
| jgi|Pospl1|53332/jgi|Pospl1|134918 | 18 | 3 | 3 | 1 | 0 | 2 | 749 | − | Chitinase |
| jgi|Pospl1|134894/jgi|Pospl1|134907 | 20 | 5 | 0 | 1 | 0 | 0 | 656 | + | β-Hexosaminidase |
| jgi|Pospl1|61331/jgi|Pospl1|112369 | 20 | 3 | 8 | 2 | 0 | 0 | 614 | + | β-Hexosaminidase |
| jgi|Pospl1|120395/n | 27 | 1 | 0 | 0 | 0 | 0 | 1038 | + | α-Galactosidase |
| jgi|Pospl1|134790/jgi|Pospl1|98662 | 27 | 4 | 3 | 2 | 5 | 3 | 898 | + | α-Galactosidase |
| jgi|Pospl1|111730/jgi|Pospl1|43189 | 28 | 12 | 0 | 0 | 7 | 0 | 671 | + | Endopolygalacturonase |
| jgi|Pospl1|117029/jgi|Pospl1|107258 | 31 | 0 | 0 | 1 | 0 | 0 | 1831 | + | α-Xylosidase |
| jgi|Pospl1|127993/jgi|Pospl1|128101 | 35 | 22 | 21 | 10 | 28 | 38 | 1661 | + | β-Galactosidase |
| jgi|Pospl1|115929/jgi|Pospl1|97540 | 37 | 7 | 1 | 8 | 8 | 7 | 740 | + | Trehalase |
| jgi|Pospl1|110809/n | 43 | 0 | 1 | 1 | 3 | 5 | 107 | + | α-l-Arabinofuranosidase |
| jgi|Pospl1|115593/jgi|Pospl1|134925 | 47 | 14 | 12 | 5 | 13 | 11 | 783 | + | α-Mannosidase |
| jgi|Pospl1|127046/jgi|Pospl1|100251 | 51 | 3 | 0 | 0 | 2 | 1 | 866 | − | α-l-Arabinofuranosidase |
| jgi|Pospl1|94557/n | 51 | 4 | 0 | 0 | 2 | 0 | 596 | − | α-l-Arabinofuranosidase |
| jgi|Pospl1|116267/jgi|Pospl1|108648 | 55 | 8 | 5 | 5 | 11 | 18 | 1501 | + | Glucan 1,3-β-glucosidase |
| jgi|Pospl1|119394/jgi|Pospl1|105490 | 55 | 16 | 21 | 14 | 19 | 39 | 1624 | + | Glucan 1,3-β-glucosidase |
| jgi|Pospl1|126692/jgi|Pospl1|111332 | 79 | 21 | 27 | 19 | 27 | 56 | 1272 | + | Glycosidase |
| jgi|Pospl1|112047/jgi|Pospl1|116992 | 92 | 47 | 47 | 28 | 55 | 90 | 1415 | + | α-1,2-Mannosidase |
| jgi|Pospl1|48716/jgi|Pospl1|62385 | 92 | 2 | 3 | 3 | 7 | 13 | 1090 | + | α-1,2-Mannosidase |
GH, glycoside hydrolase. Family numbers are according to CAZy (http://www.cazy.org).
UniProt/Swiss-Prot alignment score.
The presence of signal peptide was determined using the JGI database (http://genome.jgi-psf.org/Pospl1/Pospl1.download.html) and SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/). +, present; −, absent.
n, no allele found. Heterologously expressed proteins are shown in bold font.
The two GH5 proteins, which we named PpCel5A (model 115648/108962) and PpCel5B (model 103675/117690), are 51% and 53% identical, respectively, to a previously characterized Cryptococcus sp. GH5 endoglucanase (NCBI accession no. ABP02069.1) (45). The GH12 enzyme (PpCel12A; model 121191/112685) is 70% identical to a Fomitopsis palustris GH12 endoglucanase (NCBI accession no. BAF49602.1) (41). The GH10 enzyme (PpXyn10A; model 134783) is 58% identical to a Phanerochaete chrysosporium GH10 endoxylanase (NCBI accession no. ABZ88799.1) (8) and 43% identical to the Cellulomonas fimi GH10 enzyme Cex, which is primarily a xylanase but also exhibits glucanase activity (NCBI AAA56792.1) (49).
In a recent study, transcripts encoding all four of the above P. placenta GHs were shown to occur in shake flask cultures of P. placenta grown on ball-milled aspen wood, but some of the proteins were not detected unambiguously (48). The likely explanation is that the enzymes were produced, but at levels below the detection limit of the LC/MS-MS procedure used. Nonetheless, one potential problem with this earlier experimental design is that ball-milling disrupts the lignin barrier that otherwise prevents enzymatic saccharification of the cellulose in wood (35), and consequently it is unclear whether the biodegradative system expressed by P. placenta in submerged culture on ball-milled wood is representative of the system required to degrade intact lignocellulose. From our results, it is now clear that PpCel5A, PpCel5B, PpCel12A, and PpXyn10A are among the GHs that P. placenta secretes during brown rot of solid aspen wood.
