Abstract
Broad-host-range plasmids are known to spread genes between distinct phylogenetic groups of bacteria. These genes often code for resistances to antibiotics and heavy metals or degradation of pollutants. Although some broad-host-range plasmids have been extensively studied, their evolutionary history and genetic diversity remain largely unknown. The goal of this study was to analyze and compare the genomes of 12 broad-host-range plasmids that were previously isolated from Norwegian soils by exogenous plasmid isolation and that encode mercury resistance. Complete nucleotide sequencing followed by phylogenetic analyses based on the relaxase gene traI showed that all the plasmids belong to one of two subgroups (β and ε) of the well-studied incompatibility group IncP-1. A diverse array of accessory genes was found to be involved in resistance to antimicrobials (streptomycin, spectinomycin, and sulfonamides), degradation of herbicides (2,4-dichlorophenoxyacetic acid and 2,4-dichlorophenoxypropionic acid), and a putative new catabolic pathway. Intramolecular transposition of insertion sequences followed by deletion was found to contribute to the diversity of some of these plasmids. The previous observation that the insertion sites of a Tn501-related element are identical in four IncP-1β plasmids (pJP4, pB10, R906, and R772) was further extended to three more IncP-1β plasmids (pAKD15, pAKD18, and pAKD29). We proposed a hypothesis for the evolution of these Tn501-bearing IncP-1β plasmids that predicts recent diversification followed by worldwide spread. Our study increases the available collection of complete IncP-1 plasmid genome sequences by 50% and will aid future studies to enhance our understanding of the evolution and function of this important plasmid family.
INTRODUCTION
Plasmids play a crucial role in the evolution and adaptation of prokaryotes. They typically have mosaic genomes consisting of two distinct regions (34): a backbone region that carries genes essential for plasmid persistence and one or more accessory regions that encode host-beneficial traits. The so-called broad-host-range (BHR) plasmids are of particular importance in rapid bacterial adaptation to changing environments, because they can transfer to and replicate in phylogenetically diverse bacteria at high frequencies and because they often carry host-beneficial traits such as resistances to antibiotics or heavy metals or degradation of organic compounds. In light of the recent emergence of multidrug-resistant pathogens as an acute human health crisis, the study of the diversity and evolutionary history of BHR plasmids has become imperative.
The most promiscuous plasmids of Gram-negative bacteria, able to transfer themselves and replicate in bacteria from two or more classes of Proteobacteria, belong to incompatibility groups IncP, IncU, and IncW (7, 11) and the recently defined PromA group (39). Complete sequences of 25 IncP-1, 4 IncW, 4 PromA, and 2 IncU plasmids are currently available in GenBank. With 25 members, the IncP-1 plasmids are the best-represented and best-studied group of BHR plasmids. This group, initially consisting of only two subgroups, α and β (16, 33), has been expanded over the last 5 years to include at least three (γ, δ, and ε) (2, 13, 41) and possibly five (19; D. Sen, M. L. Bauer, L. M. Rogers, G. A. Van der Auwera, C. J. Brown, and E. M. Top, unpublished data) more subgroups.
In the mid-1990s, a set of plasmids was isolated from Norwegian soils after the soil samples were first treated in the laboratory with mercuric chloride (HgCl2) (8, 9). Two approaches to exogenous plasmid isolation were used, biparental and triparental matings. Biparental matings typically aim at capturing plasmids that encode a specific phenotypic trait by mixing the indigenous bacterial community from an environmental sample with a plasmid-free recipient strain, allowing conjugation to occur, and selecting for recipients that acquired the specific trait (here mercury resistance). Triparental matings involve one additional parental population, i.e., a donor strain carrying a nonconjugative, mobilizable plasmid (Tra− Mob+), and plasmids are captured based solely on their ability to transfer the mobilizable plasmid and themselves into the recipient. In both cases, the transconjugants are then examined for the presence of captured conjugative plasmids. Of the so-called pAKD plasmids (pAKD1 to pAKD34) captured with these methods, 19 had been found to have a broad host range based on their ability to transfer and replicate in different classes of Proteobacteria, and all were shown to confer resistance to mercuric chloride. With the exception of one plasmid, which hybridized to a probe for the IncP-1β subgroup, lack of hybridization of these plasmids with DNA probes known to detect specific incompatibility groups suggested that they belonged to new groups of BHR plasmids (8, 9).
To increase our understanding of the genetic diversity of BHR plasmids, we determined the complete nucleotide sequences of 13 of these 19 BHR plasmids from three Norwegian soils. Based on genomic comparisons and phylogenetic analyses, we found that in contrast to the earlier hybridization results, all plasmids belong to three subgroups of the IncP-1 group (β, δ, and ε). Since we have already published a detailed analysis of the IncP-1δ plasmid pAKD4 (27), this study presents a comparative analysis of the remaining 12 pAKD plasmids and a preliminary characterization of their accessory regions. Since there are so far only 26 completely sequenced IncP-1 plasmids available to the public, this study increases this group by nearly 50%. The data not only reveal new accessory genes but also help to define the interesting molecular events that took place during the evolution of these plasmids.
MATERIALS AND METHODS
Bacterial strains and plasmids used.
Table 1 provides a list of bacterial strains and control plasmids used in this study. The pAKD plasmids were kindly provided by K. Drønen (8, 9).
Table 1.
Bacterial strains and plasmids used in this study
Plasmid DNA extractions.
All plasmids were extracted from the strains in which they were received; thus, pAKD4, pAKD17, and pAKD18 were extracted from Pseudomonas putida UWC1 and the other plasmids were extracted from Escherichia coli HB101. Plasmid DNA to be used for sequence determination was isolated using the plasmid midikit (Qiagen, Valencia, CA) according to the manufacturer's instructions for low-copy-number plasmids. Plasmid DNA extractions for additional analyses were obtained using the alkaline lysis method (23). Standard methods were used for restriction analysis and gel electrophoresis (23). In all cases, EcoRI digestion of the plasmid DNA that was sequenced yielded the same restriction patterns as those observed previously by Drønen et al. (8, 9).
Sequencing and annotation.
