Abstract
Lactobacillus reuteri 121 uses the glucosyltransferase A (GTFA) enzyme to convert sucrose into large amounts of the α-d-glucan reuteran, an exopolysaccharide. Upstream of gtfA lies another putative glucansucrase gene, designated gtfB. Previously, we have shown that the purified recombinant GTFB protein/enzyme is inactive with sucrose. Various homologs of gtfB are present in other Lactobacillus strains, including the L. reuteri type strain, DSM 20016, the genome sequence of which is available. Here we report that GTFB is a novel α-glucanotransferase enzyme with disproportionating (cleaving α1→4 and synthesizing α1→6 and α1→4 glycosidic linkages) and α1→6 polymerizing types of activity on maltotetraose and larger maltooligosaccharide substrates (in short, it is a 4,6-α-glucanotransferase). Characterization of the types of compounds synthesized from maltoheptaose by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), methylation analysis, and 1-dimensional 1H nuclear magnetic resonance (NMR) spectroscopy revealed that only linear products were made and that with increasing degrees of polymerization (DP), more α1→6 glycosidic linkages were introduced into the final products, ranging from 18% in the incubation mixture to 33% in an enriched fraction. In view of its primary structure, GTFB clearly is a member of the glycoside hydrolase 70 (GH70) family, comprising enzymes with a permuted (β/α)8 barrel that use sucrose to synthesize α-d-glucan polymers. The GTFB enzyme reaction and product specificities, however, are novel for the GH70 family, resembling those of the GH13 α-amylase type of enzymes in using maltooligosaccharides as substrates but differing in introducing a series of α1→6 glycosidic linkages into linear oligosaccharide products. We conclude that GTFB represents a novel evolutionary intermediate between the GH13 and GH70 enzyme families, and we speculate about its origin.
INTRODUCTION
Glucansucrase (GS) (or glucosyltransferase [GTF]) enzymes (EC 2.4.1.5) of lactic acid bacteria (LAB) use sucrose to synthesize a diversity of α-glucans with α1→6 (dextran; found mainly in Leuconostoc), α1→3 (mutan; found mainly in Streptococcus), alternating α1→3 and α1→6 (alternan; reported only in Leuconostoc mesenteroides), and α1→4 (reuteran; synthesized by GTFA and GTFO from Lactobacillus reuteri strains) glycosidic bonds (1, 14, 16, 23, 34).
The first glycoside hydrolase 70 (GH70) family 3-dimensional (3D) structures, recently elucidated (9, 38), showed that the catalytic domains of GS enzymes possess a (β/α)8 barrel structure similar to that of members of the GH13 family, confirming earlier secondary-structure predictions (4, 21). The core of the proteins belonging to the GH13 family comprises 8 β-sheets alternated with 8 α-helices. In GS enzymes, however, this (β/α)8 barrel structure is circularly permuted (21). Also, the four conserved regions (regions I to IV) identified in members of the α-amylase family GH13 (31) are present in glucansucrases. However, as a consequence of the circular permutation, region I occurs C-terminally to regions II to IV in glucansucrase enzymes.
Upstream of the gtfA gene from L. reuteri 121 we identified a second glucansucrase-like gene (gtfB) (15, 16). However, after cloning and expression in Escherichia coli, this GTFB enzyme was inactive with sucrose (15). In parallel investigations, we identified various genes encoding putative GTFB homologs in other Lactobacillus strains. This emergence of more GTFB homologs, indicating that they occur more widely, prompted us to investigate the activity, reaction specificity, and product specificity of the L. reuteri 121 GTFB enzyme in more detail.