Although it must be kept in mind that LC/MS-MS peptide counting provides only a very rough measure of relative protein quantities (32), the relatively high counts we obtained for PpCel5B suggest that it may be one of the more abundant extracellular enzymes produced by P. placenta on aspen wood (Table 2). In addition, the results suggest that there was little temporal variation between relative levels of the secreted proteins over the 8-week experiment. We had originally expected that hemicellulases might be expressed earlier than cellulases, because most brown rot fungi depolymerize hemicelluloses before cellulose when they degrade wood (7), but our LC/MS-MS data appear more consistent with simultaneous expression of all GHs throughout the decay process.
Heterologous expression of P. placenta cellulases.
We amplified cDNAs for the four GHs, using specific primers and total RNA extracted from wood-grown cultures of P. placenta. For PpCel5A, this cDNA encoded a protein identical to that predicted for allelic variant 115648, although for reasons that are still unclear, the cDNA differed from the DNA sequence in the P. placenta genome database at the third position of five codons. For PpXyn10A, the cDNA was 21 nucleotides shorter than the predicted JGI sequence encoding protein model 134783, but this difference is attributable to an error in intron prediction by the gene model. For PpCel5B and PpCel12A, the cDNAs exactly matched the predicted coding regions of the genes for protein models 103675 and 121191, respectively.
We expressed all four GHs in P. pastoris and purified them by ion exchange and gel permeation chromatography. In addition, we expressed and purified a previously described Cel5 endoglucanase (GtCel5A) from the brown rot fungus Gloeophyllum trabeum (5), which is 45% identical to PpCel5B. SDS-PAGE analysis of the enzymes indicated that they were apparently homogeneous and also glycosylated to various extents as inferred from the following molecular mass discrepancies: PpCel5A, 50 kDa observed versus 34 kDa predicted; PpCel5B, 45 kDa versus 34 kDa; PpCel12A, 33 kDa versus 24 kDa; PpXyn10A, 49 kDa versus 37 kDa; GtCel5A, 40 kDa versus 36 kDa. Enzymatic deglycosylation of all the heterologously expressed GHs reduced their molecular masses, giving SDS-PAGE bands that migrated close to the predicted values (data not shown).
Determination of GH activities.
First, we assayed activities of the five GHs on several soluble substrates commonly used to assess GH activity. For comparison, we included the core enzyme (i.e., the catalytic domain) of Cel5A from H. jecorina (HjCel5A), because it has been previously studied and resembles the GH5 brown rot cellulases we were investigating (42). The HjCel5A core enzyme sequence is 68% identical to that of GtCel5A, 47% identical to that of PpCel5B, and 42% identical to that of PpCel5A. All of these enzymes exhibited activity on 4-nitrophenylcellobioside (4-NPC), the substrate we had employed to screen P. pastoris transformants during heterologous expression experiments. All of the Cel5 enzymes as well as PpCel12A cleaved CMC rapidly, thus exhibiting typical endoglucanase activity. PpXyn10A cleaved xylan rapidly, consistent with its assignment as a xylanase (Table 3).
Table 3.
Specific activities of glycoside hydrolases investigated
| Enzyme | Sp act (mean ± SE) on substrate (μmol reducing sugar/min/μmol enzyme)a |
|||||
|---|---|---|---|---|---|---|
| 4-NPCase | Xylan | CMC | rPASC | Filter paper | Avicel | |
| PpCel5A | 35.4 ± 3.4 | ND | 2,452 ± 314 | 20.3 ± 3.5 | 0.16 ± 0.02 | 0.05 ± 0.02 |
| PpCel5B | 20.2 ± 1.5 | ND | 2,664 ± 158 | 51.2 ± 3.1 | 0.21 ± 0.04 | 0.10 ± 0.01 |
| PpCel12A | 0.8 ± 0.1 | ND | 984 ± 223 | 14.0 ± 1.4 | 0.42 ± 0.05 | 0.12 ± 0.03 |
| PpXyn10A | 22.7 ± 8.3 | 15,744 ± 1,377 | ND | 8.5 ± 1.7 | 0.18 ± 0.12 | 0.10 ± 0.01 |
| GtCel5A | 1.0 ± 0.2 | ND | 2,749 ± 052 | 76.4 ± 3.7 | 0.75 ± 0.06 | 0.21 ± 0.07 |
| HjCel5A core | 4.7 ± 1.4 | ND | 3,305 ± 183 | 64.2 ± 2.4 | 0.56 ± 0.06 | 0.22 ± 0.05 |
ND, not determined. Errors are 95% confidence intervals.
