Abstract
The rumen, the foregut of herbivorous ruminant animals such as cattle, functions as a bioreactor to process complex plant material. Among the numerous and diverse microbes involved in ruminal digestion are the ruminal protozoans, which are single-celled, ciliated eukaryotic organisms. An activity-based screen was executed to identify genes encoding fibrolytic enzymes present in the metatranscriptome of a bovine ruminal protozoan-enriched cDNA expression library. Of the four novel genes identified, two were characterized in biochemical assays. Our results provide evidence for the effective use of functional metagenomics to retrieve novel enzymes from microbial populations that cannot be maintained in axenic cultures.
INTRODUCTION
To process fibrous plant materials, the rumen harbors a collection of diverse microorganisms, including bacteria, archaea, fungi, and protozoa (reviewed in references 33 and 45). While the diversity and functions of the thousands of microbial species of this unique ecosystem (32) are interesting from both the evolutionary and functional perspectives, the rumen also represents a rich resource of enzymes for converting lignocellulosic feedstocks into biofuel (35, 43) and other applications (19). A range of inexpensive, robust enzymes with a broad range of specificities will likely be required for efficient industrial processing of highly complex plant polysaccharides. Identification of such enzymes that microorganisms use to break down plant materials has been greatly facilitated by metagenomics (42), both in the form of activity-based screens (20, 52) and also through increasingly powerful, high-throughput genomic DNA sequencing approaches (28, 57). As evident in numerous studies (28, 39, 41), metagenomics has proven to be particularly effective for identification of carbohydrate-active genes of fiber-adherent bacterial species of the rumen.
In addition to bacteria and archaea, the rumen also hosts eukaryotic species, namely, anaerobic fungi and ciliate protozoans (reviewed in reference 33). Addressing the function of ruminal protozoans, in particular, has been a challenge due to the difficulty of maintaining these organisms in axenic cultures (55). Thus, assessing the diversity and dynamics of ruminal protozoans has been addressed historically in morphogenic studies (reviewed in reference 12) and by molecular phylogenetics (e.g., by using 18S rDNA markers [47]). Ruminal protozoans are known to contribute to fiber degradation in their hosts (21), and determination and characterization of their ability to directly process plant material have been addressed by diverse strategies, such as direct, biochemical detection of specific fibrolytic enzymes (e.g., cellulases) in extracts derived from individual protozoan species (38, 54), by molecular cloning studies to directly identify genes encoding enzymes capable of degrading cellulose or hemicellulose (49, 50) and, most recently, by sequencing of protozoan-derived expressed sequence tag (EST) libraries (41). Early studies to establish the capacity of protozoan species to express their own enzymes for degradation of plant material included that of Howard et al. (29), who demonstrated that Epidinium ecaudatum indeed contains fibrolytic enzyme activity. Similarly, Bailey et al. (3) demonstrated the presence of both a hemicellulase and a xylobiase in E. ecaudatum by using purified cell extracts. More recently, Clayet et al. (8), using gel filtration of E. ecaudatum extracts, identified at least 10 distinct enzyme activities for plant cell wall degradation; their fractions contained a range of enzymes with glycoside hydrolase (GH) activities, including two distinct carboxymethylcellulases with molecular masses of 23 and 45 kDa (8).
Altogether, about a dozen protozoan fibrolytic genes have been identified in activity-based molecular screens; a comparable number have been identified from informatics-based studies (41), predominantly in ovine and bovine rumen systems. The protozoan enzyme genes characterized to date are diverse, both in terms of the individual GH domains (27) utilized as well as the combinatorial domain organization of proteins that contain them. GH domains are modular by design; they exist in individual polypeptides in variable copy numbers and variable association with other, noncatalytic modules (e.g., carbohydrate binding domains [reviewed in reference 27]). The described rumen protozoan-derived GH domain genes primarily encode single or dual GH5 domains (cellulase superfamily) (49, 50, 53) or GH10 or GH11 domains (xylanase-related domains) (4, 14, 15). The combinatorial complexity of fibrolytic genes thus far detected in ruminal ciliates speaks to the potentially diverse utilization of GH modules within the entire ruminal protozoan population (4, 14, 15, 41, 49, 50, 53). Yet, due in part to the importance of demonstrating the existence of fibrolytic genes in a given protozoan species, enzyme cloning studies have largely been conducted using mono-faunated animals, in which the host ruminant is inoculated with a single ciliate species. Therefore, to investigate the potential diversity of fibrolytic enzymes in a total ciliate population, an activity-based metagenomics screen was conducted of the metatranscriptome of protozoans in the rumen fluid derived from a single, fistulated cow.
MATERIALS AND METHODS
Materials.
Carboxymethyl cellulose (CMC), cellulose (fibrous, medium), galactan, laminarin from Laminaria digitata, mannan from Saccharomyces cerevisiae, and xylan from beechwood were obtained from Sigma-Aldrich. AZCL-HE-cellulose, AZCL-xylan (oat), arabinan (sugar beet), β-glucan (oat, medium viscosity), wheat arabinoxylan (medium viscosity), and xyloglucan from tamarind seed (amyloid) were obtained from Megazyme, Inc. Reagents for the reducing sugar assay, ammonium iron(III) sulfate dodecahydrate, 3-methyl-2-benzothiazolinone hydrazone hydrochloride hydrate, and sulfanic acid, were obtained from Sigma-Aldrich. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was obtained from Gold Biotechnology.
Rumen sample collection, protozoa purification, and mRNA purification.