Here we show that the GTFB enzyme uses maltooligosaccharides (MOS), e.g., maltoheptaose (with α1→4 glycosidic linkages only), as substrates to synthesize oligosaccharides up to a degree of polymerization (DP) of at least 14. During this disproportionation/polymerization process, GTFB introduces α1→6 glycosidic linkages (18%) into the final mixture of products. Furthermore, we show that incubation of GTFB with a large amylose type of donor substrate (amylose-V) and smaller saccharides (glucose, maltose) as acceptor substrates results in the synthesis of larger saccharides with both α1→6 and α1→4 glycosidic linkages. Finally, the biochemical characterization of GTFB as a 4,6-α-glucanotransferase enzyme instead of a glucansucrase is discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Escherichia coli TOP10 (Invitrogen, Carlsbad, CA) was used as the host for cloning purposes. Plasmid pET15b (Novagen, Madison, WI) was used for expression of the (mutant) gtfB genes in E. coli BL21 Star (DE3) (Invitrogen, Taastrup, Denmark). E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB) medium (2). E. coli strains containing recombinant plasmids were cultivated in LB medium with 100 μg/ml ampicillin. Agar plates were made by adding 1.5% agar to the LB medium.
Amino acid sequence alignment of GTFB from L. reuteri and phylogenetic-tree construction.
Multiple amino acid sequence alignments of GTFB and known glucansucrases and putative 4,6-α-glucanotransferases from lactic acid bacteria were made with the ClustalW interface in MEGA, version 4 (www.megasoftware.net), with gap-opening and extension penalties of 10 and 0.2, respectively. The same program was used to construct a phylogenetic tree of Lactobacillus, Leuconostoc, and Streptococcus glucansucrases and 4,6-α-glucanotransferases. Amino acid sequences were acquired from the CAZy (Carbohydrate-Active Enzymes) database (www.cazy.org). A bootstrap test of phylogeny was performed by the neighbor-joining method using 500 replicates.
Molecular techniques.
General procedures for gene cloning, E. coli DNA transformations, DNA manipulations, and agarose gel electrophoresis have been described previously (27). Restriction endonuclease digestions and ligations with T4 DNA ligase were performed as recommended by the enzyme suppliers (New England BioLabs, Beverly, MA; Roche Biochemicals, Basel, Switzerland). Primers were obtained from Eurogentec, Seraing, Belgium. Sequencing was performed by GATC Biotech (Constance, Germany). DNA was amplified by PCR on a DNA Thermal Cycler, model PTC-200 (MJ Research, Waltham, MA) using Pwo DNA polymerase (Roche Biochemicals) or Expand High Fidelity polymerase (Fermentas, St. Leon-Rot, Germany). Plasmid DNA was isolated from E. coli using a GenElute plasmid extraction kit (Sigma, St. Louis, MO).
Construction of plasmids.
Appropriate primer pairs and template DNA were used to create two different expression constructs with C-terminal His tags: one for the complete GTFB protein (1,587 amino acids), constructed in three separate PCRs using the method previously described for L. reuteri 121 GTFA (see below) (16), and one for a variant lacking the N-terminal variable region of GTFB (889 amino acids).
To facilitate future mutagenesis and nucleotide sequencing, gtfB was divided and cloned in three parts. The first of the two PstI restriction sites present (at bp 1385 and bp 1751) was altered using the megaprimer method (28) and primers BpstIfor (5′-GTAAGTCGTTACTCAGCAGATGCTAATGG-3′), containing a mutated PstI restriction site (underlined; boldface letter represents a changed base resulting in a silent mutation), and BpstIrev (5′-GGTCAGTAAATCCACCGTTATTAATTGG-3′). In a subsequent PCR, the amplified product (420 bp) was used as a (reverse) primer together with Bfor (5′-GCAATTGTCGACCATGGATACAAATACTGGTGATCAGCAAACTGAACAGG-3′), containing SalI (italicized) and NcoI (boldface) restriction sites. The resulting 1,700-bp product was digested with SalI and PstI and was ligated into the corresponding sites of pBluescript II SK(+), yielding pBSP1600. The amplified 420-bp product was also used as a forward primer together with BrevBamHI (5′-GGACTGTTATCACTATTATTATTTCCGGCC-3′) 70 bp downstream of a BamHI restriction site. The resulting product (∼1,500 bp) was digested with PstI and BamHI and was ligated into the corresponding sites of pBluescript II SK(+), yielding pBPB1000. The third fragment was obtained using primers BforBamHI (5′-CGCTATGTAATTGAACAGAGTATTGCTGC-3′) 200 bp upstream of a BamHI restriction site and BRevHis (5′-CCTCCTTTCTAGATCTATTAGTGATGGTGATGGTGATGGTTGTTAAAGTTTAATGAAATTGCAGTTGG-3′), containing XbaI (italicized) and BglI (boldface) restriction sites and a 6×His tag (underlined). The resulting 2,300-bp product was digested with BamHI and XbaI and was ligated into the corresponding sites of pBluescript II SK(+), yielding pBBX2300. The complete gene was assembled as follows. pBPB1000 was digested with PstI and BamHI, and the resulting fragment was ligated into pBSP1600 restricted with the same restriction enzymes, yielding pBSB2600 (containing the first and second fragment). Subsequently, plasmid pBBX2300 was digested with BamHI and SacII (present on the plasmid; used instead of XbaI), and the fragment was ligated into pBSB2600, yielding pBSS4900, containing the full-length gtfB gene. This plasmid was digested with NcoI and BglI, and the gtfB gene was ligated into the NcoI and BamHI sites of pET15b, yielding pET15B-GTFB.