We next determined the activities of the heterologously expressed GHs on typical cellulose preparations. The enzymes all slowly hydrolyzed Avicel, which is largely crystalline cellulose I, as well as filter paper, which is a composite of crystalline cellulose I and amorphous cellulose (60) (Table 3). The somewhat higher activities we obtained on filter paper probably reflect selective hydrolysis of its more accessible amorphous regions. All of the GHs exhibited relatively high activities on phosphoric acid-swollen cellulose (PASC), which is generally more susceptible than crystalline cellulose to hydrolysis by endoglucanases and consists partially of cellulose II (33, 58). Although cellulose II is not a native form of the polymer, PASC is generally accepted as an adequate model for naturally occurring amorphous cellulose (52). Table 3 shows activities of our fungal GHs on PASC that had been treated beforehand with NaBH4 to reduce its aldehyde end groups (rPASC). These activities were essentially the same as those we obtained on nonreduced PASC (data not shown) and were in the range generally found for endoglucanases (46, 61). Interestingly, the GH5s as a group exhibited an inverse correlation between activity on rPASC versus activity on 4-NPCase. The reason remains to be determined, but it is possible that the more cellulolytic Cel5s depend on stabilizing a substrate residue in their +2 subsites, which are occupied in the case of cellulose but remain empty in the case of 4-NPC (27).
Quantitative analyses of the sugars produced from rPASC by the P. placenta and G. trabeum GHs showed that they all yielded cellobiose and glucose as the principal products, with small quantities of cellotriose produced in some cases but no larger oligomers (Table 4). The sugar analyses gave yields in new reducing ends that were 17 to 27% lower than those obtained with PAHBAH assays. The slightly higher values obtained in the latter case are likely attributable to our use of a glucose standard curve, which overestimates the number of reducing ends when the actual products include cello-oligomers rather than glucose alone (59).
Table 4.
Chromatographic determinations of rPASC saccharification by glycoside hydrolases investigated
| Enzyme | Sugar produced (mol%)b |
Glucosyl units solubilized (mean ± SE, mol%)a | Reducing ends solubilized (mean ± SE, mol%)a | ||
|---|---|---|---|---|---|
| G1 | G2 | G3 | |||
| PpCel5A | 32.3 | 67.7 | 0.0 | 3.7 ± 0.2 | 2.2 ± 0.1 |
| PpCel5B | 34.5 | 65.5 | 0.0 | 6.7 ± 0.2 | 4.0 ± 0.1 |
| PpCel12A | 22.9 | 66.6 | 10.5 | 2.9 ± 0.1 | 1.5 ± 0.1 |
| PpXyn10A | 30.1 | 51.6 | 18.2 | 0.6 ± 0.3 | 0.3 ± 0.1 |
| GtCel5A | 47.4 | 43.5 | 9.1 | 14.0 ± 0.6 | 8.8 ± 0.4 |
Errors are 95% confidence intervals.
G1, glucose; G2, cellobiose; G3, cellotriose.
Assessment of processivity.
The processivity of GHs when they cleave cellulose is often assessed empirically by measuring the ratio of soluble to insoluble reducing ends (i.e., scissions) produced during enzymatic hydrolysis of filter paper at a low extent of saccharification. Investigations by Wilson's group have shown that cellobiohydrolases typically give soluble/insoluble ratios greater than 10, processive endoglucanases give ratios around 6 to 10, and nonprocessive endoglucanases give ratios around 1 to 2 in this assay (22).
We performed this assay with our heterologously expressed brown rot GHs and with HjCel5A core enzyme on filter paper. The results showed that none of these GHs yielded soluble/insoluble reducing end ratios above 2 on this substrate (Table 5). Previously published work on HjCel5A core enzyme reported soluble/insoluble ratios around 7 on filter paper (42), and we found ratios around 10 for GtCel5A on Avicel (5). These findings originally led us to hypothesize that some fungal GH5s might exhibit processivity, but it is evident that all the GHs we investigated here should be classified as nonprocessive. A possible explanation for the high ratios we and others found earlier is that they contained errors in the determination of new insoluble reducing ends, which are difficult to quantify accurately in cellulose because the starting substrate already contains a high level of preexisting reducing ends.
Table 5.