The DNA sequences of all plasmids were determined by shotgun sequencing at the DOE Joint Genome Institute (Walnut Creek, CA). Briefly, after construction of ∼3-kb clone libraries, Sanger technology was used to determine the DNA sequences of 384 clones in both directions. These sequences were then assembled using PGA (42), and gaps were closed by primer walking. Automatic annotation was carried out by the J. Craig Venter Institute Annotation Service (http://www.jcvi.org/cms/research/projects/annotation-service/overview/) followed by manual annotation by the authors.
Bioinformatics analyses and software.
GenBank (http://ncbi.nlm.nih.gov) was searched for similar sequences using BLAST (1). Blastp searches of all hypothetical proteins were carried out against the Interpro database (http://www.ebi.ac.uk/interpro/), a database of protein families, domains, regions, repeats, and sites in which identifiable features found in known proteins can be applied to new protein sequences. The alignment figure of the pAKD plasmids was generated using Mauve (6), ClustalW (36), and TRAPPIST (G. Van der Auwera, unpublished data). The TRAPPIST toolbox was used to perform multiple alignments with Mauve, produce refined pairwise alignments between segments using ClustalW based on the Mauve output, and generate the graphical representation shown in Fig. 2. To infer the phylogenetic relationship of the pAKD plasmids with other sequenced IncP-1 plasmids, amino acid sequences of the relaxase protein (TraI) of 26 IncP-1 plasmids were aligned in ClustalX and this alignment was used to guide the alignment of nucleotide sequences in Tranalign (22). A maximum likelihood phylogeny based on this alignment was inferred using RAxML (30). Accession numbers of the IncP-1 plasmids included in the phylogenetic analysis are as follows: pQKH54 (NC_008055), pEST4011 (NC_005793), pIJB1 (DQ065837), pSP21 (CP002153), pB5 (CP002151), pB11 (CP002152), RK2 (NC_001621), pBS228 (NC_008357), pTB11 (NC_006352), pKJK5 (NC_008272), pA81 (NC_006830), pCNB (NC_010935), pB4 (NC_003430), pAOVO02 (NC_008766), pA1 (NC_007353), pB10 (NC_004840), pJP4 (NC_005912), pBP136 (NC_008459), pUO1 (NC_005088), pTP6 (NC_007680), Burkholderia cepacia AMMD plasmid 1 (pAMMD_1) (NC_008385), pADP-1 (NC_004956), pB3 (NC_006388), R751 (NC_001735), pB8 (NC_007502), and pAKD4 (GQ983559).
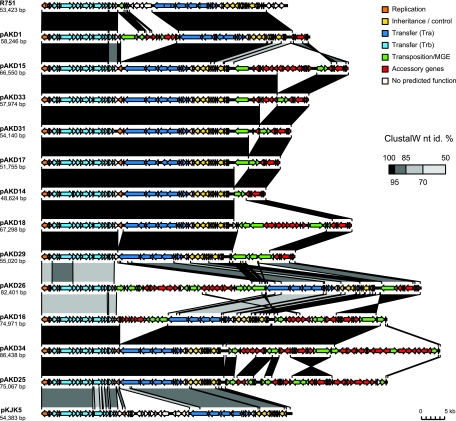
Fig. 2.
Alignment of 12 pAKD plasmids showing their evolutionary relationship to each other and to the well-known IncP-1 plasmids R751 and pKJK5. Coding regions are shown as colored arrows; putative functions are indicated by the color key (top right). The degree of similarity between plasmids (percent nucleotide identity of alignments performed using ClustalW) is indicated by grayscale-shaded regions; the darker the shading between two segments, the higher their similarity, as shown in the heat key (middle right).
Copy number analysis.
SYBR-green-based quantitative PCR (qPCR) was used to compare copy numbers of pAKD16 and pAKD34 as described previously (29). The plasmids were transferred to EC100 (Invitrogen, Carlsbad, CA). The cells were grown overnight in LB supplemented with HgCl2 (2.5 μg/ml) at 37°C and then transferred into fresh LB medium and grown into the exponential phase. The copy number of a 90-bp region within the trbB gene was used to calculate plasmid copy number, while chromosomal copy number was calculated for a 167-bp region within the atpB gene of the EC100 chromosome, close to oriC. The control vector containing a 1:1 ratio of plasmid and chromosomal genes was constructed by cloning trbB-trbE and atpB into pHSG399 (32). Primers used for qPCR were trbBF, 5′-CTCGTTTGAAGTCGCTTGTT-3′; trbBR, 5′-TTTCCTGGATACGACGAGAG-3′; atpBF, 5′-GTCGGTCCAGGTCTTCATTT-3′; and atpBR, 5′-TGCACACGGTAATCTGGAAT-3′. The ratio of plasmid to chromosomal gene copies was used as a proxy for plasmid copy number.
Antimicrobial resistance tests.
Plasmid pAKD1 was tested for conferring resistance to streptomycin, spectinomycin, and sulfonamide. Because the original host, E. coli HB101, was resistant to streptomycin and spectinomycin, the plasmid was first transferred from E. coli HB101 to E. coli CV601::gfp (supplied by K. Smalla) using a biparental mating as described previously (27). Selection for transconjugants was performed on LBA medium supplemented with 5 μg/ml of mercuric chloride and 150 μg/ml of rifampin. The presence of pAKD1 in transconjugants was confirmed after plasmid DNA extraction from randomly selected colonies using the alkaline lysis method and visualization of plasmid DNA on a 0.75% agarose gel (23). E. coli CV601::gfp(pAKD1) was tested for resistance to the three antimicrobial compounds. Plates were prepared with Luria-Bertani medium, solidified with 15 g/liter agar, and supplemented with 25, 50, and 100 μg/ml of streptomycin; 25, 50, and 100 μg/ml of spectinomycin; or 200 μg/ml of the sulfonamide sulfathiazole. Overnight cultures of E. coli CV601::gfp with and without pAKD1 were streaked onto plates, incubated at 37°C, and observed for colony growth.
Test for degradation of 2,4-D.