A 5′-truncated gtfB gene was constructed using primers GTFBcore (5′-GATGCATCCATGGGCAGCTCATGAGAAACTTGGTTGCAAAACCTAATA-3) (with the NcoI restriction site in boldface) and BrevBamHI, with pET15b-GTFB as the template. The resulting PCR product was digested with NcoI and BamHI and was ligated into the corresponding sites of pET15b-GTFB, yielding pET15b-GTFB-dN.
Site-directed mutagenesis of putative nucleophilic catalytic residues of GTFB.
Plasmid pBBX2300 (see above) was used as the template for mutagenesis. The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to construct the D1015N (putative nucleophile) mutant using primer 5′-GGTTTCCGAGTTAATGCTGCTGATA-3′ (changed bases shown in boldface) and the appropriate complementary primer. After mutagenesis, the resulting fragment was digested with BamHI and SacII and ligated in the corresponding sites of pet15b-GTFB, yielding pET15b-GTFB*D1015N.
Expression and purification of GTFB.
An overnight culture of E. coli BL21 Star (DE3) harboring (mutant) GTFB (15) was diluted 1/100. Cells were grown to an optical density at 600 nm (OD600) of 0.4 and were induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG); after 4 h of growth, cells were harvested by centrifugation (10 min at 4°C and 10,000 × g). Cell extracts were prepared by sonication, and the (mutant) GTFB proteins were purified by Ni2+-nitrilotriacetic acid (NTA) and anion-exchange chromatography, as described previously for L. reuteri 121 GTFA (reuteransucrase) (18), with the following modification: for anion-exchange chromatography, a 1-ml HiTrap Q HP column was used (GE Healthcare, Uppsala, Sweden).
pH and temperature optima.
The GTFB pH and temperature optima were determined by measuring qualitatively on thin-layer chromatography (TLC) plates (see below) the amounts of saccharides synthesized from 25 mM maltotetraose after overnight incubation (data not shown).
Incubation of MOS and other saccharide substrates with GTFB.
GTFB (90 nM) was incubated separately overnight with 25 mM sucrose (Acros), raffinose (Sigma), turanose (Sigma), palatinose (Sigma), panose (Sigma), isomaltopentaose, isomaltohexaose (Sigma), MOS with different degrees of polymerization (G2 to G7), or 0.25% amylose-V (Avebe, Foxhol, The Netherlands) in 25 mM sodium acetate buffer, pH 4.7, containing 1 mM CaCl2 at 37°C.
Donor and acceptor substrate studies.
Purified GTFB (90 nM in 25 mM sodium acetate buffer, pH 4.7, containing 1 mM CaCl2) was incubated overnight at 37°C with the donor substrate (0.25% amylose-V) and 25 mM acceptor substrate (glucose [G1] or maltose [G2]).
TLC and high-performance anion-exchange chromatography (HPAEC).
For TLC analysis of saccharide product mixtures, 1 to 3 μl of the sample was applied to a silica gel 60 F254 plate (Merck, Darmstadt, Germany), and after drying, the plate was run for 6 h in butanol-ethanol-H2O (5:5:3 [vol/vol/vol]). Then the plate was dried, sprayed with 50% H2SO4 in methanol, and left to develop for 10 min at 110°C.
