Abstract
Staphylococcus aureus produces many virulence factors, including toxins, immune-modulatory factors, and exoenzymes. Previous studies involving the analysis of virulence expression were mainly performed by in vitro experiments using bacterial medium. However, when S. aureus infects a host, the bacterial growth conditions are quite different from those in a medium, which may be related to the different expression of virulence factors in the host. In this study, we investigated the expression of virulence factors in S. aureus grown in calf serum. The expression of many virulence factors, including hemolysins, enterotoxins, proteases, and iron acquisition factors, was significantly increased compared with that in bacterial medium. In addition, the expression of RNA III, a global regulon for virulence expression, was significantly increased. This effect was partially restored by the addition of 300 μM FeCl3 into serum, suggesting that iron depletion is associated with the increased expression of virulence factors in serum. In chemically defined medium without iron, a similar effect was observed. In a mutant with agr inactivated grown in serum, the expression of RNA III, psm, and sec4 was not increased, while other factors were still induced in the mutant, suggesting that another regulatory factor(s) is involved. In addition, we found that serum albumin is a major factor for the capture of free iron to prevent the supply of iron to bacteria grown in serum. These results indicate that S. aureus expresses virulence factors in adaptation to the host environment.
INTRODUCTION
Staphylococcus aureus is one of the major pathogens of humans; it causes various suppurative diseases, food poisoning, pneumonia, and toxic shock syndrome (11, 20). In addition, S. aureus, especially methicillin-resistant S. aureus (MRSA), often causes serious problems via nosocomial infection in hospitals (10, 16). Furthermore, community-acquired MRSA has recently emerged and has been reported to cause serious infectious diseases, sepsis, and pneumonia (3, 9).
It is well known that S. aureus produces many virulence factors, such as hemolysins, leukocidins, proteases, enterotoxins, exfoliative toxins, and immune-modulatory factors (11, 12, 21, 31). The expression of these factors is tightly regulated during growth. The agr system, known as the quorum-sensing system, is known to play a central role in the regulation of virulence factors (5, 26, 27). AgrAC is a two-component system (TCS) that consists of a histidine kinase and a response regulator. The Agr system regulates the expression of the gene coding for small RNA, known as RNA III, which is localized divergently at the agr operon and regulates the expression of many virulence factors, such as hemolysins, leukocidins, and protein A (26, 27). In addition, RNA III, known as hld, encodes delta-hemolysin. The mechanisms of RNA III regulation for the expression of several virulence factors and regulatory factors, including alpha-hemolysin, coagulase, protein A, Map, and Rot, have been demonstrated to occur at the posttranscriptional level by RNA-RNA interaction (7, 13, 17, 19, 25). However, in other virulence factors, the precise mechanism of regulation by RNA III remains unknown. Besides agr, many factors, including other TCSs (arl and sae) and transcriptional regulators (the sar family, rot, and mgr), have been demonstrated to be involved in the expression of virulence factors (5, 26, 33). To date, in regard to research on the expression of virulence factors, bacterial media, such as Trypticase soy broth (TSB), brain heart infusion (BHI) broth, and Luria-Bertani (LB) broth, have been commonly used for S. aureus cultivation. However, when S. aureus infects a host, the circumstances around bacterial cells are quite different from those in a medium, with the expression pattern of virulence factors in the host suggested to be quite different from that in culture media. Several investigations using animal models have characterized the regulation of virulence factor expression in vivo (4, 5, 24, 28). However, from the findings of in vivo experiments, it is considered that many factors, including cellular immune factors and nutrient conditions, affect the expression of virulence factors, suggesting that the mechanisms of regulation of virulence factors in vivo are very complicated. Here, we used serum for analysis of the expression of virulence factors. Serum contains many biochemical agents, such as serum proteins, carbohydrates, and ions, which are quite different from those in in vitro bacterial medium. S. aureus infection sometimes causes sepsis, so the organism has the ability to survive and spread in the bloodstream. In this study, we used S. aureus MW2 isolated from a patient who suffered fatal septicemia (1) and investigated the expression of virulence factors in serum.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. S. aureus was grown in TSB (Becton Dickinson Microbiology Systems, Cockeysville, MD). Tetracycline (TC; 10 μg/ml) was added when necessary for S. aureus. Previously constructed mutants with agr inactivated were used in the study (22). Chemically defined medium (CDM) was prepared as described elsewhere (18). Before adding metal ions, the medium was treated with Chelex-100 (Bio-Rad, Tokyo, Japan) to remove the residual metal ions completely. Then, metal ions without iron were added to the medium. We named CDM without iron CDM(−) in this study.
Table 1.
Strains used in the study
| Strain | Characteristics | Reference |
|---|---|---|
| MW2 | Clinical strain, sepsis, methicillin resistant (mec+) | 1 |
| FK73 | agrC::pCL52.1 in MW2; TCra | 22 |
| MS23002 | Clinically isolated strain; sepsis | This study |
| MS23143 | Clinically isolated strain; sepsis | This study |
| MS23218 | Clinically isolated strain; sepsis | This study |
| MS23543 | Clinically isolated strain; sepsis | This study |
| TF3453 | Clinically isolated strain; sepsis | This study |
| TF3483 | Clinically isolated strain; sepsis | This study |
| TY526 | Clinically isolated strain; impetigo | This study |
TCr, tetracycline resistance.
Culturing S. aureus in serum and chemically defined medium.
S. aureus cells grown overnight were harvested and washed with distilled water (DW) treated with Chelex-100. Then, S. aureus cells were suspended in serum or CDM(−), and a small portion of bacterial suspension (108 cells) was inoculated into 10 ml of serum or CDM(−). S. aureus cells were grown at 37°C with shaking, and bacterial cells at various phases were collected. The origin of the serum used in this study was calf, rabbit, or human, and the serum was heated at 56°C for 30 min prior to use. Calf and rabbit sera were obtained from Gibco Life Technologies Japan (Tokyo, Japan). Human serum was prepared from the blood extracted from a volunteer. When necessary, FeCl3, holotransferrin (Sigma-Aldrich, Tokyo, Japan), and hemin (Sigma-Aldrich) were added to the medium.