Rumen protozoans were harvested by a procedure based on that described in reference 36, modified as follows: approximately 3 liters of total rumen content (fluid and solids) was collected from a fistulated Holstein cow maintained at the University of Missouri Dairy Farm, Columbia, MO. The donor cow was fed a total mixed ration (TMR) of a common lactation diet that consisted of alfalfa haylage, corn silage, corn, and protein, minerals, and fat-soluble vitamins added to meet or exceed nutrient requirements. The rumen sample was collected in the morning, prior to feeding, into a prewarmed canister and transported to the lab within 30 min. Aliquots of the sample were treated in three pulses of 30 s each, with a blender, and pressed through a double layer of cheesecloth to remove bulk solids. The resulting liquid (approximately 1 liter) was supplemented to 1% maltose and 0.5% sucrose and then flocculated anaerobically for 1 h at 39°C. After aspirating the floating feed particle layer, the liquid was dialyzed against eight changes of 39°C Coleman anaerobic buffer (55) by using a home-made 10-μm-pore-size Nitex filter cloth bag (Sefar America) with gentle agitation. The volume of the resulting material was ∼50 ml and represented a concentrated mixture of protozoa whose composition was verified by microscopic observation. Total RNA was isolated from this material using the TRIzol Plus RNA purification system (Invitrogen); mRNA was then purified using the magnetic bead-based FastTrack 2.0 mRNA isolation kit (Invitrogen).
Lambda Zap-based protozoan cDNA library construction.
A Lambda Zap II-based protozoan cDNA expression library was constructed by using the Zap cDNA synthesis kit (catalog number 200401; Stratagene), starting with 5 μg of polyadenylated mRNA (for detailed protocols, the manual for this cDNA synthesis kit is available on the Agilent website). After size fractionation of total cDNA by gel exclusion chromatography using Sepharose CL-2B gel filtration medium (procured from Stratagene) in a 1-ml disposable plastic pipette, cDNA fractions ranging from 0.5 to >10 kb were pooled and ligated into the prepared lambda vector (see below) and packaged using the ZAP-cDNA Gigapack III gold cloning kit (catalog number 200450; Stratagene), according to protocols provided by the manufacturer. After titer determinations, 700,000 PFU of the primary library were amplified on plates by using standard lambda phage procedures (Stratagene Zap cDNA synthesis manual) in order to generate the secondary library, which was used for activity-based screening.
Activity-based screen for fibrolytic enzymes.
To identify candidate fibrolytic enzymes for IPTG-inducible cDNA expression, plaque-based high-throughput screening used plates containing dye-linked insoluble polysaccharide substrates (44, 51). Two classes of fibrolytic enzymes were screened: xylanases and cellulases, using AZCL-HE-cellulose and AZCL-xylan (oat), respectively (Megazyme, Inc.). The expression library was screened on petri dishes containing NZYM bottom agar supplemented with a 1× micronutrient solution (1,000× MNS; 3.0 mM H3BO3, 0.46 mM MnCl2, 0.16 mM CuSO4, 0.6 mM ZnSO4, 0.1 mM NaMoO4, 0.01 mM NiSo4, 0.01 mM CoCl2) and NZYM top agarose, supplemented with 1× MNS plus 20 mM IPTG. To identify cDNA clones encoding fibrolytic enzymes, the top agarose incorporated either AZCL-HE-cellulase or AZCL-xylan (oat) at a final concentration of 0.3% (wt/vol). For each substrate, approximately 1,000,000 PFU were screened (10,000 PFU per 150-mm plate). Over a 2- to 5-day incubation period at 37°C, 70 clones were picked that exhibited xylan-degrading activity and 10 clones were picked that exhibited cellulose-degrading activity. Positives were plaque purified in three rounds of plaque purification and then in vivo excised to generate pBluescript DNA preparations, using procedures described in the Stratagene Zap cDNA synthesis manual. To initially characterize the positives, rescued plasmid-borne cDNAs were sequenced with the T3 promoter primer (5′-AAT-TAA-CCC-TCA-CTA-AAG-GG-3′), which flanks the 5′ end of the directionally cloned cDNA insert. Selected clones of the longer cDNA types (types 3 and 4, see below) were then completely sequenced with the T7 promoter primer (5′-TAA-TAC-GAC-TCA-CTA-TAG-GG-3′) in addition to custom, internal primers when required (data not shown). To initially assess the diversity of the cDNA collection, the CAP3 sequence assembly program (30), BLASTp (1), and ClustalW2 (6) were used. The sequences of the longest cDNAs of each type (see Results for analysis findings) were deposited in GenBank (see below).
Sequence searches, alignments, and phylogeny.
Protozoan and bacterial glycoside hydrolase sequences were collected for our analysis using BLASTp searches (31) of the GenBank nonredundant (nr) protein sequences database, using default search parameters to identify sequences with homology values of 1e−50. Protein sequences were aligned using MUSCLE 3.6 (16) with a FASTA output format and then manually edited using Jalview (7). Majority-ruled parsimonious trees were generated using the program protpars of PHYLIP (18), with maximum likelihood branch lengths calculated using TREE-PUZZLE (46). Bootstrap values were calculated using the program seqboot of the PHYLIP package. All trees were viewed and printed in a pdf format using Tree Viewer (58).
Molecular cloning for expression constructs.
To investigate the biochemical properties of representative positives (see Results section for summary of positive classes; see also Fig. S1 in the supplemental material), a single-domain gene for each substrate screened was characterized: one type 1 (identified on cellulose substrate) and one type 2 (identified on xylan substrate). The coding sequence of the longest cDNA from each class was cloned into the NcoI and XhoI sites of the multiple-cloning site of the C-terminal His6 tag expression vector, pET29a (Novagen). To accomplish this, restriction sites were added to the PCR primers and amplified DNA fragments as follows. For type I (cellulose substrate), the 5′ primer sequence was GGG-CCA-TGG-CTT-TGG-GCT-TAA-TTT-CAA-TTTC (the NcoI site is underlined and the first codon of the cDNA is indicated by bold text) and the 3′ PCR primer sequence was GGG-CTC-GAG-TTT-GGA-AAC-AGC-GGC-TTT-GTA-AG (XhoI site is underlined). For type 2, the 5′ primer sequence was GGG-CCA-TGG-CTT-TAA-ATT-ATG-TAT-CAT-CTA-ATA-ATT-TTC (NcoI site is underlined and the first codon of the cDNA is indicated by bold text) and the 3′ PCR primer sequence was GGG-CTC-GAG-TGC-TCC-AGC-AAC-TTG-CAT-AAT (XhoI site is underlined). Coding sequences were amplified using the Platinum PCR supermix high-fidelity kit (Invitrogen) or Phusion high-fidelity DNA polymerase (New England BioLabs) under the conditions recommended by the manufacturers. The resulting amplicon DNAs were purified using the Wizard PCR DNA purification system (Promega) and cloned into pET29a, which was prepared by standard molecular biology techniques. Miniprep (Promega) DNA for each type was sequenced with T7 promoter and T7 terminator primers to verify the absence of PCR-derived mutations. The resulting expression constructs, and proteins derived from them, are designated here type 1-7.1 and type 2-8.6.