Apparent extents of saccharification and soluble/insoluble reducing end ratios for glycoside hydrolases investigateda
| Enzyme | Reducing ends produced on substrate |
|||||
|---|---|---|---|---|---|---|
| rPASC |
Filter paper |
|||||
| Sol (mol %) | Insol (mol %) | Ratio | Sol (mol %) | Insol (mol %) | Ratio | |
| PpCel5A | 2.6 ± 0.5 | 1.2 ± 2.3 | 2.3 ± 0.9 | 0.31 ± 0.03 | 0.18 ± 0.25 | 1.1 ± 0.9 |
| PpCel5B | 6.6 ± 0.4 | 1.8 ± 0.4 | 3.6 ± 0.9 | 0.40 ± 0.08 | 0.35 ± 0.02 | 1.1 ± 0.2 |
| PpCel12A | 1.8 ± 0.2 | 1.1 ± 0.0 | 1.6 ± 0.2 | 0.80 ± 0.09 | 0.99 ± 0.29 | 0.7 ± 0.3 |
| PpXyn10A | 1.1 ± 0.8 | 0.9 ± 1.9 | 1.3 ± 1.0 | 0.34 ± 0.23 | 0.54 ± 0.23 | 0.5 ± 0.2 |
| GtCel5A | 9.9 ± 0.5 | 2.6 ± 0.3 | 3.8 ± 0.6 | 1.41 ± 0.11 | 0.75 ± 0.09 | 1.8 ± 0.3 |
| HjCel5A core | 8.3 ± 0.3 | 2.6 ± 0.1 | 3.2 ± 0.2 | 1.05 ± 0.11 | 0.76 ± 0.30 | 1.4 ± 0.4 |
Determined in PAHBAH assays. Errors are 95% confidence intervals.
Assessment of randomness.
Although the above-described experiments show that the brown rot cellulases we examined are not processive endoglucanases, they do not reveal whether they act strictly via random endo attack. To address this question, we developed a computer model to calculate the soluble/insoluble reducing end ratios that would result if cellulose were hydrolyzed randomly. That is, the model assumes that all linkages in cellulose and cello-oligomers are attacked with equal probability and that the probability of the enzyme processing is zero. It also assumes that rPASC has a number average DP (DPn) of 200, that filter paper has a DPn of 1,850 (59), that the cello-oligomers produced by hydrolysis are soluble if they have a DP of ≤6, and that cellobiose (DP = 2) is not hydrolyzed.
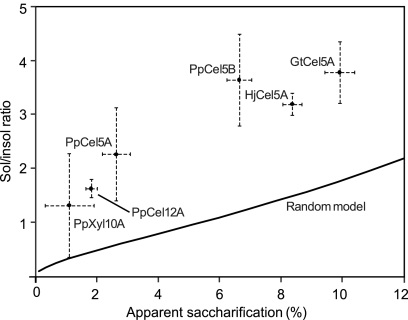
The results of the computer simulation for rPASC are shown in Fig. 1, in which the soluble/insoluble reducing end ratio is plotted against the apparent percent saccharification, which we define as (moles of soluble reducing ends produced/moles of anhydroglucose in the initial cellulose) × 100. Also plotted in Fig. 1 are the actual values we obtained in rPASC assays using the heterologously expressed P. placenta and G. trabeum GHs as well as H. jecorina Cel5A (Table 5). It is apparent that all the GHs in this study gave soluble/insoluble ratios that were significantly higher, at the 95% confidence level, than those predicted by a random cleavage model. Results for filter paper exhibited a similar discrepancy (data not shown).
Fig. 1.
Comparison of the trend in soluble/insoluble reducing sugar ratios modeled by computer for completely random hydrolysis of rPASC (plotted line) versus experimental values obtained for glycoside hydrolases investigated (points with error bars). Error bars indicate 95% confidence intervals.
There are three mechanisms that could explain why the production of soluble reducing ends by a cellulase exceeds the extent expected from random scission: (i) hydrolysis of soluble cello-oligomers is rapid compared to hydrolysis of insoluble cellulose, (ii) hydrolyses are clustered within particular regions of the cellulose, or (iii) once the enzyme catalyzes a hydrolysis, there is an increased probability that it will remain bound to the cellulose and catalyze an adjacent hydrolysis. We consider each of these possibilities below.
Rapid hydrolysis of oligomers.