Quantitative analysis of gene expression.
Twenty-seven major virulence factors, including toxins, immune-modulatory factors, exoenzymes, and iron acquisition factors were selected, and their expression was analyzed by quantitative PCR. Total RNA was extracted from bacterial cells with a FastRNA Pro Blue kit (MP Biomedicals, OH) in accordance with the manufacturer's protocol. One microgram of total RNA was reverse transcribed to cDNA using a first-strand cDNA synthesis kit (Roche, Tokyo, Japan). Using cDNA as template DNA, quantitative PCR was performed with a LightCycler system (Roche, Tokyo, Japan). Primers were constructed and used to determine the optimal conditions for analysis of expression. The amount of gyrA was used as an internal control. The primers are shown in Table S1 in the supplemental material.
Fractionation of calf serum.
Fifteen milliliters of calf serum (CS) was applied to a CentriprepYM-50 centrifugal filter unit (Millipore, Tokyo, Japan) to remove molecules larger than 50 kDa. After centrifugation at 1,500 rpm, 15 ml of the flowthrough fraction was applied to a CentriprepYM-3 centrifugal filter unit (Millipore, Tokyo, Japan) to remove molecules larger than 3 kDa. After centrifugation, 1 ml of the flowthrough fraction was applied to a SepPack cartridge (Waters, Tokyo, Japan) to remove lipids and peptides. Finally, 3 kinds of solutions, (i) CS with molecules larger than 50 kDa removed; (ii) CS with molecules larger than 3 kDa removed; and (iii) CS with molecules larger than 3 kDa, lipids, and peptides removed, were prepared.
Purification of albumin from calf serum.
Five milliliters of CS was applied to ToyoPearl AF-Blue HC-650 (Tosoh, Tokyo, Japan; 15 by 200 mm) equilibrated with 50 mM potassium phosphate buffer (pH 7.0) (buffer 1). The column was washed with 50 mM potassium phosphate buffer (pH 7.5) (buffer 2) until unbound proteins passed through. Bound proteins were then eluted with a linear gradient from buffer 1 to 50 mM potassium phosphate buffer (pH 7.5) containing 1.5 M KCl (buffer 3) at a flow rate of 0.5 ml/min. Each collected fraction (5 ml each) was resolved by SDS-polyacrylamide gel electrophoresis in a 7.5% gel and then stained with Coomassie brilliant blue. The fractions containing albumin were collected and desalted with a CentriprepYM-50 centrifugal filter unit to remove KCl. Finally, the protein concentration was determined.
RESULTS
Expression of virulence factors in serum.
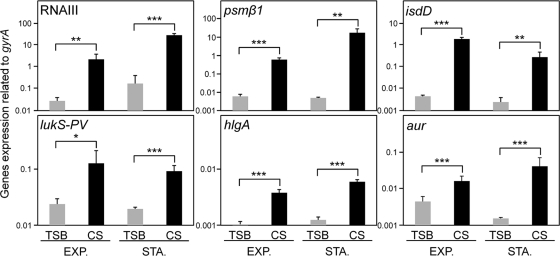
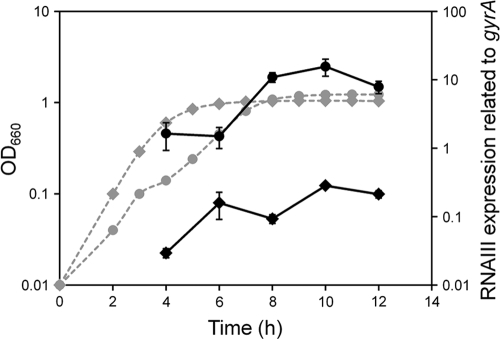
To compare the expression of virulence factors in S. aureus cells grown in TSB and the cells in CS, we first checked the growth curve using both media and found similar rates of growth. Then, we collected the cells at various phases, using the same optical density for the culture with TSB or CS. The expression levels of most virulence factors, such as hemolysins, leukocidins, cell surface adhesins, and exoenzymes, were increased significantly in CS, while that of protein A was reduced in CS (Fig. 1 and Table 2). In addition, factors responsible for iron acquisition, such as isdD, sbnF, and sirA, were expressed significantly in CS. The higher expression levels of most virulence factors in CS were found to be significant at the stationary and exponential phases (Fig. 1). Since RNA III, comprising the gene (hld) encoding delta-hemolysin, is considered to be a global regulon responsible for many virulence factors, we investigated its expression during growth in TSB and CS. RNA III expression in CS was significantly higher than that upon growth in TSB from early exponential phase to stationary phase (Fig. 2). During growth, RNA III expression in CS was almost 10- to 100-fold higher than that in TSB. The expression pattern of RNA III in CS during growth had a tendency similar to that in TSB, showing expression that gradually increased until the stationary phase.
Fig. 1.
Expression of virulence factors of S. aureus grown in serum. S. aureus MW2 cells cultured in TSB and CS were collected at exponential (EXP.) and stationary (STA.) phases. Then, total RNA was extracted by the method described in Materials and Methods. The expression of virulence factors was determined by quantitative PCR. The data shown represent means and standard deviations of the means (SD) of measurements performed in triplicate. The asterisks indicate significant increases compared with TSB as determined by Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Table 2.