For HPAEC analysis, appropriate dilutions of enzyme reaction mixtures were dissolved in 90% dimethyl sulfoxide. A commercial mixture of MOS (DP1 to DP7) and a debranched waxy maize starch solution containing a broad mixture of oligosaccharides of known compositions were used as standards. Separation was achieved on a CarboPac PA1 anion-exchange column (250 mm by 4 mm) coupled to a CarboPac1 guard column (both from Dionex, Amsterdam, The Netherlands). Eluent A was 0.1 M NaOH; eluent B was 0.6 M sodium acetate in 0.1 M NaOH; and the gradient used was eluent A (1 ml/min) at 95% (10 min), 65% (10 min), 55% (30 min), 35% (4 min), 0% (7 min), and 95% (14 min). Detection was performed with an ED40 electrochemical detector (Dionex) with an Au working electrode and an Ag/AgCl reference electrode with a sensitivity of 300 nC. The pulse program used was as follows: +0.1 V (0 to 0.41 s), −2.0 V (0.41 to 0.43 s), +0.6 V (0.43 to 0.44 s), and −0.10 V (0.44 to 0.50 s); integration time, 0.20 to 0.40 s. Data were integrated using a TotalChrom (Perkin-Elmer) data integration system.
Production and analysis of saccharides from maltoheptaose incubation with GTFB.
Purified GTFB (90 nM) was incubated for 7 days with 150 mM maltoheptaose (G7) (Sigma) under the conditions described under “Incubation of MOS and other saccharide substrates with GTFB” above. The saccharides produced were separated by treatment with 96% ethanol into two fractions (33): a supernatant fraction (O1) and a precipitate fraction (P2).
(i) Methylation analysis.
Fractions O1 and P2 were permethylated using methyl iodide and sodium methylsulfinylmethanide in dimethyl sulfoxide at room temperature (15). After hydrolysis with 2 M trifluoroacetic acid (2 h, 120°C), the partially methylated monosaccharide mixtures generated were reduced with NaBD4 (2 h, room temperature). The workup involved neutralization with acetic acid and removal of boric acid by coevaporation with methanol. Partially methylated alditols were peracetylated with acetic anhydride-pyridine, 1:1 (vol/vol) (3 h, 120°C), yielding mixtures of partially methylated alditol acetates, which were analyzed by gas-liquid chromatography–electron impact mass spectrometry (GLC-EI-MS).
(ii) NMR spectroscopy.
One-dimensional 1H nuclear magnetic resonance (NMR) spectra of the incubation sample and fraction P2 were recorded on a Bruker DRX 500 spectrometer (Bijvoet Center, Department of NMR Spectroscopy, Utrecht University) at a probe temperature of 300 K. Samples were exchanged once with 99.9 atom% D2O, lyophilized, and dissolved in 650 μl D2O. 1H chemical shifts (δ) are expressed in parts per million by reference to internal acetone (δ 2.225). The 1H NMR spectra were recorded with a spectral width of 5 kHz in 16 kHz complex data sets and were zero filled to 32 kHz. A water-eliminated Fourier transform (WEFT) pulse sequence was applied to suppress the HOD signal (7). When necessary, a fifth-order polynomial baseline correction was applied. NMR data were processed using software originally developed by J. A. van Kuik (Bijvoet Center, Department of Bio-Organic Chemistry, Utrecht University).
(iii) MS.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) experiments with the incubation sample and fractions O1 and P2 were performed on a Voyager DE Pro mass spectrometer (Applied Biosystems, Nieuwerkerk aan de IJssel, The Netherlands) equipped with a nitrogen laser (wavelength, 337 nm; pulse width, 3 ns) (Bijvoet Center, Department of Bio-Organic Chemistry, Utrecht University). Positive-ion-mode spectra were recorded in the reflector mode at an accelerating voltage of 24 kV, using an extraction delay of 90 ns, with a resolution of 5,000 to 9,000 (full width at half-maximum intensity [FWHM]). The acquisition mass range was 500 to 3,000 Da. Samples were prepared by mixing on the target 1 μl of aqueous saccharide solutions with 1 μl 2,5-dihydroxybenzoic acid (10 mg/ml) in 40% aqueous acetonitrile as the matrix solution. GLC-EI-MS was performed on a Fisons Instruments GC 8060/MD 800 system (Interscience BV, Breda, The Netherlands) equipped with an AT-1 column (30 m by 0.25 mm; Alltech, Uden, The Netherlands) by using a temperature gradient of 140 to 240°C at 4°C/min (11) (Bijvoet Center, Department of Bio-Organic Chemistry, Utrecht University).