The tfd gene-bearing plasmids pAKD25 and pAKD26 were tested for their ability to confer degradation of 2,4-dichlorophenoxyacetic acid (2,4-D) to their bacterial hosts. Biparental matings were used to transfer both plasmids from E. coli HB101 to Cupriavidus pinatubonensis JMP228n (37) as previously described (27). Transconjugants were selected on LBA plates supplemented with 5 μg/ml of mercuric chloride and 100 μg/ml of nalidixic acid, and plasmid presence was confirmed as described above. Single colonies of JMP228n(pAKD25) and JMP228n(pAKD26) were isolated and grown overnight in LB. Cells were pelleted and washed with MMO medium (31), and 107 cells were inoculated in liquid MMO medium supplemented with 250, 500, and 1000 μg/ml of 2,4-D. JMP228n harboring pEMT1::Km (37) was used as a positive control, and plasmid-free JMP228n was used as a negative control. All cultures were grown at 30°C with constant shaking at 200 rpm. Turbidity of cultures was monitored daily for 6 days.
Test for degradation of 2,4-dichlorophenoxypropionic acid.
Plasmid pAKD34 was transferred to JMP228n by biparental mating as described above. Transconjugants were selected on LBA plates supplemented with 5 μg/ml of mercuric chloride and 100 μg/ml of nalidixic acid (31), and plasmid presence was confirmed as described above. A single colony of JMP228n(pAKD34) was inoculated in LB overnight, cells were pelleted and washed with MMO medium (31), and 107 cells were inoculated in liquid MMO medium supplemented with 250 μg/ml of racemic 2,4-dichlorophenoxypropionic acid (also called dichlorprop; Sigma-Aldrich, San Diego, CA). Turbidity of cultures was monitored for 5 days. Triplicate measurements of optical density (at 600 nm) and dichlorprop absorbance (at 280 nm) were recorded on days 2 and 5 for JMP228n(pAKD34) and plasmid-free JMP228n with a spectrophotometer. To measure absorbance, 1 ml of culture was centrifuged and the supernatant was transferred to a UV cuvette. Dilutions of a known concentration of dichlorprop solution in MMO were used as standards to calculate concentrations of dichlorprop in unknown samples.
Antimicrobial resistance tests.
Plasmid pAKD1 was tested for conferring resistance to streptomycin, spectinomycin, and sulfonamide. Because the original host, E. coli HB101, was resistant to streptomycin and spectinomycin, the plasmid was first transferred from E. coli HB101 to E. coli CV601::gfp (supplied by K. Smalla) using a biparental mating as described previously (27). Selection for transconjugants was performed on LBA medium supplemented with 5 μg/ml of mercuric chloride and 150 μg/ml of rifampin. The presence of pAKD1 in transconjugants was confirmed after plasmid DNA extraction from randomly selected colonies using the alkaline lysis method and visualization of plasmid DNA on a 0.75% agarose gel (23). E. coli CV601::gfp(pAKD1) was tested for resistance to the three antimicrobial compounds. Plates were prepared with Luria-Bertani medium, solidified with 15 g/liter agar, and supplemented with 25, 50, and 100 μg/ml of streptomycin; 25, 50, and 100 μg/ml of spectinomycin; or 200 μg/ml of the sulfonamide sulfathiazole. Overnight cultures of E. coli CV601::gfp with and without pAKD1 were streaked onto plates, incubated at 37°C, and observed for colony growth.
Nucleotide sequence accession numbers.
Complete nucleotide sequences of 12 pAKD plasmids have been submitted to GenBank, and their accession numbers are as follows: pAKD1 (JN106164), pAKD14 (JN106165), pAKD15 (JN106166), pAKD16 (JN106167), pAKD17 (JN106168), pAKD18 (JN106169), pAKD25 (JN106170), pAKD26 (JN106171), pAKD29 (JN106172), pAKD31 (JN106173), pAKD33 (JN106174), and pAKD34 (JN106175).
RESULTS AND DISCUSSION
General sequence features.
The complete nucleotide sequences of 12 plasmids previously isolated from three Norwegian soils (8, 9) were determined by Sanger sequencing. All plasmids have regions for replication (trfA, ssb, oriV), conjugative DNA transfer (tra, oriT), mating-pair formation (trb), and stable inheritance and central control (ctl). Based on their nucleotide sequences and gene content, they are typical IncP-1 plasmids, with accessory genes situated between oriV and trfA and/or between the tra and trb regions. This result was surprising given the previous report of lack of hybridization of all but pAKD1 to IncP-1 plasmid-specific probes (repPα and repPβ), leading to the earlier conclusion that they were novel BHR plasmids (8, 9). General features of these newly sequenced plasmids are listed in Table 2.
Table 2.
General features of the pAKD plasmids
| Plasmid | Soil of origina | Isolation methodb | Size (bp) | IncP-1 subgroup | Phenotypec |
|---|---|---|---|---|---|
| pAKD1 | StendS | BM | 58,246 | β | Hgr Spr Smr Sur |
| pAKD4d | StendS | BM | 56,803 | δ | Hgr |
| pAKD14 | Ås | BM | 48,624 | β | Hgr |
| pAKD15 | Ås | BM | 66,550 | β | Hgr |
| pAKD16 | Ås | TM | 74,971 | ε | Hgr |
| pAKD17 | Ås | BM | 51,755 | β | Hgr |
| pAKD18 | Ås | BM | 67,298 | β | Hgr |
| pAKD25 | Garpestad | BM | 75,067 | ε | Hgr |
| pAKD26 | Garpestad | BM | 82,401 | β | Hgr |
| pAKD29 | StendS | BM | 55,020 | β | Hgr |
| pAKD31 | Ås | BM | 54,140 | β | Hgr |
| pAKD33 | Ås | BM | 57,974 | β | Hgr |
| pAKD34 | Ås | BM | 86,438 | ε | Hgr |
Soil of origin—agricultural fields growing cereal and vegetables in StendS and Garpestad and only cereal in Ås.
Phenotype: phenotype confirmed. Hgr, mercury resistance; Smr, streptomycin resistance; Spr, spectinomycin resistance; Sur, sulfonamide resistance.
Previously published (27).
Phylogenetic and comparative genomic analysis of the pAKD plasmid backbones.