Expression of virulence factors of S. aureus grown in TSB, CS and CS with 300 μM FeCl3
| GeneIDa | Gene name | Function | TSB expressionb | CS |
CS + 300 μM FeCl3 |
||
|---|---|---|---|---|---|---|---|
| Expressionb | Fold change (TSB)c | Expressionb | Fold change (CS)d | ||||
| RNA III | Global regulon | 1.41 | 135.30 | 95.96* | 4.75 | 0.04+ | |
| MW1044 | hla | Hemolysin | 1.72 | 8.66 | 5.03 | 2.55 | 0.29 |
| MW2342 | hlgA | Hemolysin | 0.25 | 18.76 | 75.84* | 5.08 | 0.27+ |
| MW1942 | lukM | Leukocidin | 4.63 | 13.02 | 2.82 | 5.70 | 0.44 |
| MW1767 | lukD | Leukocidin | 0.21 | 2.19 | 10.48* | 0.90 | 0.41++ |
| MW1379 | lukS-PV | Leukocidin | 1.73 | 4.60 | 2.65* | 1.83 | 0.40 |
| psmα | Phenol-soluble modulin | 0.88 | 7.93 | 9.00** | 2.52 | 0.32+ | |
| MW1056 | psmβ1 | Phenol-soluble modulin | 1.44 | 73.03 | 50.84* | 11.01 | 0.15+ |
| MW0759 | sec4 | Enterotoxin | 5.16 | 173.97 | 33.72* | 26.58 | 0.15+ |
| MW1889 | sea | Enterotoxin | 2.41 | 6.58 | 2.73* | 2.59 | 0.39+ |
| MW2558 | aur | Protease | 0.87 | 7.29 | 8.40* | 2.73 | 0.37+ |
| MW0932 | sspA | Protease | 0.69 | 5.75 | 8.39* | 1.81 | 0.31+ |
| MW1755 | splA | Protease | 0.25 | 7.32 | 29.17* | 1.96 | 0.27+ |
| MW0297 | geh | Lipase | 1.21 | 9.66 | 7.96* | 2.43 | 0.25+ |
| MW0206 | coa | Coagulase | 3.51 | 10.67 | 3.04* | 3.63 | 0.34+ |
| MW1885 | sak | Staphylokinase | 5.16 | 25.47 | 4.93* | 10.25 | 0.40+ |
| MW2341 | sbi | IgG binding | 46.74 | 79.02 | 1.69 | 26.37 | 0.33+ |
| MW0084 | spa | Protein A | 2347.95 | 370.07 | 0.16** | 193.49 | 0.52 |
| MW0130 | capG | Capsule | 0.18 | 12.84 | 72.84* | 2.24 | 0.17+ |
| MW2586 | icaA | Intercellular adhesin | 0.14 | 4.20 | 30.88* | 1.75 | 0.42+ |
| MW0516 | sdrC | Fibrinogen binding | 0.22 | 1.19 | 5.33 | 0.63 | 0.53 |
| MW0764 | clfA | Clumping factor | 9.15 | 95.86 | 10.48* | 29.36 | 0.31+ |
| MW2421 | fnb | Fibronectin binding | 3.10 | 13.42 | 4.33 | 5.86 | 0.44 |
| MW1884 | SCIN | Complement inhibitor | 9.86 | 12.29 | 1.25 | 9.33 | 0.76 |
| MW0088 | sirA | Iron acquisition | 0.78 | 26.91 | 34.59* | 5.28 | 0.20+ |
| MW0094 | sbnF | Iron acquisition | 0.05 | 1.41 | 28.75* | 0.33 | 0.23+ |
| MW1014 | isdD | Iron acquisition | 0.32 | 10.36 | 32.66* | 1.60 | 0.15+ |
GeneID in S. aureus strain MW2.
Expression ratio to gyrA (×100) at exponential phase.
Significant changes of expression of S. aureus grown in CS compared with TSB as determined by Student's t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Significant changes of expression of S. aureus grown in CS containing 300 μM FeCl3 compared with CS as determined by Student's t test; +, P < 0.05; ++, P < 0.01; +++, P < 0.001.
Fig. 2.
Expression of RNA III during growth. S. aureus MW2 cells cultured in TSB (diamonds) and CS (circles) were collected at various growth phases. The expression of RNA III was determined by quantitative PCR by the method described in Materials and Methods. The data shown represent means ± SD of measurements performed in triplicate. The expression of RNA III and bacterial growth are represented by solid lines and dashed lines, respectively.
Next, we investigated the expression levels of RNA III in sera from calf, rabbit, and human to determine whether our observation was specific for CS. We found that the expression patterns in rabbit and human sera were quite similar to those in CS, showing higher expression of virulence factors, including RNA III, than in TSB (Table 3).
Table 3.
RNA III, hla, and isdD expression of S. aureus grown in human and rabbit sera
| Gene name | TSB expressiona | HSd |
HS + 300 μM FeCl3 |
RSe |
RS + 300 μM FeCl3 |
||||
|---|---|---|---|---|---|---|---|---|---|
| Expressiona | Fold change (TSB)b | Expressiona | Fold change (HS)c | Expressiona | Fold change (TSB)b | Expressiona | Fold change (RS)c | ||
| RNAIII | 6.1 | 260.0 | 42.5*** | 66.5 | 0.3+ | 143.8 | 23.5*** | 18.0 | 0.1+++ |
| hla | 0.4 | 7.0 | 18.1*** | 2.6 | 0.4+++ | 1.3 | 3.3*** | 0.6 | 0.4+++ |
| isdD | 0.5 | 85.4 | 176.2*** | 8.6 | 0.1+++ | 5.0 | 10.3*** | 0.8 | 1.7+++ |
Expression ratio to gyrA (×100) at exponential phase.
Significant changes of expressions of S. aureus grown in HS or RS compared with TSB as determined by Student's t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Significant changes of expressions of S. aureus grown in HS or RS containing 300 μM FeCl3 compared with HS or RS as determined by Student's t test; +, P < 0.05; ++, P < 0.01; +++, P < 0.001.
HS, human serum.
RS, rabbit serum.
Identification of the factor responsible for the expression of virulence factors in serum.
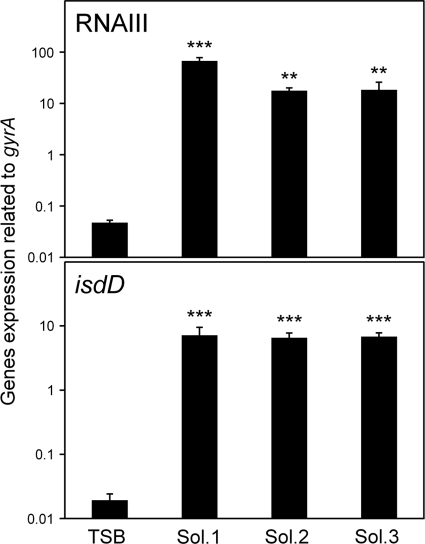
Since the expression levels of many virulence factors were increased in CS, we tried to identify the factor(s) responsible for these increased expression levels. At first, we used three solutions prepared from CS, (i) CS with proteins larger than 50 kDa removed (solution 1); (ii) CS with proteins larger than 3 kDa removed (solution 2); and (iii) CS with proteins, peptides, and lipids removed (solution 3). The expression of RNA III in all solutions tested was higher than that in TSB, indicating that the factor(s) was not among proteins, peptides, or lipids (Fig. 3).