Expression and purification of recombinant enzymes.
The type 1-7.1 and type 2-8.6 pET29a constructs were transformed into Escherichia coli BL21(DE3) cells (Invitrogen), and expression cultures for each construct were grown at 37°C in 500 ml LB broth containing 30 μg/ml kanamycin. The cultures were grown to an optical density at 600 nm (OD600) of 0.6 to 0.8, at which point expression was induced by the addition of IPTG to a final concentration of 1 mM. Cultures were then grown at 37°C for an additional 3 h. Bacterial cells were harvested by centrifugation (10,000 × g at 4°C for 10 min) and resuspended in 20 ml of equilibration/wash solution (50 mM sodium phosphate buffer [pH 7.0], 300 mM NaCl) supplemented with 1× Complete, EDTA-free protease inhibitor cocktail (Roche) and phenylmethylsulfonyl fluoride to a final concentration of 1 mM. Cell resuspensions were lysed using a French press, and the resulting lysates were centrifuged at 10,000 × g at 4°C for 10 min, to remove cell debris. Recombinant proteins were then affinity purified by using Talon metal affinity resin (Clontech), according to the manufacturer's protocols. The purified enzymes were eluted in a single-step elution with equilibration/wash buffer supplemented with 150 mM imidazole. Proteins were then concentrated and imidazole removed with an ultrafiltration membrane (Vivaspin-20 column; GE Healthcare). Enzyme purity was confirmed by SDS-PAGE (see Fig. S2 in the supplemental material), and the protein concentration was determined using the Quick Start Bradford dye reagent (Bio-Rad).
Enzyme assays.
The activity of each enzyme was initially confirmed in a simple, colorimetric assay using the same insoluble substrates that were used in the plate screening procedure. Specifically, 1 mg of the respective substrate (AZCL-HE-cellulose or AZCL-xylan) was suspended in 1 ml of protein purification equilibration buffer (300 mM NaCl, 50 mM sodium phosphate [pH 7.0]), at 37°C for 30 min. Reactions were initiated by adding 50 μl of purified enzyme (0.5 to 2.2 mg/ml protein), and the release of solubilized dye was visually validated. Optimal pH conditions were then preliminarily determined in assays using 550 μl of 50 mM Britton-Robinson buffer (5) plus 400 μl 0.2% AZCL-labeled substrates. Reactions were initiated by adding 50 μl of purified enzyme solution (10 μg protein/μl). After incubation at 37°C for 1 h, supernatant absorbance at 590 nm was determined (see Fig. S3A and S4A in the supplemental material). More-precise pH optimal activity determinations were made by measuring the release of reducing sugars from polysaccharides by using the 3-methyl-2-benzothiazolinone hydrazone reagent (MBTH) (2). Assays were run with either 1% CMC in 50 mM sodium acetate buffer or with 1% xylan in 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer. Assays to determine optimal temperature, substrate specificities, and enzymatic activities were performed in 250 μl of 1% polysaccharide solutions buffered with either 50 mM MES [2-(N-morpholino)ethanesulfonic acid (pH 6.0)] or 50 mM MOPS (pH 7.4). Reactions were initiated by adding 10 μl (22 μg protein/ml) of purified enzyme. After incubation, aliquots were added to the MBTH reagent. The quantities of released reducing sugars were determined using glucose, mannose, galactose, arabinose, and xylose as standards. The apparent Km and Vmax values were determined by fitting the rate data to the Michaelis-Menten equation (KaleidaGraph, version 3.6; Synergy Software). Enzyme activity was assessed by measuring the release of reducing sugars over polysaccharide concentration ranges of 0.05 to 10 mg/ml. Triplicates were collected for each time point at each substrate concentration used throughout the analyses.
Polysaccharide analysis using HPLC.
Solutions of xylan or β-glucan (250-μl volume at 1.0 mg/ml) were treated with the type 1-7.1 or type 2-8.6 proteins (10 μl of solution at 22 μg protein/ml) for 10 and 60 min. Negative controls included xylan and β-glucan substrates without enzyme amendments. Samples were incubated at 45°C, and reactions were terminated by adding 10 μl of 0.5 M NaOH. The 25-μl reaction mixtures were then injected onto a DX-500 high-performance liquid chromatography (HPLC) instrument (Dionex) equipped with a 250- by 4-mm CarboPac-1 column (Dionex) at a solvent flow rate of 1 ml min−1. The gradient system utilized 100 mM NaOH as solvent A and 100 mM NaOH, 1 M NaO-acetate as solvent B. The gradient was run at 100% solvent A for 15 min, followed by a linear gradient to 100% solvent B over 60 min. Detection was by pulsed amperometry using an ED40 electrochemical detector (Dionex) (25).
Nucleotide sequence accession numbers.
The sequences of the longest cDNAs of each type were deposited in GenBank and assigned accession numbers as follows: type 1, JN635693; type 2, JN635694; type 3, JN635695; type 4, JN635696.
RESULTS
Classification of protozoan metagenomic cDNAs.
Sixty-three clones positive for glycoside hydrolase activity were sequenced: 60 were identified on xylan substrate and three were identified on cellulose substrate. Sequencing of the 5′ end of each cDNA generated approximately 800 bp of sequence for each clone; analysis of these sequences permitted classification of the cDNAs into four types, which are discussed in detail below. Because 8 of the 63 cDNAs likely represent aberrant clones (see discussion below and Fig. S1 in the supplemental material), they were omitted in our overviews, which were thus restricted to 55 clones (Table 1 and Fig. 1).
Table 1.