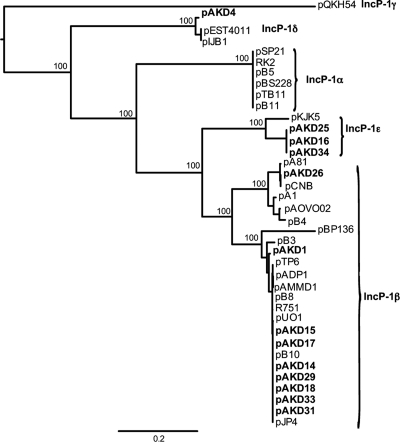
To analyze the phylogenetic relationship of the pAKD plasmids with 26 known IncP-1 plasmids, a maximum likelihood tree was inferred using RAxML (30) based on an alignment of the relaxase gene, traI (Fig. 1). This gene was chosen as a representative backbone gene since the relaxase protein has recently been proposed as a universal protein encoded on mobilizable plasmids (10). The tree shows that the 12 newly sequenced pAKD plasmids (shown in bold in Fig. 1) are diverse and belong to two of the five IncP-1 subgroups: IncP-1β and IncP-1ε. This same main tree topology was found when using the replication initiation gene trfA1 or several other backbone genes (data not shown). These plasmids are clearly distinct from the IncP-1δ plasmid pAKD4, which was isolated from one of the three soils in the same study and described previously (27).
Fig. 1.
Phylogenetic tree showing the relationship of the 12 pAKD plasmids described in this study and pAKD4 (27) (bold) with other completely sequenced IncP-1 plasmids. A maximum likelihood tree was inferred from nucleotide sequences of the relaxase gene, traI. Bootstrap values of the deep branches are shown to the left of each node. The scale bar represents the probability of nucleotide substitutions per site. Accession numbers for the plasmids used to construct the phylogenetic tree are given in Materials and Methods.
While eight pAKD plasmids (pAKD1, pAKD14, pAKD15, pAKD17, pAKD18, pAKD29, pAKD31, and pAKD33) cluster with the prototype IncP1-β plasmid R751 in the phylogenetic tree, pAKD26 falls within a separate smaller IncP1-β clade that contains multidrug resistance plasmid pB4, catabolic plasmids pCNB-1 and pA81, cryptic plasmid pA1, and so-far-uncharacterized plasmid pAOVO02, which encodes a putative oxidoreductase (Fig. 1). These two clades within the IncP-β subgroup have recently been designated β-1 and β-2 (18). Moreover, the IncP1-β plasmids from the two subclades are different not only in backbone gene sequence but also in gene content within the ctl region (kfrC to klcA). The plasmid alignment shown in Fig. 2 clearly indicates the lower sequence and gene content similarity between plasmid pAKD26 and the eight R751-like pAKD plasmids.
The phylogenetic tree also indicates that pAKD16, pAKD25, and pAKD34 belong to the recently proposed IncP-1ε group. This subgroup is so far represented by only a single, completely sequenced, published plasmid, prototype pKJK5, and a partially sequenced catabolic plasmid, pEMT3 (2, 12, 37). The addition of these three new IncP-1ε plasmid genome sequences increases the diversity of IncP-1ε plasmids, since their backbone genes are considerably divergent in sequence from those of pKJK5 (Fig. 1 and 2).
Of the three IncP-1ε plasmids, pAKD16 and pAKD34 are 100% identical in the entire backbone region except for their origins of replication (oriV). The oriV of pAKD34 appeared to have undergone a tandem duplication of 480 bp, resulting in 20 iterons instead of the 10 found on pAKD16. Since iterons are known to negatively regulate plasmid copy number (5) and have been directly implicated in IncP-1 copy number control (35), we tested whether pAKD34 had a lower copy number than that of pAKD16 in E. coli. As a proxy for plasmid copy number, we determined the ratio of plasmid-carried trbB to chromosomal atpB in exponential phase by qPCR. However, these ratios were not significantly different (0.39 ± 0.04 for pAKD34 and 0.38 ± 0.06 for pAKD16). The low values for both plasmids could be in part due to the presence of multiple chromosome replication forks in exponential phase, since the atpB gene is close to oriC. The findings suggest that the extra iterons of pAKD34 did not detectably affect its copy number.
Deletions in accessory regions of five IncP-1β plasmids.
We found deletion variants when comparing the accessory regions between oriV and trfA of five IncP-1β plasmids, pAKD14, pAKD15, pAKD17, pAKD31, and pAKD33, which were isolated from the same soil and have identical backbones. As shown in Fig. 3, they all share the same IS21-like element with intact ends and a remnant of a Tn501-like mercury resistance transposon in this accessory region, yet they differ in the number of genes present upstream of these mobile genetic elements. Plasmid pAKD15 has the longest region, composed of an IS1071-like element and a cluster of genes forming a putative novel catabolic pathway, while the other four plasmids seem to be deletion derivatives of pAKD15. While the sequence flanking the deletion at one end is always conserved and corresponds to the right inverted repeat (IRR) of the IS21-like element, the other end is variable between plasmids (Fig. 3). We therefore hypothesize that the plasmids were generated from pAKD15 or a similar ancestral plasmid by DNA rearrangements due to intramolecular transposition of IS21. Since we consider these five plasmids to be variants of each other, we will from here on refer to the most complete plasmid, i.e., pAKD15, as the representative of this group.
Fig. 3.
Alignment of accessory regions of five pAKD plasmids indicating deletions of various sizes relative to pAKD15. Arrows represent ORFs showing direction of transcription. The plasmids shown here have 100% identity along their backbone sequences but have deletions in their accessory regions, shown as black lines. All deletions are flanked by IS21 at the 3′ end, while sequences at the 5′ end are variable. The mercury resistance genes are carried on a Tn501-like transposon that is not intact. Also shown here is the putative novel catabolic pathway of pAKD15 (Table 3). Intergenic regions are not drawn to scale.
Deletions in the accessory region of IncP-1ε plasmids.