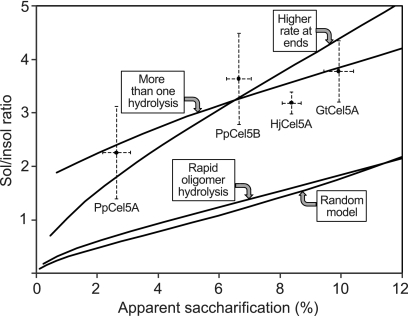
A preferential, rapid hydrolysis of soluble cello-oligomers having a DP of ≤6 would increase the number of soluble reducing ends without any real change in the molar quantity of anhydroglucose liberated as soluble material. To investigate this possibility, we adjusted our computational model to assume that all soluble oligomers were immediately hydrolyzed to a mixture consisting of 33% glucose and 67% cellobiose, which corresponds closely to the soluble sugar production we found for PpCel5A and PpCel5B (Table 4). As shown in Fig. 2, this assumption shifts the curves for predicted soluble/insoluble ratios only slightly. This counterintuitive similarity of the two curves results because rapid hydrolysis of oligomers increases not only the soluble/insoluble ratio (y axis value) but also the apparent percent saccharification (x axis value). We conclude that the generation of additional soluble reducing ends via preferential hydrolysis of cello-oligomers cannot explain why the experimental ratios we obtained using rPASC as the substrate are so much larger than those predicted by a random hydrolysis model.
Fig. 2.
Computer-modeled trends in soluble/insoluble reducing sugar ratios for rPASC hydrolysis. The random model from Fig. 1 is compared with models that incorporate immediate hydrolysis of cello-oligomers produced (rapid oligomer hydrolysis), pseudoprocessive attack within four residues of chain ends at a rate 65 times that of internal attack (higher rate at ends), and processive attack with a P value of 0.73 (more than one hydrolysis). Experimental values for the GH5 enzymes investigated are included for comparison.
Clustering of hydrolyses.
If some regions of the cellulose are poorly accessible to the enzymes, any given number of hydrolyses will be grouped closer to each other on the more accessible subset of polymer, thus increasing the probability that a soluble fragment with a DP of ≤6 is produced. This is likely part of the explanation for the higher-than-expected soluble/insoluble ratios we observed for our GHs on filter paper (data not shown), because this form of cellulose contains both crystalline and noncrystalline regions, and the latter may have incurred a disproportionately high fraction of the hydrolyses that occurred. PASC, by contrast, is relatively amorphous (60), but as usually prepared it retains some residual crystallinity (26). Therefore, we cannot rule out the possibility that our rPASC preparation may have contained relatively inaccessible regions that resulted in clustering of the GH-catalyzed scissions we observed.
However, an alternative type of clustering could also explain the high soluble/insoluble ratios we obtained with rPASC: because of its relatively low DP, it has a high content of end groups, and preferential repeated attack by endoglucanases within six sugar residues of these ends would result in a high proportion of soluble versus insoluble hydrolysis products. This mode of exo attack has been termed pseudoprocessive, because it proceeds via multiple associations between the enzyme and cellulose rather than a single binding event (31). To consider this possibility, we modified our computer model to allow for an increased likelihood of reaction near the ends of chains. The simulation showed that the soluble/insoluble ratios we obtained with our Cel5 endoglucanases on rPASC can be accounted for if the rate of enhancement for hydrolysis of bonds near the ends is set higher than the rate for more internal bonds. For example, Fig. 2 depicts the soluble/insoluble ratio trend expected if the hydrolysis rate within four sugar residues of the chain ends is set to 65 times the rate for residues further from the ends. Whether brown rot endoglucanases actually work in this fashion remains to be determined, but pseudoprocessivity could partially mitigate the inherent inefficiency of a nonprocessive cellulase system.
Multiple hydrolyses per association.
A limited degree of true processivity could also explain why our endoglucanases released more soluble sugars from rPASC than predicted by a random hydrolysis model. In this case, once the enzyme has catalyzed a hydrolysis, it would have to remain associated while moving two glucose units further along the cellulose chain, even though it contains no CBM. Translation by two glucose units would be required, because cellulose has a 2-fold screw axis and would not be in the proper orientation to associate with the enzyme after translation by only one glucose unit.
To consider this possibility, we modified our computer model to include a probability, P, that any given hydrolysis is followed by an additional hydrolysis that releases cellobiose. The kinetic chain length N, which is the average number of hydrolysis events per enzyme association, is related to P by the equation N = 1/(1 − P). Application of this model shows that it could account for the soluble/insoluble ratios of around 2 to 4 that our brown rot Cel5 endoglucanases produced on rPASC (Fig. 2), provided they exhibit processivity around a P value of 0.73 and an N value of 3.7. By contrast, the highly processive cellobiohydrolases produce soluble/insoluble ratios above 15 on rPASC (42), which corresponds to a P value of >0.97 and an N value of >33 (data not shown). That is, the soluble/insoluble ratios our Cel5 enzymes gave on rPASC could indicate some processivity, but cellobiohydrolases are an order of magnitude more processive.
Conclusion.