Summary of metagenomic screen positivesa
| cDNA type | Substrate | CDS length (aa) | GH domain(s) | Pfam | Best match | Species | % identity/% similarity | Total no. of cDNAs sequenced | No. of cDNAs with unique 5′ ends |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Cellulose | 498 | GH5 | 00150 | CAH69214 | E. ecaudatum | 83/91 | 3 | 1 |
| 2 | Xylan | 346 | GH10 | 00331 | CAL91981 | E. ecaudatum | 82/91 | 1 | 1 |
| 3 | Xylan | 351 | GH11, GH11 | 00457 | CAL91983 | E. ecaudatum | 67/74 | 2 | 1 |
| 4 | Xylan | 462 | GH11, GH11 | 00457 | CAL91983 | E. ecaudatum | 70/78 | 49 | 13 |
Four types of cDNAs were recovered from the activity-based screens, which utilized either a cellulose- or xylan-based dye-linked substrate. The length of the coding region (CDS), in amino acids (aa), of the longest cDNA for the given type is shown. The indicated GH domain(s) was detected by BLASTp homology search; the Pfam assignment for the respective GH domain is also reported. Best match indicates the GenBank (protein) accession number for the best hits, which were all derived from E. ecaudatum.
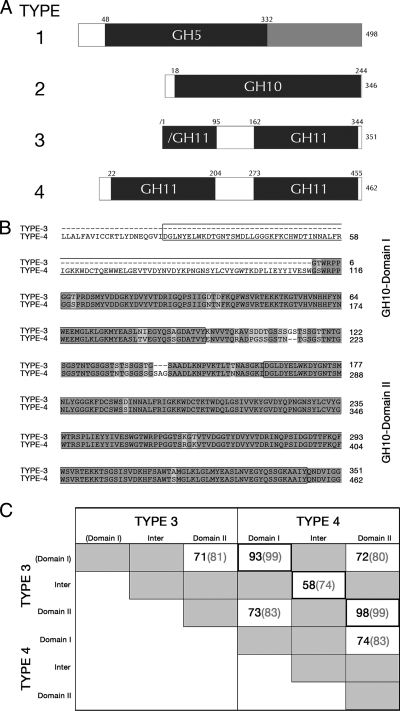
Fig. 1.
Summary of metagenomic screen positives. (A) Overall domain organization of the polypeptide encoded by the longest cDNA of each type. The numbers adjacent to the diagrams indicate amino acid residues. The type 1 cDNA encodes a protein with a single, N-terminal GH5 domain and a C-terminal domain of unknown function. The type 2 cDNA encodes a protein with a single GH10 domain. The type 3 (partial) cDNA encodes a protein with a partial N-terminal GH11 domain and a second C-terminal GH11 domain, whereas the type 4 cDNA encodes a protein with two GH11 domains. (B) Alignment between type 3 and 4 protein sequences, with the GH11 domains indicated by boxes. (C) Domain comparison between type 3 and 4 proteins. Domain I and domain II refer to the first and second GH11 domains, as indicated in panel B. The first number indicates the percent identity, whereas the second number (in parentheses) indicates the percent similarity. “Inter” refers to the interdomain sequence.
Cellulase positives.
The three type 1 cDNAs were isolated on cellulose indicator plates. Each type 1 cDNA encoded a single GH5 domain-containing protein (cellulase superfamily) (26); all three cDNA sequences were identical, suggesting that they were independent isolates of the same, amplified cDNA.
Xylanase positives.
Type 2 through type 4 cDNAs were isolated on xylan indicator plates. Sequence analysis of these positives with xylanase activity sorted into three distinct classes (Fig. 1A): the type 2 cDNA encodes a protein with a single GH10 domain. The type 3 cDNA encodes a partial, N-terminal GH11 domain in addition to a second, full-length GH11 domain; thus, it is unlikely to be a full-length cDNA. The type 4 cDNAs encode a protein with two complete GH11 domains. Among the type 4 clones, DNA sequencing identified 13 different 5′ ends. The longest cDNA encodes an open reading frame (ORF) that contains two GH11 domains, whereas the shortest cDNAs encode at least the C-terminal GH11 domain. In addition to these intact type 4 cDNAs, eight cDNAs likely represent aberrant, deletion forms of the full-length type 4 cDNA (see Fig. S1 in the supplemental material).
The sequences of the two-domain GH11-positive types (type 3 and type 4) were then compared through DNA and polypeptide alignments, which indicated that they represent highly similar, yet distinct genes. The DNA alignments (data not shown) between the overlapping 1,118 bp of the two cDNAs showed 94.6% identity, with nine gaps, most of which were in the 3′-untranslated regions of the two cDNAs. A ClustalW protein alignment (Fig. 1B) indicated an overall identity between the two proteins of 92.5%, with two gaps. The best conservation (Fig. 1C) was between the first GH11 domain (domain I) of each protein and between the second GH11 domain (domain II) of each protein. In contrast, comparing the first GH11 domain of each protein to the GH11 domain of other protein revealed lower conservation, which may indicate a diversification of substrate specificity between the two domains.