Similarly to the deletion variants found among the IncP-1β pAKD plasmids, we found a deletion variant among the IncP-1ε plasmids. The closely related plasmids pAKD16 and pAKD34 were isolated from the same soil (Ås) but using different procedures (triparental and biparental mating, respectively [Table 2]). The main differences between the plasmids are the following: (i) pAKD16 is missing nearly 20 kb of accessory genes present on pAKD34 between oriV and trfA and (ii) pAKD16 has a 10-kb transposon between the tra and trb regions that is not present on pAKD34. First, the loss of the 20-kb fragment may have occurred through recombination between two direct repeat sequences of tnpAISPps1 (orf57 and orf77 of pAKD34). It is more likely that the difference is a deletion in pAKD16 rather than an insertion in pAKD34 because the deleted region is part of a dichloropropionic acid degradation pathway. De novo addition of pathway genes is less likely than partial loss of an existing pathway. Second, the 10-kb transposon between tra and trb of pAKD16 is identical to Tn6048 in the genome of Cupriavidus metallidurans CH34. We postulate that, like plasmid pMOL98 (40), pAKD16 also captured Tn6048 from the recipient used in the triparental mating, which was strain AE815, a derivative of CH34. Thus, although exogenous plasmid isolation is an invaluable tool to obtain plasmids from uncultured bacteria, the potential of gene exchange between plasmid and recipient genome should be considered.
Mercury resistance transposons of the pAKD plasmids.
Mercury resistance genes were found on all 12 pAKD plasmids, as expected since all plasmids are known to confer mercuric chloride resistance (Table 2) (8, 9). The plasmids differ, however, in the transposons that carry the mercury resistance genes and in the location of these within the plasmid backbones. First, a Tn501 family transposon is located between oriV and trfA of the eight IncP-1β plasmids closely related to prototype R751 (pAKD1, pAKD15 and its four derivatives, pAKD18, and pAKD29); on pAKD1, however, the transposon may have inserted during an independent transposition event because of its reverse orientation and high nucleotide sequence divergence in the mer genes (7%). Second, another distinctly different Tn501 family transposon was found between tra and trb of the divergent IncP-1β plasmid pAKD26 and between oriV and trfA of the three IncP-1ε plasmids. The latter transposon differs from the former in the presence of an additional mer gene (merC) and high nucleotide sequence divergence (13%). These transposons were different from the Tn5053-like transposon found on the previously characterized IncP-1δ plasmid pAKD4 from the same plasmid collection (27). The different combinations of plasmid backbones and transposons clearly suggest various independent acquisition events.
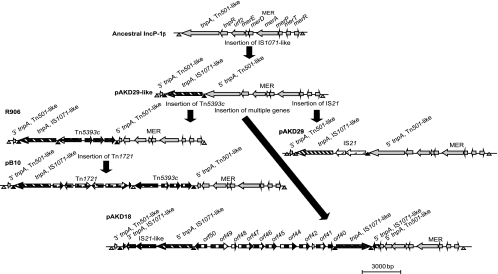
Hypothetical evolutionary pathway for the Tn501 group of IncP-1β plasmids.
The eight plasmids carrying the first type of Tn501-like transposon (all except pAKD1) are closely related to the four IncP-1β plasmids pB10, R906, R772, and pJP4 based not only on high nucleotide sequence identity (99 to 100%) in their backbone genes but also on the occurrence of a Tn501-like mercury transposon in exactly the same site (20, 25, 28). There are two possible hypotheses to explain this observation: (i) all plasmids share a recent common ancestor that already contained the Tn501-like transposon and, thus, that transposon inserted once into an ancestor of these plasmids, which subsequently gained different accessory genes, such as a Tn5393c transposon in pB10 or the tfd genes in pJP4, or (ii) the sequence between oriV and trfA in IncP-1β plasmids serves as a hot spot for the insertion of Tn501-like transposons, and this occurred multiple times in these seven plasmids. The high level of sequence identity found between the merA genes provides strong support for the first hypothesis. We therefore propose the following model for the evolution of some of these plasmids, as illustrated in Fig. 4. The ancestral IncP-1β plasmid with a Tn501-like insertion between oriV and trfA acquired an IS1071-related element in tnpATn501. This resulted in the formation of a pAKD29-like plasmid with 273 bp of the 3′ end of tnpA removed from its 5′ end. The pAKD29-like plasmid then underwent rearrangements to give rise to the following: (i) R906, through transposition of the streptomycin resistance carrying transposon Tn5393c into tnpATn501, at a site upstream of the previous insertion; (ii) pB10, formed from an R906-like plasmid by the acquisition of a tetracycline resistance transposon, Tn1721, which disrupted tnpAIS1071; (iii) pAKD29, formed from the pAKD29-like plasmid described above, through the acquisition of an IS21 family element within its IS1071-related element; and (iv) pAKD18, formed from the pAKD29-like plasmid by the insertion of multiple genes, orf50 to orf40. Plasmid R772 was probably not formed from the pAKD29-like ancestral plasmid, since the 3′ end of tnpATn501 is much longer (638 bp) and does not have an IS1071-like element near it. Formation of R772 by the insertion and subsequent disruption of the Tn501-like element by multiple Tn21-like elements has been recently hypothesized (20). Thus, although R772 is very similar to R906, pB10, and pJP4, it may have been formed through a different pathway. Because plasmids pAKD15 and pJP4 are missing tnpAR genes of Tn501, depriving us of crucial information regarding the context of insertion, it is difficult to hypothesize their evolutionary pathway. In any case, such high backbone conservation among these plasmids is surprising, considering that the plasmids were isolated from different parts of the world over a period of 30 years. This could be explained by a very recent (but at least 30-year-old) worldwide spread of these plasmids, with insufficient time for multiple mutations to accumulate, or strong purifying selection acting on the entire plasmid backbone, preventing sequence divergence.
Fig. 4.
Hypothetical evolutionary pathway for the Tn501 group of IncP-1β plasmids.
Catabolic pathways encoded on four plasmids.
Four of the 12 pAKD plasmids contain multiple catabolic genes, even though none was isolated from soil based on the ability to confer degradation of an organic compound. In line with several previously published IncP-1 plasmids (26, 38), at least three plasmids encode gene products known to be involved in the degradation of chlorinated aromatic compounds: (i) 2,4-dichlorophenoxyacetic acid (2,4-D) degradation (tfd genes on pAKD25 and pAKD26), (ii) chlorocatechol degradation (mocp genes on pAKD26), (iii) dichlorophenoxypropionic acid (dichlorprop) degradation (sdpA and rdpA genes on pAKD34), and (iv) a possible novel pathway of unknown function on pAKD15.