By analyzing the full complement of GHs produced by P. placenta on solid wood and then expressing the likely cellulases in the data set, we have provided evidence that this brown rot basidiomycete employs no highly processive cellulases when it degrades lignocellulose. The endoglucanases we found saccharify amorphous cellulose somewhat more extensively than expected for random endo hydrolysis but do not approach the soluble sugar production typical of processive cellulases. If processive cellulases are in fact produced by P. placenta, it is safe to conclude that they do not closely resemble currently known cellulases in the CAZy database (2). It remains to be determined why most brown rot fungi apparently lack an enzyme generally needed for efficient cellulolysis, but the explanation may lie in their reliance on reactive oxygen species to initiate lignocellulose breakdown. These free radicals introduce ketone and carboxylic acid groups into cellulose (24), and there is some evidence that processive cellulases do not hydrolyze linkages efficiently between the resulting modified sugar units (36).
Supplementary Material
ACKNOWLEDGMENTS
We thank Doreen Mann for cellulose viscosity measurements, Fred J. Matt for chromatographic analyses of sugars, Dan Cullen for advice regarding P. placenta allelic relationships, and David B. Wilson for alerting us to the technical difficulties involved in assessing soluble/insoluble reducing sugar ratios.
This work was supported by a research fellowship from Geongsangnam-do Province, Republic of Korea (J.S.R.), by the U.S. Department of Energy, Los Alamos National Laboratory (grant no. DE-AI32-08NA28543) (K.E.H.), and by the U.S. Department of Energy Office of Science, Biological and Environmental Research (grant no. BER-DE-AI02-07ER64480) (K.E.H).
The proteomic data were processed and archived by the Instrument Development Laboratory at the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy's Office of Biological and Environmental Research.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Bradford M. M. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 2. Cantarel B. L., et al. 2009. The Carbohydrate-Active Enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clausen C. A. 1995. Dissociation of the multienzyme complex of the brown-rot fungus Postia placenta. FEMS Microbiol. Lett. 127:73–78 [Google Scholar]
- 4. Cohen R., Suzuki M. R., Hammel K. E. 2004. Differential stress-induced regulation of two quinone reductases in the brown rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 70:324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen R., Suzuki M. R., Hammel K. E. 2005. Processive endoglucanase active in crystalline cellulose hydrolysis by the brown rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 71:2412–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cowling E. B. 1961. Comparative biochemistry of the decay of sweetgum sapwood by white-rot and brown-rot fungi. USDA technical bulletin 1258. U.S. Government Printing Office, Washington, DC [Google Scholar]
- 7. Curling S. F., Clausen C. A., Winandy J. E. 2002. Relationships between mechanical properties, weight loss, and chemical composition of wood during incipient brown-rot decay. Forest Prod. J. 52:34–39 [Google Scholar]
- 8. Decelle B., Tsang A., Storms R. K. 2004. Cloning, functional expression and characterization of three Phanerochaete chrysosporium endo-1,4-beta-xylanases. Curr. Genet. 46:166–175 [DOI] [PubMed] [Google Scholar]
- 9. Eng. J. K., McCormack A. L., Yates J. R. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989 [DOI] [PubMed] [Google Scholar]
- 10. Eriksson K.-E., Blanchette R. A., Ander P. 1990. Microbial and enzymatic degradation of wood and wood components. Springer, Berlin, Germany [Google Scholar]
- 11. Farr D. F., Bills G. F., Chamuris G. P., Rossman A. Y. 1989. Fungi on plants and plant products in the United States. American Phytopathological Society, St. Paul, MN [Google Scholar]
- 12. Flournoy D. S., Kirk T. K., Highley T. L. 1991. Wood decay by brown-rot fungi: changes in pore structure and cell wall volume. Holzforschung 45:383–388 [Google Scholar]
- 13. Ghose T. K. 1987. Measurement of cellulase activities. Pure Appl. Chem. 59:257–268 [Google Scholar]
- 14. Gilbertson R. L., Ryvarden L. 1986. North American polypores. Fungiflora, Oslo, Norway [Google Scholar]