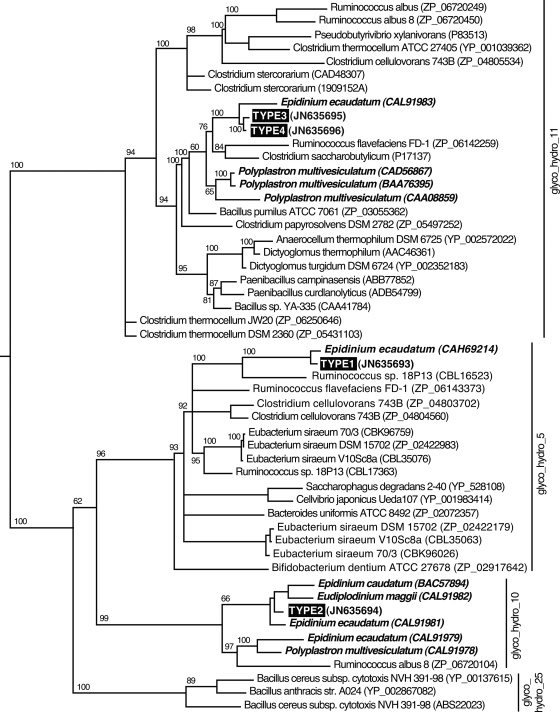
A phylogenetic analysis on the full-length peptide sequences of each positive type was performed (Fig. 2). The phylogenetic tree was generated by maximum parsimony analysis of the retrieved amino acid sequences and the closest related sequences in the NCBI protein database. The closest related amino acid sequences for all of the searched sequences originated from another protozoan. The 498-residue type 1-7.1 (cellulose substrate) ORF shares 83% identity and 91% similarity over 496 residues with a GH5-containing protein sequence (CAH6914) identified from E. ecaudatum. In addition to the GH5-homologous domain (residues 64 to 335, identified as Pfam00150), this ORF also contains a C-terminal 163-amino-acid sequence (residues 336 to 498) with no significant hit from either BLASTp or psi-Blast searches. Additional analysis would be required to determine the potential role of this novel domain in carbohydrate binding, noncatalytic stabilization, etc. The type 2-8.6 sequence (putative xylanase) is most closely related to a GH10 domain-containing protein from E. ecaudatum, whereas the dual-domain type 3 and 4 proteins are most closely related to a different dual GH11 domain sequence, also identified in E. ecaudatum. Thus, the phylogenetic analysis suggested that the amino acid sequences identified in this study were of protozoan, not bacterial, origin. In further support of this, cDNAs for the shorter types (types 1 to 3), as well as the full-length sequence of the type 4 cDNA, have poly(A) tracts; types 1 to 3 also had a typical eukaryotic upstream polyadenylation signal (AATAAA). As has been reported for numerous other ruminal protozoan genes (14, 15, 17, 37), codon usage analysis (data not shown) indicated a strong bias for A and T nucleotides in the first and third positions, as was also reflected by the G+C content (32 to 36%) of the nucleotide sequences of all four types analyzed. Notably, the closest nonprotozoan homolog for each type included Ruminococcus species, a group that includes anaerobic, cellulolytic bacteria.
Fig. 2.
Phylogenetic topology of rumen protozoan and bacterial glycoside hydrolases. A majority-ruled parsimony tree with maximum likelihood branch lengths was calculated using full-length amino acid sequences. Bootstrap values of 1,000 independent trees larger than 60 are labeled on each branch. Major clades are delimited by solid horizontal lines. Sequences identified in this study are shown in white in a black box. Protozoan sequences are shown in bold italics. GenBank accession numbers for each sequence are given within parentheses.
Biochemical characterization of type 1-7.1.
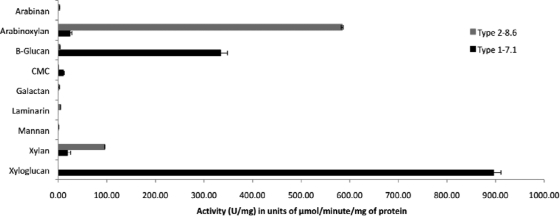
The type 1-7.1-positive sample was identified as a possible cellulase due to its activity on cellulose indicator plates and its GH5 domain homology. While many characterized cellulases are typically active against carboxymethyl cellulose (an artificial substrate), the recombinant enzyme derived from our library had 85 times higher activity against xyloglucan (896.06 ± 14.98 U/mg) than against CMC (10.45 ± 2.03 U/mg) and 32 times the activity against β-glucan (334.29 ± 13.92 U/mg) (Fig. 3). It also exhibited minimal activity against arabinoxylan (24.39 ± 3.64 U/mg) and xylan (19.09 ± 6.49 U/mg). Furthermore, no activity was detected against arabinan, galactan, laminarin, or mannan (Fig. 3). When the type 1-7.1 amino acid sequence was compared to its closest BLASTp match (CAH69214) (48), a cellulase identified in E. ecaudatum, it exhibited additional differences. The pH optimum for the cellulase from E. ecaudatum is reported to be 8.3 (53), whereas the optimum for type 1-7.1 is 5.9 (Table 2; see also Fig. S3A and B in the supplemental material). The apparent Km, Vmax, and Kcat values and the Kcat/Km ratio for β-glucan are 0.83 mg/ml, 97.7 μmol/min/mg, 7.4 s−1, and 8.9 ml mg−1 s−1, and for xyloglucan the values are 0.19 mg/ml, 179.1 μmol/min/mg, 13.6 s−1, and 71.6 ml mg−1 s−1, respectively (Table 3). The Km value for xyloglucan was slightly lower than the Km value for β-glucan, indicating a slightly higher affinity for this substrate. Moreover, the Vmax value for xyloglucan was 2.3 times higher than for β-glucan. The enzyme turnover rate, Kcat, and the catalytic efficiency (Kcat/Km) were also higher for xyloglucan. It is possible that this enzyme might be considered a specific xyloglucanase, because its activity against xyloglucan is more than 10 times higher than its activity against CMC (24). However, further analyses are required for confirmation. Xyloglucanases have been also identified in fungi and bacteria (22, 23, 24, 40, 56). Due to its broad pH and temperature tolerances (Table 2; see also Fig. S3A and C in the supplemental material), our xyloglucanase could be useful in industrial degradation of hemicelluloses from plant biomass.
Fig. 3.
Substrate specificity analysis for type 1-7.1 and type 2-8.6 recombinant proteins. Substrate specificity assays were performed in triplicate in 250 μl of 1% polysaccharide solution buffered with either 50 mM MES (pH 6.0; optimum conditions for type 1-7.1) or 50 mM MOPS (pH 7.4; optimum conditions for type 2-8.6). Reactions were initiated by adding 10 μl (22 g of protein/ml) of purified enzyme.
Table 2.
Basic biochemical properties of type 1-7.1 and type 2-8.6 recombinant proteins
| Property | Value for protein type |
|
|---|---|---|
| 1-7.1 | 2-8.6 | |
| GH family | 5 | 10 |
| Molecular mass (kDa) | 56.0 | 39.2 |
| pI (calculated) | 4.56 | 6.35 |
| pH activity range | 5.0–8.0 | 5.0–9.0 |
| pH optimum | 5.9 | 7.4 |
| Temp activity range (°C) | 4–60 | 25–60 |
| Temp optimum (°C) | 50 | 45 |
Table 3.