Fig. 3.
Expression of RNA III and isdD in S. aureus grown in various fractions of serum. S. aureus MW2 cells cultured in TSB; CS with molecules larger than 50 kDa removed (solution [Sol.] 1); CS with molecules larger than 3 kDa removed (solution 2); and CS with molecules larger than 3 kDa, lipids, and peptides removed (solution 3) were collected at the exponential phase. The expression levels of RNA III and isdD were determined by quantitative PCR by the method described in Materials and Methods. The data shown represent means and SD of measurements performed in triplicate. The asterisks indicate significant increases compared with TSB as determined by Dunnett's test: **, P < 0.01; ***, P < 0.001.
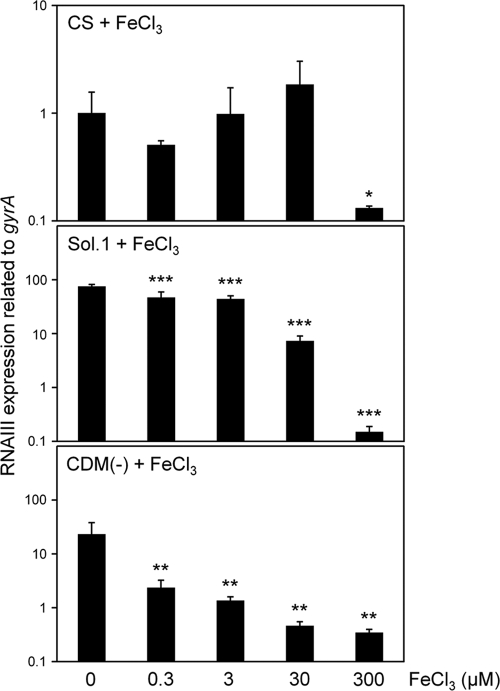
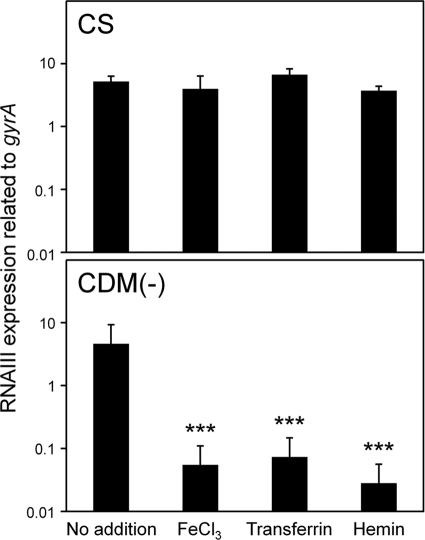
It is generally known that serum contains little (free) iron, so we hypothesized that iron depletion in serum increased the expression levels of various virulence factors. Indeed, the isdD gene was expressed significantly in CS and CS fractionates (solutions 1 to 3) compared with that in TSB (Table 2 and Fig. 1 and 3). In addition, other iron acquisition genes were expressed significantly in solutions 1 to 3 (data not shown). The addition of 300 μM FeCl3 to CS decreased the expression levels of many virulence factors, including RNA III, significantly (Table 2). In particular, the expression of RNA III showed a decrease of about 25-fold after the addition of 300 μM FeCl3 to the CS. Then, we investigated the effects of various concentrations of FeCl3 on the expression of RNA III. FeCl3 at concentrations below 30 μM had no effect on the reduction of RNA III expression in CS, while in CS with proteins larger than 50 kDa removed (solution 1), 0.3 μM FeCl3 decreased the expression of RNA III and concentrations of FeCl3 above 30 μM decreased its expression significantly (Fig. 4). We further investigated the effects of other iron sources, holotransferrin and hemin, on the expression of RNA III in CS (Fig. 5). The addition of holotransferrin or hemin containing 3 μM Fe(III) or Fe(II) had no effect on its expression. Owing to the solubility of these reagents in CS, we could not investigate the effects of high concentrations of holotransferrin and hemin on RNA III expression. In addition, we investigated the effects of various iron sources on the expression of RNA III and iron acquisition factors in solutions 1 to 3 and found that all 3 μM iron sources, including FeCl3, decreased their expression levels significantly (see Fig. S1 in the supplemental material).
Fig. 4.
Effect of FeCl3 on the expression of RNA III. S. aureus MW2 cells cultured in CS, CS with molecules larger than 50 kDa removed (solution 1), and CDM(−) with or without various concentrations of FeCl3 were collected at the exponential phase. The expression of RNA III was determined by quantitative PCR by the method described in Materials and Methods. The data shown represent means and SD of measurements performed in triplicate. The asterisks indicate significant decreases compared with the medium with no addition of FeCl3 as determined by Dunnett's test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Fig. 5.
Effect of hemin and holotransferrin on the expression of RNA III. S. aureus MW2 cells cultured in CS and CDM(−) with or without 3 μM FeCl3, hemin, or holotransferrin were collected at the exponential phase. The expression of RNA III was determined by quantitative PCR by the method described in Materials and Methods. The data shown represent means and SD of measurements performed in triplicate. The asterisks indicate significant decreases compared with the medium with no addition of iron sources as determined by Dunnett's test: ***, P < 0.001.
Virulence factor expression of clinical isolates grown in serum.