- 15. Goodell B., et al. 1997. Low molecular weight chelators and phenolic compounds isolated from wood decay fungi and their role in the fungal biodegradation of wood. J. Biotechnol. 53:133–162 [Google Scholar]
- 16. Green F., Larsen M. J., Winandy J. E., Highley T. L. 1991. Role of oxalic acid in incipient brown-rot decay. Mat. Org. 26:191–213 [Google Scholar]
- 17. Hammel K. E., Kapich A. N., Jensen K. A., Ryan Z. C. 2002. Reactive oxygen species as agents of wood decay by fungi. Enzyme Microb. Technol. 30:445–453 [Google Scholar]
- 18. Hastings W. K. 1970. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57:97–109 [Google Scholar]
- 19. Herr D., Baumer F., Dellweg H. 1978. Purification and properties of an extracellular endo-1,4-β-glucanase from Lenzites trabea. Arch. Microbiol. 117:287–292 [Google Scholar]
- 20. Hibbett D. S., Donoghue M. J. 2001. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst. Biol. 50:215–242 [PubMed] [Google Scholar]
- 21. Irbe I., et al. 2006. On the changes of pinewood (Pinus sylvestris L.) chemical composition and ultrastructure during the attack by brown-rot fungi Postia placenta and Coniophora puteana. Int. Biodeter. Biodegr. 57:99–106 [Google Scholar]
- 22. Irwin D. C., Spezio M., Walker L. P., Wilson D. B. 1993. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42:1002–1013 [DOI] [PubMed] [Google Scholar]
- 23. Jeffries T. W., Yang V. W., Davis M. W. 1998. Comparative study of xylanase kinetics using dinitrosalicylic, arsenomolybdate, and ion chromatographic assays. Appl. Biochem. Biotechnol. 70:257–265 [DOI] [PubMed] [Google Scholar]
- 24. Kirk T. K., Ibach R., Mozuch M. D., Conner A. H., Highley T. L. 1991. Characteristics of cotton cellulose depolymerized by a brown-rot fungus, by acid, or by chemical oxidants. Holzforschung 45:239–244 [Google Scholar]
- 25. Kleman-Leyer K. M., Kirk T. K. 1994. Three native cellulose-depolymerizing endoglucanases from solid-substrate cultures of the brown-rot fungus Meruliporia (Serpula) incrassata. Appl. Environ. Microbiol. 60:2839–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuo C. H., Lee C. K. 2009. Enhancement of enzymatic saccharification of cellulose by cellulose dissolution pretreatments. Carbohydr. Polym. 77:41–46 [Google Scholar]
- 27. Lo Leggio L., Larsen S. 2002. The 1.62 angstrom structure of Thermoascus aurantiacus endoglucanase: completing the structural picture of subfamilies in glycoside hydrolase family 5. FEBS Lett. 523:103–108 [DOI] [PubMed] [Google Scholar]
- 28. Mansfield S. D., Saddler J. N., Gubitz G. M. 1998. Characterization of endoglucanases from the brown rot fungi Gloeophyllum sepiarium and Gloeophyllum trabeum. Enzyme Microb. Technol. 23:133–140 [Google Scholar]
- 29. Martínez A. T., et al. 2011. Selective lignin and polysaccharide removal in natural fungal decay of wood as evidenced by in situ structural analyses. Environ. Microbiol. 13:96–107 [DOI] [PubMed] [Google Scholar]
- 30. Martinez D., et al. 2009. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. U. S. A. 106:1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Medve J., Karlsson J., Lee D., Tjerneld F. 1998. Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol. Bioeng. 59:621–634 [PubMed] [Google Scholar]
- 32. Nesvizhskii A. I., Vitek O., Aebersold R. 2007. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4:787–797 [DOI] [PubMed] [Google Scholar]
- 33. Northolt M. G., et al. 2001. The structure and properties of cellulose fibres spun from an anisotropic phosphoric acid solution. Polymer 42:8249–8264 [Google Scholar]
- 34. Powell J. C., Lever M. 1972. New automated procedure for colorimetric determination of glucose. Biochem. Med. 6:543–547 [DOI] [PubMed] [Google Scholar]
- 35. Ralph J., et al. 2006. Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J. Biol. Chem. 281:8843–8853 [DOI] [PubMed] [Google Scholar]
- 36. Rättö M., Ritschkoff A.-C., Viikari L. 1997. The effect of oxidative pretreatment on cellulose degradation by Poria placenta and Trichoderma reesei cellulases. Appl. Microbiol. Biotechnol. 48:53–57 [Google Scholar]
- 37. Ritschkoff A.-C., Buchert J., Viikari L. 1992. Identification of carbohydrate degrading enzymes from the brown-rot fungus Gloeophyllum trabeum. Mat. Org. 27:19–29 [Google Scholar]
- 38. Reference deleted.