Kinetic data for type 1-7.1 and type 2-8.6 recombinant proteins
| Type and substrate | Apparent Km (mg/ml) | Vmax (μmol/min/mg) | Kcat (s−1) | Catalytic efficiency (Kcat/Km [ml/mg/s]) |
|---|---|---|---|---|
| Type 1-7.1 | ||||
| β-Glucan | 0.83 | 97.7 | 7.4 | 8.9 |
| Xyloglucan | 0.19 | 179.1 | 13.6 | 71.6 |
| Type 2-8.6 | ||||
| Xylan | 6.95 | 14.5 | 1.1 | 0.2 |
| Arabinoxylan | 0.14 | 117.3 | 8.9 | 63.6 |
Biochemical characterization of type 2-8.6.
The closest BLASTp match for type 2-8.6 was also a GH10 domain-containing enzyme. Typically, members of this GH tend to have low pI values (9). In contrast, type 2-8.6 has a calculated pI value of 6.35 (Table 2), a value that is higher than those reported for GH10 enzymes but a lower value than is typically observed for GH11 enzymes (which tend to be high values [9]). Although the enzyme had detectable activity against xylan (95.62 ± 0.38 U/mg), it possessed a higher activity against arabinoxylan (584.39 ± 2.07 U/mg) (Fig. 3). The apparent Km, Vmax, Kcat, and Kcat/Km values for xylan are 6.95 mg/ml, 14.5 μmol/min/mg, 1.1 s−1, and 0.2 ml mg−1 s−1, and for arabinoxylan they are 0.14 mg/ml, 117.3 μmol/min/mg, 8.9 s−1, and 63.6 ml mg−1 s−1, respectively (Table 2). While enzymes that possess activity against arabinoxylan have been characterized in bacteria, fungi, and plants (11), no protozoan enzymes have been identified to date. To confirm the complete enzymatic hydrolysis of arabinoxylan, further studies need to be completed to determine the reaction products and structure of the enzyme (10).
Mode of action of the type 1-7.1 and type 2-8.6 enzymes.
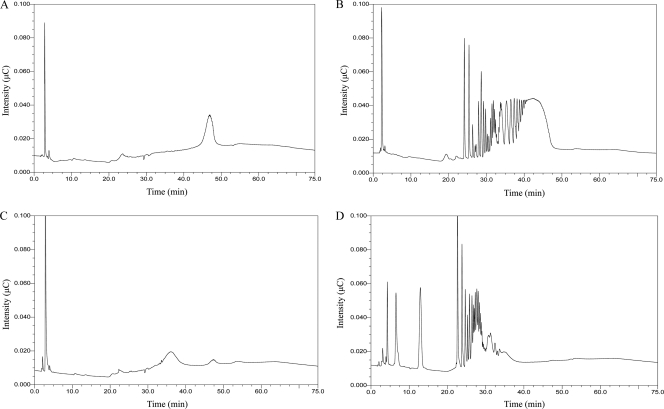
Fibrolytic enzymes are essential for the digestion of cellulosic biomass in the ruminant diet. A suite of enzymes is required to produce a variety of free sugars, (exoenzymes), as well as oligosaccharides (endoenzymes) for metabolism by rumen bacteria and subsequent digestion by the ruminant host. The HPLC analysis of hydrolysis products resulting from exposure of β-glucan to type 1-7.1 and xylan exposure to type 2-8.6 indicated that each tested enzyme employs an endo-type of cleavage (Fig. 4). Compared to chromatographs of β-glucan (Fig. 4A) and xylan (Fig. 4C), each of the treated substrates had multiple peaks prior to the polysaccharide peaks (Fig. 4B and D), indicating that a range of oligosaccharides was generated by the actions of the endoglycoside hydrolases. It should be noted that similar chromatographs were obtained for both the 10- and 60-min exposures. If the enzymes were exo-acting, the primary products would be either glucose (from β-glucan) or xylose (from xylan). Both glucose and xylose formed a single peak at 4 min on the chromatographs (data not shown). There was no peak at the 4-min point for glucose when β-glucan was exposed to type 1-7.1, yet multiple peaks were present as the chromatographic run proceeded (Fig. 4B). There was a peak at 4 min, indicating xylose production after xylan exposure to type 2-8.6, as well as multiple peaks at later time points, indicating the presence of oligoxylose chains (Fig. 4D). These data are consistent with the fact that many GH5 and GH10 enzymes are known to be endo-β-1,4-glucanases and endo-β-1,4-xylanases (9).
Fig. 4.
HPLC results for cleavage product analysis of type 1-7.1 and type 2-8.6 recombinant proteins. (B and D) HPLC results for hydrolysis products of β-glucan after exposure to type 1-7.1 (B) and xylan after exposure to type 2-8.6 (D) for 60 min. (A and C) Polysaccharides β-glucan (A) and xylan (C), without addition of enzyme, were used as controls. The glucose and xylose standard peaks were detected at 4 min (data not shown). Intensity is reported in microcoulombs (μC).
DISCUSSION
An activity-based metagenomic screen was executed with the aim of assessing the diversity of fibrolytic enzymes encoded by the metatranscriptome of protozoans present in bovine rumen fluid. Using just two substrates, a cellulose and a hemicellulose, four genes with diverse GH domains and modular organizations were identified. Phylogenetic analysis of these genes revealed the closest homologs to be protozoan and the closest nonprotozoan homologs to be most closely related to Gram-positive bacteria. These observations support the hypothesis that lignocellulose-degrading genes were acquired by protozoans from ruminal bacteria by horizontal gene transfer (15, 41). There was close homology of the positive sequences obtained in our study (Fig. 1B) to E. ecaudatum genes identified in a previous protozoan EST sequencing study (41). The putative cellulases and xylanases identified by Ricard et al. (41) were all derived from one (the Entodiniophorphids) of the two major groups of rumen ciliates. The fact that each of the gene types identified in our bovine rumen screens had potential homologs in protozoan species originating from sheep rumen raises the possibly that the genes represent orthologs derived from related ciliate species in the two different hosts. The fact that none of our positives exhibited novel domain organization (relative to other identified ciliate GH genes) may indicate that ruminal ciliates acquired only a limited repertoire of bacterial fibrolytic genes.