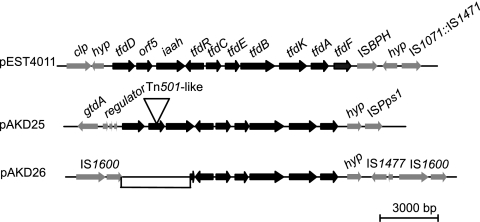
The two plasmids that carry 2,4-D degradation (tfd) genes, pAKD25 and pAKD26, were isolated from the same agricultural soil and yet belong to different IncP-1 subgroups, IncP-1ε and IncP-1β, respectively (Fig. 1). They carry these genes at different sites: between oriV and trfA on pAKD25 and between trb and tra on pAKD26. The identical gene organization and sequence identity of the tfd genes suggest that the two gene clusters are closely related to each other and to those found on the IncP-1δ plasmids pEST4011 (41) (Fig. 5) and pIJB1 (21) and less similar to the tfdI and tfdII clusters of the prototype 2,4-D catabolic plasmid pJP4. Both plasmids appear to lack tfdF, which encodes chloromaleylacetate reductase, the last plasmid-encoded step in the 2,4-D pathway. Moreover, the tfd clusters on both plasmids are disrupted by different insertions. On pAKD25, the insertion of a Tn501-like mercury transposon disrupted the open reading frame (ORF) between tfd and iaah. On pAKD26, an IS1600 element transposed into the tfdR end of the cluster, deleting a segment spanning tfdD, an uncharacterized ORF, and part of iaah. TfdD has been shown to be essential for the conversion of 2,4-dichloro-cis,cis-muconate to 2-chlorodiene lactone in the pJP4-encoded pathway. A simple growth assay indicated that plasmids pAKD25 and pAKD26 did not allow C. pinatubonensis JMP228n to utilize 2,4-D as sole carbon source when inoculated in liquid MMO supplemented with 2,4-D. No increase in turbidity was observed after 6 days, but both strains grew well in the same medium supplemented with acetate. In contrast, JMP228n(pEMT1::Km), previously shown to degrade 2,4-D (37), grew well with either acetate or 2,4-D. This lack of functionality of the tfd operons is likely due to the absence of tfdF from both plasmids and tfdD on pAKD26.
Fig. 5.
Comparison of tfd gene regions from pAKD25 and pAKD26 with pEST4011. The black arrows show tfd genes, while the gray arrows show flanking sequences. The tfd gene order is conserved among all three plasmids, but pEST4011 has the most complete tfd gene cluster, while both pAKD25 and pAKD26 have been interrupted and are missing tfdF. clp, chloride channel protein; hyp, hypothetical protein; gtdA, gentisate 1,2-dioxygenase; iaah, indole acetamide hydrolase. Intergenic regions are not drawn to scale.
The IncP-1β plasmid pAKD26 has a second chlorocatechol degradation cluster which shows 99% nucleotide sequence identity to a corresponding region of Bordetella petrii strain DSM 12804 (AM902716) and plasmid pA81 (15). On pA81, this cluster is called mocp and is involved in chlorocatechol degradation. Based on the very high sequence, we also designated the pAKD26 genes mocp. The two chlorocatechol degradation clusters on pAKD26 (tfd and mocp) have no sequence similarity to each other, suggesting that they have different evolutionary origins and were not generated by intramolecular duplication. Apparently, MocpB and MocpD were unable to substitute for the functionally equivalent proteins TfdD and TfdF, respectively.
The accessory region of pAKD34 between oriV and trfA contains genes that may be involved in degradation of the common herbicide dichlorprop, based on comparison with plasmid pMC1 from Delftia acidovorans (24). This region of pAKD34 is approximately 45 kb in size, of which a 20-kb segment shows 99% identity in nucleotide sequence to the corresponding sequence of pMC1. It encodes two dioxygenases, a transporter, a decarboxylase, a bacterioferritin, and a regulator. Just like on pMC1, the two dioxygenase-encoding genes, rdpA and sdpA, are also separated by a Tn501-like mercury resistance transposon that inserted into the selA gene downstream of sdpA on MC1, suggesting that the 20-kb fragments on the two plasmids share a recent common ancestor. A preliminary degradation assay showed no change in the concentration of dichlorprop or cell density in cultures of JMP228n(pAKD34) for at least 6 days, suggesting that the pathway is not functional in this host and needs further investigation.
The 24,640-bp-long accessory region of pAKD15 showed intermittent similarity when queried against nucleotide databases at NCBI, suggesting that this collection of genes had not been sequenced before. Additionally, preliminary characterization by similarity searches against the Interpro database showed that this region might encode a catabolic pathway. Components of this novel catabolic pathway are summarized in Table 3 (17). An identical pathway was also detected on another IncP-1β plasmid, pKSP18, isolated from the rhizosphere of sugar beets in Wales, United Kingdom (K. Smalla, H. Heuer, and E. M. Top, unpublished data). The presence of these two plasmids isolated from different countries suggests that this gene cluster encodes a catabolic pathway and does not just represent a random collection of genes. Further studies should enable us to determine what catabolic compounds are degraded by this pathway.
Table 3.