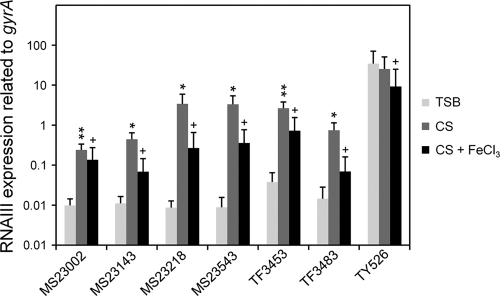
We investigated the expression of RNA III in CS using 7 clinical isolates and found that, with the exception of one strain, TY526, all the strains showed significant increase of RNA III expression in CS (Fig. 6). Furthermore, the addition of 300 μM FeCl3 to CS resulted in decreased RNA III expression. Other factors, such as psmβ1 and icaA, also showed results similar to those of RNA III (data not shown).
Fig. 6.
RNA III expression in clinical isolates grown in serum. S. aureus clinical isolates cultured in TSB, CS, and CS with 300 μM FeCl3 were collected at the exponential phase. The expression of RNA III was determined by quantitative PCR by the method described in Materials and Methods. The data shown represent means and SD of measurements performed in triplicate. The asterisks indicate significant increases compared with TSB as determined by Student's t test: *, P < 0.05; **, P < 0.01. The pluses indicate significant decreases compared with the medium with no addition of FeCl3 as determined by Student's t test: +, P < 0.05.
Expression of virulence factors in chemically defined medium.
Since iron availability is restricted in serum, we also investigated the effects of iron on the expression of virulence factors in chemically defined medium. Table 4 shows the results of the expression of virulence factors grown in the presence or absence of 3 μM FeCl3 in CDM(−). In CDM(−), S. aureus could still grow, although the growth rate was decreased slightly compared with that in CDM(−) with iron. This growth in CDM(−) is due to the presence of iron inside bacteria cultured in TSB as precultured cells. Since iron acquisition genes were increased significantly in CDM(−) (Table 4), it is considered that the concentration of iron in bacteria was significantly low. In a result similar to that with CS, the expression of many factors, including RNA III, hla, psm, sec4, capG, isdD, sirA, and sbnF, was increased significantly compared with that in TSB, and that of spa was decreased. However, the increased levels of expression of virulence factors were different in CS and CDM(−). The increases in the levels of RNA III, psm, sec4, and isdD in CDM(−) were higher than in CS, while those of others were lower than in CS. When 3 μM FeCl3 was added to CDM(−), the expression of almost all factors was drastically decreased. In addition, the expression of RNA III was significantly reduced when 0.3 μM FeCl3 was added to CDM(−) (Fig. 4). Furthermore, the addition of holotransferrin and hemin containing 3 μM Fe(III) or Fe(II) resulted in a significant decrease of RNA III (Fig. 5).
Table 4.
Expression of virulence factors of S. aureus grown in TSB, CDM(−), and CDM(−) with 3 μM FeCl3
| GeneIDa | Gene name | Function | TSB expressionb | CDM(−) |
CDM(−) + 3 μM FeCl3 |
||
|---|---|---|---|---|---|---|---|
| Expressionb | Fold change (TSB)c | Expressionb | Fold change [CDM(−)]d | ||||
| RNAIII | Global regulon | 1.41 | 1942.26 | 1377.53** | 31.66 | 0.02+++ | |
| MW1044 | hla | Hemolysin | 1.72 | 5.76 | 3.35* | 1.80 | 0.31 |
| MW2342 | hlgA | Hemolysin | 0.25 | 0.86 | 3.48* | 0.25 | 0.29+ |
| MW1942 | lukM | Leukocidin | 4.63 | 9.31 | 2.01* | 2.80 | 0.30+++ |
| MW1767 | lukD | Leukocidin | 0.21 | 0.54 | 2.57* | 0.23 | 0.43++ |
| MW1379 | lukS-PV | Leukocidin | 1.73 | 2.71 | 1.56 | 0.95 | 0.35+++ |
| psmα | Phenol-soluble modulin | 0.88 | 111.72 | 126.74* | 2.97 | 0.03+ | |
| MW1056 | psmβ1 | Phenol-soluble modulin | 1.44 | 2380.99 | 1657.47** | 46.09 | 0.02++ |
| MW0759 | sec4 | Enterotoxin | 5.16 | 177.22 | 34.35*** | 21.86 | 0.12++ |
| MW1889 | sea | Enterotoxin | 2.41 | 4.67 | 1.94** | 1.56 | 0.34+ |
| MW2558 | aur | Protease | 0.87 | 1.24 | 1.43* | 0.40 | 0.32++ |
| MW0932 | sspA | Protease | 0.69 | 1.40 | 2.04** | 0.56 | 0.40++ |
| MW1755 | splA | Protease | 0.25 | 0.82 | 3.25* | 0.35 | 0.42++ |
| MW0297 | geh | Lipase | 1.21 | 5.26 | 4.33*** | 1.72 | 0.33++ |
| MW0206 | coa | Coagulase | 3.51 | 7.29 | 2.08** | 3.90 | 0.53+ |
| MW1885 | sak | Staphylokinase | 5.16 | 5.86 | 1.14 | 1.50 | 0.26+ |
| MW2341 | sbi | IgG binding | 46.74 | 40.99 | 0.88 | 15.80 | 0.39+ |
| MW0084 | spa | Protein A | 2347.95 | 143.31 | 0.06** | 93.82 | 0.65+ |
| MW0130 | capG | Capsule | 0.18 | 2.25 | 12.75* | 0.58 | 0.26++ |
| MW2586 | icaA | Intercellular adhesin | 0.14 | 0.21 | 1.55 | 0.13 | 0.60++ |
| MW0516 | sdrC | Fibrinogen binding | 0.22 | 0.18 | 0.79 | 0.05 | 0.29+++ |
| MW0764 | clfA | Clumping factor | 9.15 | 54.36 | 5.94* | 19.21 | 0.35+++ |
| MW2421 | fnb | Fibronectin biding | 3.10 | 5.06 | 1.63** | 8.27 | 1.63 |
| MW1884 | SCIN | Complement inhibitor | 9.86 | 53.69 | 5.45** | 13.62 | 0.25+++ |
| MW0088 | sirA | Iron acquisition | 0.78 | 13.99 | 17.98* | 0.17 | 0.01+ |
| MW0094 | sbnF | Iron acquisition | 0.05 | 8.23 | 167.55** | 0.04 | 0.00++ |
| MW1014 | isdD | Iron acquisition | 0.32 | 14.91 | 46.98* | 0.25 | 0.02+ |
GeneID in S. aureus strain MW2.