One strength of activity-based screening is its ability to directly recover genes encoding biocatalysts for specific substrates (e.g., cellulose). In our current study, two enzymes were identified, expressed, and biochemically characterized. The putative xyloglucanse possesses a high specific activity toward tamarind xyloglucan (896.06 U/mg of protein) (Fig. 3). In comparison, previously characterized xyloglucanses from fungal sources ranged in their activity from 45 to 98 U/mg of protein (24). Our xyloglucanse also had a lower apparent Km value (0.19 mg/ml) (Table 3) than a xyloglucan-specific endo-β-1,4-glucanase gene (xeg5A), isolated from bovine rumen microflora and expressed in E. coli (Km, 3.61 mg/ml) (56) and one isolated from the fungus Aspergillus aculeatus (Km, 3.6 mg/ml) (40). Similarly, our putative arabinoxylanase had a lower apparent Km (0.14 mg arabinoxylan/ml) (Table 3) than a GH10 xylanase isolated from Penicillium funiculosum (3.7 mg/ml) (34). In addition, our putative arabinoxylanase had a higher specific activity (584.39 U/mg) (Fig. 3) than the P. funiculosum xylanase (106.4 U/mg) (34) on arabinoxylan, as well as a higher specific activity against beechwood xylan (type 2-8.6, 95.62 U/mg; P. funiculosum xylanase, 60.1 U/mg) (34).
The powerful and rapid, activity-based metagenomics approach does have its own technical and efficiency obstacles (as discussed in reference 52), however, as evidenced by our recovery of hybrid/deletion cDNA clones. Thus, more direct, sequence-based metagenomics (e.g., single-cell-based genomic sequencing [57]), in combination with molecular phylogenetics (13, 48) may represent the more attractive technique for characterizing the population dynamics, functions, and fibrolytic genes of ciliate ruminal protozoa.
Supplementary Material
ACKNOWLEDGMENT
We thank the University of Missouri Mizzou Advantage Program for funding.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anthon G. E., Barrett D. M. 2002. Determination of reducing sugars with 3-methyl-2-benzothiazolinonehydrazone. Anal. Biochem. 305:287–289 [DOI] [PubMed] [Google Scholar]
- 3. Bailey R. W., Clarke R. T., Wright D. E. 1962. Carbohydrases of the rumen ciliate Epidinium ecaudatum (Crawley). Biochem. J. 83:517–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bera-Maillet C., Devillard E., Cezette M., Jouany J. P., Forano E. 2005. Xylanases and carboxymethylcellulases of the rumen protozoa Polyplastron multivesiculatum, Eudiplodinium maggii and Entodinium sp. FEMS Microbiol. Lett. 244:149–156 [DOI] [PubMed] [Google Scholar]
- 5. Britton H. T. S., Robinson R. A. 1931. Universal buffer solutions and the dissociation constant of veronal. J. Chem. Soc. 1931:1456–1462 [Google Scholar]
- 6. Chenna R., et al. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clamp M., Cuff J., Searle S. M., Barton G. J. 2004. The Jalview Java alignment editor. Bioinformatics 20:426–427 [DOI] [PubMed] [Google Scholar]
- 8. Clayet F., Senaud J., Bohatier J. 1992. Chromatographic separation of some cell wall polysaccharide-degrading enzymes of the sheep rumen ciliate Epidinium caudatum. Ann. Zootech. 41:81 [Google Scholar]
- 9. Collins T., Gerday C., Feller G. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29:3–23 [DOI] [PubMed] [Google Scholar]
- 10. Correia M. A., et al. 2011. Structure and function of an arabinoxylan-specific xylanase. J. Biol. Chem. 286:22510–22520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Courtin C. M., Delcour J. A. 2002. Arabinoxylans and endoxylanases in wheat flour bread-making. J. Cereal Sci. 35:225–243 [Google Scholar]
- 12. Dehority B. A. 1993. Laboratory manual for classification and morphology of ruminal ciliate protozoa. CRC Press, Boca Raton, FL [Google Scholar]
- 13. Deng W., Xi D., Mao H., Wanapat M. 2008. The use of molecular techniques based on ribosomal RNA and DNA for rumen microbial ecosystem studies: a review. Mol. Biol. Rep. 35:265–274 [DOI] [PubMed] [Google Scholar]
- 14. Devillard E., et al. 2003. Characterization of XYN10B, a modular xylanase from the ruminal protozoan Polyplastron multivesiculatum, with a family 22 carbohydrate-binding module that binds to cellulose. Biochem. J. 373:495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Devillard E., et al. 1999. A xylanase produced by the rumen anaerobic protozoan Polyplastron multivesiculatum shows close sequence similarity to family 11 xylanases from gram-positive bacteria. FEMS Microbiol. Lett. 181:145–152 [DOI] [PubMed] [Google Scholar]
- 16. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eschenlauer S. C., et al. 1998. Phylogenetic position and codon usage of two centrin genes from the rumen ciliate protozoan, Entodinium caudatum. FEMS Microbiol. Lett. 166:147–154 [DOI] [PubMed] [Google Scholar]
- 18. Felsenstein J. 2000. PHYLIP (Phylogeny Inference Package). Department of Genetics, University of Washington, Seattle, WA [Google Scholar]
- 19. Fernandez-Arrojo L., Guazzaroni M. E., Lopez-Cortes N., Beloqui A., Ferrer M. 2010. Metagenomic era for biocatalyst identification. Curr. Opin. Biotechnol. 21:725–733 [DOI] [PubMed] [Google Scholar]
- 20. Ferrer M., et al. 2005. Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ. Microbiol. 7:1996–2010 [DOI] [PubMed] [Google Scholar]
- 21. Gijzen H. J., Lubberding H. J., Gerhardus M. J. T., Vogels G. D. 1988. Contribution of rumen protozoa to fibre degradation and cellulase activity in vitro. FEMS Microbiol. Lett. 53:35–43 [Google Scholar]
- 22. Gilbert H. J., Stalbrand H., Brumer H. 2008. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr. Opin. Plant Biol. 11:338–348 [DOI] [PubMed] [Google Scholar]
- 23. Gloster T. M., et al. 2007. Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J. Biol. Chem. 282:19177–19189 [DOI] [PubMed] [Google Scholar]