- 39. Sadana J. C., Patil R. V. 1988. 1,4-β-d-Glucan cellobiohydrolase from Sclerotium rolfsii. Methods Enzymol. 160:307–314 [Google Scholar]
- 40. Sakamoto R., Arai M., Murao S. 1989. Reduced cellulose as a substrate of cellulases. Agric. Biol. Chem. 53:1407–1409 [Google Scholar]
- 41. Shimokawa T., Shibuya H., Nojiri M., Yoshida S., Ishihara M. 2008. Purification, molecular cloning, and enzymatic properties of a family 12 endoglucanase (EG-II) from Fomitopsis palustris: role of EG-II in larch holocellulose hydrolysis. Appl. Environ. Microbiol. 74:5857–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ståhlberg J., Johansson G., Pettersson G. 1993. Trichoderma reesei has no true exo-cellulase: all intact and truncated cellulases produce new reducing end groups on cellulose. Biochim. Biophys. Acta 1157:107–113 [DOI] [PubMed] [Google Scholar]
- 43. Suurnäkki A., et al. 2000. Trichoderma reesei cellulases and their core domains in the hydrolysis and modification of chemical pulp. Cellulose 7:189–209 [Google Scholar]
- 44. Technical Association of the Pulp and Paper Industry 1992. Viscosity of pulp (capillary viscometer method), T230 om-89. TAPPI test methods 1992–1993. TAPPI Press, Atlanta, GA [Google Scholar]
- 45. Thongekkaew J., Ikeda H., Masaki K., Iefuji H. 2008. An acidic and thermostable carboxymethyl cellulase from the yeast Cryptococcus sp. S-2: purification, characterization and improvement of its recombinant enzyme production by high cell-density fermentation of Pichia pastoris. Protein Expression Purif. 60:140–146 [DOI] [PubMed] [Google Scholar]
- 46. Tomme P., Kwan E., Gilkes N. R., Kilburn D. G., Warren R. A. J. 1996. Characterization of CenC, an enzyme from Cellulomonas fimi with both endo- and exoglucanase activities. J. Bacteriol. 178:4216–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valásková V., Baldrian P. 2006. Degradation of cellulose and hemicelluloses by the brown rot fungus Piptoporus betulinus: production of extracellular enzymes and characterization of the major cellulases. Microbiology 152:3613–3622 [DOI] [PubMed] [Google Scholar]
- 48. Vanden Wymelenberg A., et al. 2010. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl. Environ. Microbiol. 76:3599–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. White A., Withers S. G., Gilkes N. R., Rose D. R. 1994. Crystal structure of the catalytic domain of the β-1,4-glycanase Cex from Cellulomonas fimi. Biochemistry 33:12546–12552 [DOI] [PubMed] [Google Scholar]
- 50. Wolter K. E., Highley T. L., Evans F. J. 1980. A unique polysaccharide-degrading and glycoside-degrading enzyme complex from the wood-decay fungus Poria placenta. Biochem. Biophys. Res. Commun. 97:1499–1504 [DOI] [PubMed] [Google Scholar]
- 51. Wolters D. A., Washburn M. P., Yates J. R. 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73:5683–5690 [DOI] [PubMed] [Google Scholar]
- 52. Wood T. M. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 160:19–25 [Google Scholar]
- 53. Yatzkan E., Yarden O. 1995. Inactivation of a single type-2a phosphoprotein phosphatase is lethal in Neurospora crassa. Curr. Genet. 28:458–466 [DOI] [PubMed] [Google Scholar]
- 54. Yelle D. J., Ralph J., Lu F. C., Hammel K. E. 2008. Evidence for cleavage of lignin by a brown rot basidiomycete. Environ. Microbiol. 10:1844–1849 [DOI] [PubMed] [Google Scholar]
- 55. Yelle D. J., Wei D. S., Ralph J., Hammel K. E. 2011. Multidimensional NMR analysis reveals truncated lignin structures in wood decayed by the brown rot basidiomycete Postia placenta. Environ. Microbiol. 13:1091–1100 [DOI] [PubMed] [Google Scholar]
- 56. Yoon J. J., Cha C. J., Kim Y. S., Son D. W., Kim Y. K. 2007. The brown-rot basidiomycete Fomitopsis palustris has the endo-glucanases capable of degrading microcrystalline cellulose. J. Microbiol. Biotechnol. 17:800–805 [PubMed] [Google Scholar]
- 57. Zabel R. A., Morrell J. J. 1992. Wood microbiology: decay and its prevention. Academic Press, San Diego, CA [Google Scholar]
- 58. Zhang Y. H. P., Cui J. B., Lynd L. R., Kuang L. R. 2006. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 7:644–648 [DOI] [PubMed] [Google Scholar]
- 59. Zhang Y. H. P., Lynd L. R. 2005. Determination of the number-average degree of polymerization of cellodextrins and cellulose with application to enzymatic hydrolysis. Biomacromolecules 6:1510–1515 [DOI] [PubMed] [Google Scholar]
- 60. Zhang Y. H. P., Lynd L. R. 2004. Toward an aggregated understanding of enzymatic hydrolysis of cellulose: noncomplexed cellulase systems. Biotechnol. Bioeng. 88:797–824 [DOI] [PubMed] [Google Scholar]
- 61. Zhou W. L., Irwin D. C., Escovar-Kousen J., Wilson D. B. 2004. Kinetic studies of Thermobifida fusca Cel9A active site mutant enzymes. Biochemistry 43:9655–9663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.