Expression ratio to gyrA (×100) at exponential phase.
Significant changes of expression of S. aureus grown in CDM(−) compared with TSB as determined by Student's t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Significant changes of expression of S. aureus grown in CDM(−) containing 3 μM FeCl3 compared with CDM(−) as determined by Student's t test; +, P < 0.05; ++, P < 0.01; +++, P < 0.001.
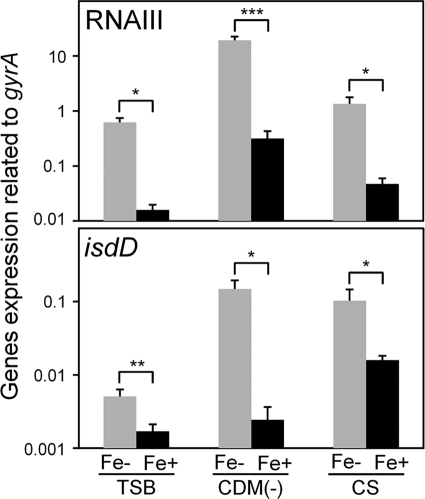
We also investigated the expression of virulence factors grown in TSB treated with Chelex-100 (see Table S2 in the supplemental material). The expression of many virulence factors, including RNA III, was increased compared with that in TSB without Chelex-100, showing an expression pattern similar to those grown in CS or CDM(−), although some factors showed different expression patterns under the three conditions (see Table S2 in the supplemental material). In Fig. 7, the amounts of expression of RNA III and isdD in TSB with Chelex-100, CDM(−), and CS can be seen to be higher than in TSB, CDM(−) with 3 μM FeCl3, and CS with 300 μM FeCl3. The expression patterns of virulence factors were similar under the three iron-depleted conditions [TSB with Chelex-100, CDM(−), and CS] or conditions with iron supplied [TSB, CDM(−) with 3 μM FeCl3, and CS with 300 μM FeCl3]. In addition, the expression patterns were quite different under iron-depleted and iron-supplied conditions, although the expression levels for some factors were different under the three conditions (Fig. 7 and Tables 2 and 4; see Fig. S2 in the supplemental material).
Fig. 7.
Expression of RNA III and isdD in iron-depleted and -supplied media. S. aureus MW2 cells cultured in iron-depleted media [TSB with Chelex-100, CDM(−), and CS] or in iron-supplied media [TSB, CDM(−) with 3 μM FeCl3, and CS with 300 μM FeCl3] were collected at the exponential phase. The expression of RNA III and isdD was determined by quantitative PCR by the method described in Materials and Methods. The data shown represent means and SD of measurements performed in triplicate. The asterisks indicate significant decreases compared with iron-limited medium as determined by Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Expression of virulence factors in a mutant with agr inactivated.
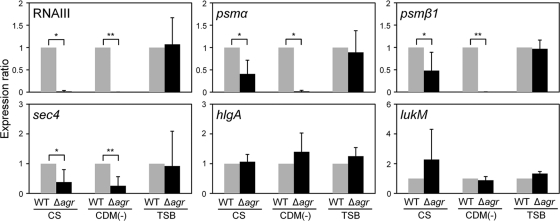
We investigated the expression of virulence factors in a mutant with agr inactivated grown in CS or CDM(−) (Fig. 8; see Table S3A and B in the supplemental material). Among the factors that were expressed significantly in CS, the expression of RNA III, psm, and sec4 in the mutant with agr inactivated was not increased, while the expression of other factors was still increased almost as much as that of the wild type grown in CS (see Table S3A in the supplemental material). We also investigated the expression of virulence factors in the agr mutant grown in CDM(−) (see Table S3B in the supplemental material) and found results similar to those with CS.
Fig. 8.
Effect of agr inactivation on the expression of virulence factors. S. aureus MW2 and the mutant with agr inactivated were cultured in TSB, CS, and CDM(−). These cells were collected at the exponential phase. The expression of virulence factors was determined by quantitative PCR by the method described in Materials and Methods. The data shown represent means and SD of measurements performed in triplicate. The asterisks indicate significant decreases compared with the wild type as determined by Student's t test: *, P < 0.05; **, P < 0.01.
Effects of serum albumin on the expression of virulence factors in CDM with iron.
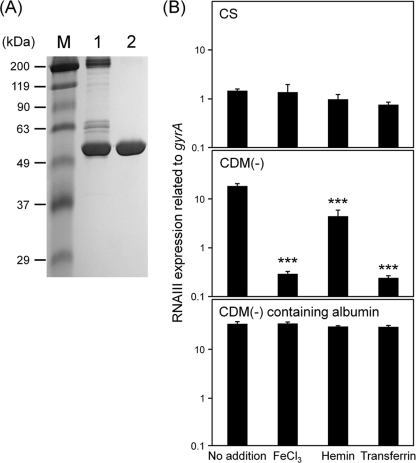
Serum albumin was purified from CS by affinity chromatography. The purified albumin showed a single band in a 7.5% polyacrylamide gel (Fig. 9A). In CDM(−) containing purified albumin at 20 mg/ml (almost a 50% concentration of the albumin in serum), the addition of a low concentration of FeCl3 (3 μM) did not decrease the expression of RNA III, showing a result similar to that with CS (Fig. 9B). This effect was also found when holotransferrin or hemin was added to CDM(−) containing serum albumin. Furthermore, other virulence factors, including iron acquisition genes, were not decreased (or the decrease was suppressed) by addition of 3 μM iron sources, including FeCl3 in CDM(−) containing albumin, compared with those with 3 μM iron sources in CDM(−) (see Fig. S2 in the supplemental material). The expression of isdD in CDM(−) containing albumin was decreased by the addition of 3 μM FeCl3, but its level of decrease (a 2-fold decrease) was significantly lower than that (a 53-fold decrease) in CDM(−) with 3 μM FeCl3.