- 24. Grishutin S. G., et al. 2004. Specific xyloglucanases as a new class of polysaccharide-degrading enzymes. Biochim. Biophys. Acta 1674:268–281 [DOI] [PubMed] [Google Scholar]
- 25. Hausalo T. 1995. Analysis of wood and pulp carbohydrates by anion exchange chromatography with pulse amperometric detection, p. 131–136 Proceedings of the 8th International Symposium on Wood Pulping Chemistry, vol. III Helsinki, Finland Tappi Press, Norcross, GA [Google Scholar]
- 26. Henrissat B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Henrissat B., Davies G. J. 2000. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 124:1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hess M., et al. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467 [DOI] [PubMed] [Google Scholar]
- 29. Howard B. H., Jones G., Purdom M. R. 1960. The pentosanases of some rumen bacteria. Biochem. J. 74:173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang X., Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karlin S., Altschul S. F. 1990. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. U. S. A. 87:2264–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim M., Morrison M., Yu Z. 2011. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 76:49–63 [DOI] [PubMed] [Google Scholar]
- 33. Krause D. O., et al. 2003. Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol. Rev. 27:663–693 [DOI] [PubMed] [Google Scholar]
- 34. Lafond M., et al. 2011. GH10 xylanase D from Penicillium funiculosum: biochemical studies and xylooligosaccharide production. Microb. Cell Fact 10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li L. L., McCorkle S. R., Monchy S., Taghavi S., van der Lelie D. 2009. Bioprospecting metagenomes: glycosyl hydrolases for converting biomass. Biotechnol. Biofuels 2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin C., Williams A. G., Michalet-Doreau B. 1994. Isolation and characteristics of the protozoal and bacterial fractions from bovine ruminal contents. J. Anim. Sci. 72:2962–2968 [DOI] [PubMed] [Google Scholar]
- 37. McEwan N. R., Eschenlauer S. C., Calza R. E., Wallace R. J., Newbold C. J. 2000. The 3′ untranslated region of messages in the rumen protozoan Entodinium caudatum. Protist 151:139–146 [DOI] [PubMed] [Google Scholar]
- 38. Michalowski T., Rybicka K., Wereszka K., Kasperowicz A. 2001. Ability of the rumen ciliate Epidinium ecaudatum to digest and use crystalline cellulose and xylan for in vitro growth. Acta Protozool. 40:203–210 [Google Scholar]
- 39. Palackal N., et al. 2007. A multifunctional hybrid glycosyl hydrolase discovered in an uncultured microbial consortium from ruminant gut. Appl. Microbiol. Biotechnol. 74:113–124 [DOI] [PubMed] [Google Scholar]
- 40. Pauly M., et al. 1999. A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9:93–100 [DOI] [PubMed] [Google Scholar]
- 41. Ricard G., et al. 2006. Horizontal gene transfer from bacteria to rumen ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics 7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Riesenfeld C. S., Schloss P. D., Handelsman J. 2004. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38:525–552 [DOI] [PubMed] [Google Scholar]
- 43. Rubin E. M. 2008. Genomics of cellulosic biofuels. Nature 454:841–845 [DOI] [PubMed] [Google Scholar]
- 44. Ruijssenaars H. J., Hartmans S. 2001. Plate screening methods for the detection of polysaccharase-producing microorganisms. Appl. Microbiol. Biotechnol. 55:143–149 [DOI] [PubMed] [Google Scholar]
- 45. Russell J. B., Rychlik J. L. 2001. Factors that alter rumen microbial ecology. Science 292:1119–1122 [DOI] [PubMed] [Google Scholar]
- 46. Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504 [DOI] [PubMed] [Google Scholar]
- 47. Shin E. C., et al. 2004. Phylogenetic analysis of protozoa in the rumen contents of cow based on the 18S rDNA sequences. J. Appl. Microbiol. 97:378–383 [DOI] [PubMed] [Google Scholar]
- 48. Sylvester J. T., Karnati S. K., Yu Z., Morrison M., Firkins J. L. 2004. Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J. Nutr. 134:3378–3384 [DOI] [PubMed] [Google Scholar]
- 49. Takenaka A., D'Silva C. G., Kudo H., Itabashi H., Cheng K. J. 1999. Molecular cloning, expression, and characterization of an endo-beta-1,4-glucanase cDNA from Epidinium caudatum. J. Gen. Appl. Microbiol. 45:57–61 [DOI] [PubMed] [Google Scholar]
- 50. Takenaka A., Tajima K., Mitsumori M., Kajikawa H. 2004. Fiber digestion by rumen ciliate protozoa. Microbes Environ. 19:203–210 [Google Scholar]
- 51. Ten L. N., Im W. T., Aslam Z., Larina L., Lee S. T. 2007. Novel insoluble dye-labeled substrates for screening inulin-degrading microorganisms. J. Microbiol. Methods 69:353–357 [DOI] [PubMed] [Google Scholar]
- 52. Uchiyama T., Miyazaki K. 2009. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr. Opin. Biotechnol. 20:616–622 [DOI] [PubMed] [Google Scholar]
- 53. Wereszka K., et al. 2004. A cellulase produced by the rumen anaerobic protozoan Epidinium ecaudatum has an unusual pH optimum. Endocytobiosis Cell Res. 15:561–569 [Google Scholar]
- 54. Williams A. G., Coleman G. S. 1985. Hemicellulose-degrading enzymes in rumen ciliate protozoa. Curr. Microbiol. 12:85–90 [Google Scholar]
- 55. Williams A. G., Coleman G. S. 1992. The rumen protozoa. Springer-Verlag, New York, NY [Google Scholar]
- 56. Wong D. D., Chan V. J., McCormack A. A., Batt S. B. 2010. A novel xyloglucan-specific endo-beta-1,4-glucanase: biochemical properties and inhibition studies. Appl. Microbiol. Biotechnol. 86:1463–1471 [DOI] [PubMed] [Google Scholar]
- 57. Woyke T., et al. 2009. Assembling the marine metagenome, one cell at a time. PLoS One 4:e5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zmasek C. M., Eddy S. R. 2001. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics 17:383–384 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.