Amino acid similarities to proteins in GenBank and domains in Interpro for ORFs of the putative pAKD15 catabolic pathwaya
| ORF no. | % amino acid similarity | Best hit, accession no. | Domain(s) | PFAM/Interpro no. |
|---|---|---|---|---|
| Orf60 | 86 | Arsenical resistance protein, YP_002943872 | NADPH-dependent FMN reductase, arsenate resistance | PF03358, IPR014063 |
| Orf59 | 40 | Protein of unknown function DUF81, YP_002947552 | Protein of unknown function DUF81 | PF01925 |
| Orf57 | 31 | Methanesulfonate monooxygenase, hydroxylase beta subunit, YP_001020117 | Aromatic-ring-hydroxylating dioxygenase beta subunit | PF00866 |
| Orf55 | 39 | Putative dioxygenase subunit, YP_004417630 | Aromatic-ring-hydroxylating dioxygenase alpha subunit, aromatic-ring-hydroxylating dioxygenase 2Fe-2S-binding site, Rieske iron-sulfur domain | IPR001663, IPR015881, IPR017941 |
| Orf54 | 48 | Periplasmic isophthalate binding receptor, BAH70270 | Bordetella uptake gene | IPR0005064/PF03401 |
| Orf53 | 43 | Transcriptional regulator, IclR family, YP_003607133 | Transcriptional regulator IcIR N-terminal, winged HTH transcription repressor DNA-binding, transcriptional regulator IcIR C terminal | PF09339/IPR005471, IPR011991, IPR014757 |
| Orf52 | 68 | Phthalate 4,5-dioxygenase, YP_002947550 | NAD-binding domain, FAD-binding domain, 2Fe-2S cluster | PF00111 |
| Orf51 | 35 | Transcriptional regulator, MarR family protein, YP_002298705 | Transcriptional regulator HTH MarR type, winged HTH transcription repressor DNA binding | IPR000835, IPR011991 |
| Orf50 | 45 | Extracytoplasmic solute receptor, YP_725900 | Bordetella uptake gene | PF03401/IPR005064 |
| Orf49 | 45 | Rieske [2Fe-2S] domain protein, ZP_03542810 | Rieske [2Fe-2S] iron-sulfur domain | PF00355/IPR017941 |
| Orf48 | 44 | FAD-dependent oxidoreductase, YP_973874 | NAD(P)-binding domain, FAD/NAD(P)-binding domain | SSF51735, SSF51905 |
| Orf47 | 34 | ABC transporter, ZP_07673750 | NMT1/THI5-like | PF09084 |
| Orf43 | 39 | GntR family transcriptional regulator, YP_675818 | Transcriptional regulator HTH GntR, GntR C terminal, winged HTH transcription repressor DNA binding | IPR000524, IPR011711, IPR011991 |
Abbreviations: FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; HTH, helix-turn-helix.
Multiple catabolic pathways were found on the pAKD plasmids. Since each of the plasmids was isolated from agricultural soils treated with herbicides, these compounds may have selected for the catabolic pathways. Based on the limited information about the soil treatment histories (K. Drønen, personal communication), a link between the chemicals used and the catabolic pathways found on the plasmids is difficult to establish and therefore speculative. For example, plasmids pAKD25 and pAKD26 with the tfd genes were both isolated from soil Garpestad, a potato field likely treated with herbicides like Sencor (metribuzin, a triazinone) and Roundup (glyphosate), but also 2,4-D and related compounds such as 2-methyl-4-chlorophenoxyacetic acid (MCPA) have been widely applied in the past. Plasmids pAKD34, containing the dichlorprop degradation genes, and pAKD15, with the novel degradation pathway, were isolated from soil Ås, which was used to grow various crops, including cereals, and was likely treated with herbicides such as Roundup, Ramrod (propachlor or 2-chloro-N-isopropylacetanilide), and Banvel (also named dicamba or 3,6-dichloro-2-methoxybenzoic acid). These chlorinated aromatic compounds or their degradation by-products may well have been substrates for the putative catabolic pathways encoded on these plasmids.
Plasmid pAKD1 encoding antibiotic resistance.
The only plasmid that encoded antibiotic resistance in addition to mercury resistance is the IncP-1β plasmid pAKD1. It confers resistance to 25 to 100 μg/ml of spectinomycin, 25 to 50 μg/ml of streptomycin, and up to 200 μg/ml of the sulfonamide sulfathiazole to E. coli CV601. As shown in Fig. 6, this plasmid carries a multidrug resistance Tn21-like transposon with a class 1 integron Thus, even though pAKD1 was isolated from an agricultural soil based on its ability to confer resistance to HgCl2 only, it encodes several antibiotic resistance determinants, suggesting that mercury treatment might have coselected for the multidrug resistance genes. This would be in line with previous observations that pollution with heavy metals can cause a rise in the abundance of drug resistance genes (3).
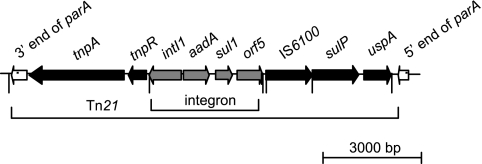
Fig. 6.
Antibiotic resistance-bearing Tn21-like transposon of pAKD1. This 11,920-bp transposon disrupted the resolvase (parA), resulting in separation of the 3′ and 5′ ends. It consists of transposition genes tnpA and tnpR, a class 1 integron, IS6100, a sulfate transporter (sulP), and a universal stress protein (uspA). The integron has an integrase (int1), an aminoglycoside adenyltransferase gene (aadA) encoding resistance to streptomycin and spectinomycin, a sulfonamide resistance gene (sul1), and a putative acetyltransferase (orf5).
In addition to our finding of new divergent IncP-1ε plasmids, a recent study defined a sixth subgroup (ζ) represented by plasmids from a marine biofilm (18), and plasmids from Neisseria gonorrhoeae were also suggested to belong to a new subgroup (19). Thus, our and other studies are showing that the IncP-1 group of promiscuous plasmids is very diverse in both backbone sequence and backbone and accessory gene content. Further work in this field is needed to understand the extent and implications of the diversity and evolutionary history of the IncP-1 and other BHR plasmid groups.
ACKNOWLEDGMENTS
This work was funded by National Science Foundation grant EF-0627988 to E.M.T. and C.J.B. and by National Institutes of Health COBRE NCRR grants P20RR16448 and P20RR016454 through INBRE and IBEST fellowships to D.S. via the BCB graduate program and in support of the bioinformatics core facility.
We are grateful to Kerrie Barry, Brian Foster, and Alla Lapidus at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI) for providing draft genome sequences, supported by the DOE Office of Science under contract no. DE-AC02-05CH11231. We also thank Karine Drønen for providing us with the 12 pAKD plasmids and information on soil treatment history, Kornelia Smalla for the recipient strains used in phenotypic tests, and JCVI for the JCVI Annotation Service, which provided us with automatic annotation data and the manual annotation tool Manatee.
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bahl M. I., Hansen L. H., Goesmann A., Sørensen S. J. 2007. The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established alpha, beta and delta sub-groups. Plasmid 58:31–43 [DOI] [PubMed] [Google Scholar]
- 3. Baker-Austin C., Wright M. S., Stepanauskas R., McArthur J. V. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176–182 [DOI] [PubMed] [Google Scholar]
- 4. Reference deleted.