Fig. 9.
Effect of serum albumin on the expression of RNA III. (A) Serum albumin from calf serum was purified using ToyoPearl AF-Blue HC-650 and then added to CDM(−). Small portions of CS and CDM(−) containing purified albumin were subjected to SDS-PAGE using a 7.5% polyacrylamide gel and then stained with Coomassie brilliant blue. M, size marker; lane 1, CS (3 μg); lane 2, CDM(−) containing purified albumin from CS (2 μg). (B) S. aureus MW2 cells cultured in CS, CDM(−), and CDM(−) containing purified serum albumin from CS, with or without 3 μM iron sources, were collected at the exponential phase. The expression of RNA III was determined by quantitative PCR by the method described in Materials and Methods. The data shown represent means and SD of measurements performed in triplicate. The asterisks indicate significant decreases compared with no addition of an iron source as determined by Dunnett's test: ***, P < 0.001.
DISCUSSION
In this study, we showed that many virulence factors, including hemolysins, leukocidins, exoenzymes, and adhesins, were expressed significantly when S. aureus was grown in CS. Since we observed similar results using calf, rabbit, and human sera, it is considered that this effect is common among mammalian sera. Our results in this study indicated that depletion of free iron is strongly associated with high expression of virulence factors in serum. Generally, the iron source in serum is transferrin, although blood contains a high concentration of Fe(II) as a heme in hemoglobin. Therefore, free iron is very rare in serum, with the concentration of free iron in serum estimated to be 10−24 M (30). Because iron is an essential element for bacteria, including S. aureus, bacteria, especially pathogenic bacteria, need to scavenge free iron or iron-binding proteins, including hemin, lactoferrin, and transferrin, from host tissues or fluids (14). To counter the sequestration of free iron, bacteria produce high-affinity iron chelators called siderophores. S. aureus produces several siderophores (staphyloferrins A and B, staphylobactin, and aureochelin) to bind iron and then transports the iron-siderophore complexes to the cytoplasm via ATP-binding cassette (ABC) transporters (2, 6, 8, 14, 32). In S. aureus, ABC transporters encoded by sirABC, sitABC, fhuCBG, and sstABCD were demonstrated to transport the complexes. Furthermore, S. aureus has a system encoded within the isd locus to scavenge iron from heme (23). In this study, we found increased expression of the factors involved in iron acquisition in serum compared with that in TSB, implying that the serum used in this study contained little iron. Therefore, it is reasonable that not only factors for iron acquisition, but also factors (toxins and exoenzymes) for the destruction of erythrocytes, leukocytes, and host tissues to capture iron, were upregulated in S. aureus under iron-restricted conditions like those in serum.
Among the factors expressed significantly in serum or CDM(−), RNA III expression was markedly enhanced. Since RNA III is thought to be a global regulator for many virulence factors (5, 26), we focused on its regulation. Generally, it has been demonstrated that RNA III expression is tightly associated with agr, a quorum-sensing system in S. aureus (5, 26). In our study, agr expression was increased significantly in serum or CDM(−) (data not shown), and this increase was suppressed by the addition of iron. In addition, in a mutant with agr inactivated, the expression of RNA III was not increased in CS. These results strongly suggested that high expression of RNA III in CS was caused by high expression of agr. In this study, we showed the expression of virulence factors in CS at the transcriptional level, not the translational level. Since the mechanisms of RNA III regulation for the expression of several virulence factors occur at the posttranscriptional level by RNA-RNA interaction, it is considered that a significant increase of RNA III affects the expression of virulence factors at the protein level. In the mutant with agr inactivated, the expression of many virulence factors, except hld, psm, and sec4, was still high in CS and CDM(−) at the transcriptional level, showing the same levels of expression as the wild type. These results indicate that factors other than agr and/or RNA III are also associated with the transcriptional regulation of virulence factors in CS.
Recently, Queck et al. reported an agr-dependent but RNA III-independent gene regulation mechanism in S. aureus (29). They demonstrated that many virulence factors, such as leukocidins, serine proteases, and protein A, were regulated by RNA III through agr but that psm expression was directly regulated by agr, not through RNA III. In our study, the expression of psm, sec4, and RNA III was drastically increased in CS and CDM(−) compared with that of other virulence factors, and this increase was not observed only in the agr mutant, showing that these 3 factors exhibited responses that differed from those of other virulence factors. Therefore, it is considered that the expression of sec4, together with psm and RNA III, was directly regulated by agr, although the mechanism of agr upregulation in serum remains unknown. To determine whether other regulators were involved in the expression of virulence factors in CS, we investigated the expression of all TCSs in CS or CDM(−). The expression of many TCSs was increased in CS compared with that in TSB (data not shown), while only agr expression was increased in CDM(−). However, we investigated the expression of several virulence factors in all TCSs, except mutants with vicRK inactivated, and found no difference between the wild type and each TCS mutant (data not shown). These results indicate that another factor(s) besides TCSs is involved in the increased expression of virulence factors, except hld, psm, and sec4 in CS.
A ferric uptake regulator (Fur) is a repressor of iron acquisition system binding to upstream regions of iron acquisition factor genes when iron is abundant (35). In S. aureus, Fur regulates the factors, including isd, involved in iron acquisition. Torres et al. reported that Fur affected the expression of cytolysins (increased expression with fur inactivation) and a subset of immune-modulatory factors (decreased with fur inactivation), although the Fur box, which is the consensus sequence for binding Fur, is not present in the upstream regions of the genes coding for their factors (34). To investigate the association of Fur with the expression of virulence factors in CS, we attempted to construct a mutant with fur inactivated but were unsuccessful for an unknown reason. Further study is needed to resolve the association of Fur with the expression of virulence factors in CS.
Of 7 clinical isolates tested, 6 strains showed a response similar to that of MW2, with increased expression of RNA III in CS; this increase was suppressed by the addition of iron, although the levels of increase were quite different among strains. However, one strain, TY526, did not increase RNA III expression in CS, although RNA III expression in TSB was quite high compared with those of other strains. These results indicate that the expression of virulence factors in CS is variable among strains.