- 5. Chattoraj D. K. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467–476 [DOI] [PubMed] [Google Scholar]
- 6. Darling A. C., Mau B., Blattner F. R., Perna N. T. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Datta N., Hedges R. W. 1972. Host ranges of R factors. J. Gen. Microbiol. 70:453–460 [DOI] [PubMed] [Google Scholar]
- 8. Drønen A. K., Torsvik V., Top E. M. 1999. Comparison of the plasmid types obtained by two distantly related recipients in biparental exogenous plasmid isolations from soil. FEMS Microbiol. Lett. 176:105–110 [Google Scholar]
- 9. Drønen A. K., Torsvik V., Goksøyr J., Top E. M. 1998. Effect of mercury addition on plasmid incidence and gene mobilizing capacity in bulk soil. FEMS Microbiol. Ecol. 27:381–394 [Google Scholar]
- 10. Garcillan-Barcia M. P., Francia M. V., de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657–687 [DOI] [PubMed] [Google Scholar]
- 11. Götz A., et al. 1996. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl. Environ. Microbiol. 62:2621–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gstalder M. E., et al. 2003. Replication functions of new broad host range plasmids isolated from polluted soils. Res. Microbiol. 154:499–509 [DOI] [PubMed] [Google Scholar]
- 13. Haines A. S., et al. 2006. Plasmids from freshwater environments capable of IncQ retrotransfer are diverse and include pQKH54, a new IncP-1 subgroup archetype. Microbiology 152:2689–2701 [DOI] [PubMed] [Google Scholar]
- 14. Heuer H., et al. 2002. Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol. Ecol. 42:289–302 [DOI] [PubMed] [Google Scholar]
- 15. Jencova V., et al. 2008. Nucleotide sequence, organization and characterization of the (halo)aromatic acid catabolic plasmid pA81 from Achromobacter xylosoxidans A8. Res. Microbiol. 159:118–127 [DOI] [PubMed] [Google Scholar]
- 16. Jobanputra R. S., Datta N. 1974. Trimethoprim R factors in enterobacteria from clinical specimens. J. Med. Microbiol. 7:169–177 [DOI] [PubMed] [Google Scholar]
- 17. Mason J. R., Cammack R. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277–305 [DOI] [PubMed] [Google Scholar]
- 18. Norberg P., Bergström M., Jethava V., Dubhashi D., Hermansson M. 2011. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pachulec E., van der Does C. 2010. Conjugative plasmids of Neisseria gonorrhoeae. PLoS One 5:e9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrovski S., Stanisich V. A. 2011. Embedded elements in the IncPβ plasmids R772 and R906 can be mobilized and can serve as a source of diverse and novel elements. Microbiology 157:1714–1725 [DOI] [PubMed] [Google Scholar]
- 21. Poh R. P., Smith A. R., Bruce I. J. 2002. Complete characterisation of Tn5530 from Burkholderia cepacia strain 2a (pIJB1) and studies of 2,4-dichlorophenoxyacetate uptake by the organism. Plasmid 48:1–12 [DOI] [PubMed] [Google Scholar]
- 22. Rice P., Longden I., Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Schleinitz K. M., Kleinsteuber S., Vallaeys T., Babel W. 2004. Localization and characterization of two novel genes encoding stereospecific dioxygenases catalyzing 2(2,4-dichlorophenoxy)propionate cleavage in Delftia acidovorans MC1. Appl. Environ. Microbiol. 70:5357–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schlüter A., et al. 2003. The 64,508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1beta group. Microbiology 149:3139–3153 [DOI] [PubMed] [Google Scholar]
- 26. Schlüter A., Szczepanowski R., Pühler A., Top E. M. 2007. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31:449–477 [DOI] [PubMed] [Google Scholar]
- 27. Sen D., et al. 2010. Comparative genomics of pAKD4, the prototype IncP-1δ plasmid with a complete backbone. Plasmid 63:98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith C. A., Thomas C. M. 1987. Comparison of the organisation of the genomes of phenotypically diverse plasmids of incompatibility group P: members of the IncP beta sub-group are closely related. Mol. Gen. Genet. 206:419–427 [DOI] [PubMed] [Google Scholar]
- 29. Sota M., et al. 2010. Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. ISME J. 4:1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 31. Stanier R. Y., Palleroni N. J., Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159–271 [DOI] [PubMed] [Google Scholar]
- 32. Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. 1987. High-copy-number and low-copy-number plasmid vectors for lacZαss-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63–74 [DOI] [PubMed] [Google Scholar]
- 33. Thomas C. M. 1981. Molecular genetics of broad host range plasmid RK2. Plasmid 5:10–19 [DOI] [PubMed] [Google Scholar]
- 34. Thomas C. M. 2000. Paradigms of plasmid organization. Mol. Microbiol. 37:485–491 [DOI] [PubMed] [Google Scholar]
- 35. Thomas C. M., Cross M. A., Hussain A. A., Smith C. A. 1984. Analysis of copy number control elements in the region of the vegetative replication origin of the broad host range plasmid RK2. EMBO J. 3:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson J., Higgins D., Gibson T. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Top E. M., Holben W. E., Forney L. J. 1995. Characterization of diverse 2,4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl. Environ. Microbiol. 61:1691–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Top E. M., Springael D., Boon N. 2002. Catabolic mobile genetic elements and their potential use in bioaugmentation of polluted soils and waters. FEMS Microbiol. Ecol. 42:199–208 [DOI] [PubMed] [Google Scholar]
- 39. Van der Auwera G. A., et al. 2009. Plasmids captured in C. metallidurans CH34: defining the PromA family of broad-host-range plasmids. Antonie Van Leeuwenhoek 96:193–204 [DOI] [PubMed] [Google Scholar]
- 40. Van Houdt R., Monchy S., Leys N., Mergeay M. 2009. New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie Van Leeuwenhoek 96:205–226 [DOI] [PubMed] [Google Scholar]
- 41. Vedler E., Vahter M., Heinaru A. 2004. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 186:7161–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao F., Hou H., Bao Q., Wu J. 2009. PGA4genomics for comparative genome assembly based on genetic algorithm optimization. Genomics 94:284–286 [DOI] [PubMed] [Google Scholar]