In addition, we found that serum albumin captured free iron to inhibit bacterial acquisition of free iron. Previously, it was reported that serum albumin had binding affinity to free iron, heme, or transferrin (15, 36). However, this is the first report to demonstrate that iron capture by serum albumin affects the expression of a virulence factor(s) in S. aureus. Serum albumin is the most abundant protein in the blood and has a structural domain for heme binding (15). Purified albumin from calf serum suppressed the increase of RNA III expression in spite of the addition of FeCl3, holotransferrin, and hemin, suggesting its affinity for these molecules. Since S. aureus causes bacteremia, it can clearly grow and survive in the bloodstream by utilizing several iron-scavenging systems. In addition, in this study, S. aureus produced a high level of cytolysins, including hemolysins, to disrupt and sequester iron from host tissues or cells. Therefore, iron scavenging between the host and parasite in blood might be one of the critical factors involved in S. aureus infection.
In conclusion, we showed that many virulence factors were expressed significantly upon growth in serum. Increased expression of agr was associated with the expression of some virulence factors, but another regulatory factor(s) is also involved. Our findings in this study suggest that the expression pattern of virulence factors of S. aureus upon infection of a host is quite different from that upon growth in bacterial media.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (no. 19041050) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Baba T., et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 2. Beasley F. C., et al. 2009. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol. Microbiol. 72:947–963 [DOI] [PubMed] [Google Scholar]
- 3. Berman D. S., Eisner W., Kreiswirth B. 1993. Community-acquired methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 329:1896. [DOI] [PubMed] [Google Scholar]
- 4. Burian M., et al. 2010. Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J. Infect. Dis. 201:1414–1421 [DOI] [PubMed] [Google Scholar]
- 5. Cheung A. L., Bayer A. S., Zhang G., Gresham H., Xiong Y. Q. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1–9 [DOI] [PubMed] [Google Scholar]
- 6. Cheung J., Beasley F. C., Liu S., Lajoie G. A., Heinrichs D. E. 2009. Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol. Microbiol. 74:594–608 [DOI] [PubMed] [Google Scholar]
- 7. Chevalier C., et al. 2010. Staphylococcus aureus RNA III binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 6:e1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dale S. E., Doherty-Kirby A., Lajoie G., Heinrichs D. E. 2004. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 72:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deleo F. R., Otto M., Kreiswirth B. N., Chambers H. F. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deurenberg R. H., et al. 2007. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 13:222–235 [DOI] [PubMed] [Google Scholar]
- 11. Foster T. J. 2004. The Staphylococcus aureus “superbug”. J. Clin. Invest. 114:1693–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foster T. J. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948–958 [DOI] [PubMed] [Google Scholar]
- 13. Geisinger E., Adhikari R. P., Jin R., Ross H. F., Novick R. P. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 261:1038–1048 [DOI] [PubMed] [Google Scholar]
- 14. Genco C. A., Dixon D. W. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1–11 [DOI] [PubMed] [Google Scholar]
- 15. Grinberg L. N., O'Brien P. J., Hrkal Z. 1999. The effects of heme-binding proteins on the peroxidative and catalytic activities of hemin. Free Radic. Biol. Med. 27:214–219 [DOI] [PubMed] [Google Scholar]
- 16. Grundmann H., Aires-de-Sousa M., Boyce J., Tiemersma E. 2006. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–875 [DOI] [PubMed] [Google Scholar]
- 17. Huntzinger E., et al. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24:824–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hussain M., Hastings J. G., White P. J. 1991. A chemically defined medium for slime production by coagulase-negative staphylococci. J. Med. Microbiol. 34:143–147 [DOI] [PubMed] [Google Scholar]
- 19. Liu Y., et al. 2011. RNAIII activates map expression by forming an RNA-RNA complex in Staphylococcus aureus. FEBS Lett. 585:899–905 [DOI] [PubMed] [Google Scholar]
- 20. Lowy F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 21. Manders S. M. 1998. Toxin-mediated streptococcal and staphylococcal disease. J. Am. Acad. Dermatol. 39:383–398 [DOI] [PubMed] [Google Scholar]
- 22. Matsuo M., et al. 2010. Distinct two-component systems in methicillin-resistant Staphylococcus aureus can change the susceptibility to antimicrobial agents. J. Antimicrob. Chemother. 65:1536–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mazmanian S. K., et al. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906–909 [DOI] [PubMed] [Google Scholar]
- 24. Montgomery C. P., et al. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J. Infect. Dis. 198:561–570 [DOI] [PubMed] [Google Scholar]
- 25. Morfeldt E., Taylor D., von Gabain A., Arvidson S. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14:4569–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Novick R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 27. Novick R. P., Geisinger E. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541–564 [DOI] [PubMed] [Google Scholar]
- 28. Que Y. A., et al. 2005. Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J. Exp. Med. 201:1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Queck S. Y., et al. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raymond K. N., Dertz E. A., Kim S. S. 2003. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U. S. A. 100:3584–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rooijakkers S. H., van Kessel K. P., van Strijp J. A. 2005. Staphylococcal innate immune evasion. Trends Microbiol. 13:596–601 [DOI] [PubMed] [Google Scholar]
- 32. Sebulsky M. T., Heinrichs D. E. 2001. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183:4994–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Somerville G. A., Proctor R. A. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 73:233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Torres V. J., et al. 2010. Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 78:1618–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xiong A., Singh V. K., Cabrera G., Jayaswal R. K. 2000. Molecular characterization of the ferric-uptake regulator, Fur, from Staphylococcus aureus. Microbiology 146:659–668 [DOI] [PubMed] [Google Scholar]
- 36. Xu X., Zhang L., Shen D., Wu H., Liu Q. 2008. Oxygen-dependent oxidation of Fe(II) to Fe(III) and interaction of Fe(III) with bovine serum albumin, leading to a hysteretic effect on the fluorescence of bovine serum albumin. J. Fluoresc. 18:193–201 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.