RESULTS
Dendrogram and alignment of GTFB.
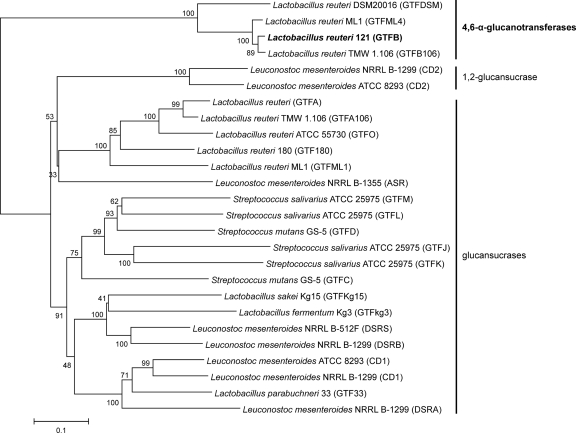
L. reuteri 121 GTFB is the first representative of a group of homologous enzymes identified in different lactic acid bacteria. Genes encoding putative GTFB homologs are present in several Lactobacillus strains, e.g., in L. reuteri ML1, where fragments of a GTFB homolog, gtfML4, were identified upstream of gtfML1, encoding a mutansucrase (15). A GTFB homolog could also be identified in the recently elucidated genome sequence of L. reuteri DSM 20016 (73% identity and 85% similarity in 883 amino acids). Finally, L. reuteri TMW1.106 also contains GTFB (GTF106B) and GTFA (GTF106A) homologs. The GTF106B enzyme showed 92% identity and 95% similarity in 1,383 amino acids with L. reuteri 121 GTFB. However, in contrast to GTFB, GTF106B hydrolyzed sucrose slowly (apparent after 27 h of incubation) (10). Screening of samples from Indonesia also revealed the catalytic part of a putative GTFB homolog in Weissella confusa MBF 8.1 (22). Phylogenetically, these GTFB enzymes cluster together but are also closely related to glucansucrases acting on sucrose as a substrate (Fig. 1) (5, 34). Nevertheless, GTFB has no detectable activity with sucrose (15). In the present study, it is shown (see below) that GTFB uses maltooligosaccharides (MOS) instead as donor and acceptor substrates and that on the basis of the analytical findings, GTFB has to be designated a (1→4)-α-d-glucan:(1→4),(1→6)-α-d-glucan α-glucanotransferase (in short, a 4,6-α-glucanotransferase).
Fig. 1.
Unrooted phylogenetic tree of GTF proteins (glucansucrases and [putative] 4,6-α-glucanotransferases) from lactic acid bacteria. Alignments and dendrogram construction were carried out (using the catalytic cores only [for example, from “WYRP” to “WVPDQ” of L. reuteri 121 GTFA]) with MEGA version 4, using the neighbor joining method. Bootstrap values (expressed as percentages) are given at the branching points. The bar corresponds to a genetic distance of 0.1 substitution per position (10% amino acid sequence difference).
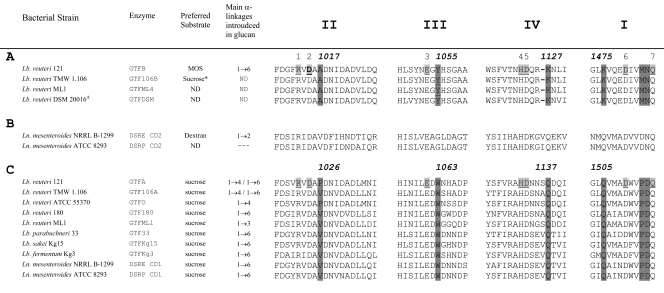
Alignments of members of this novel group of GTFB-like enzymes with typical glucansucrases showed similarities (for GTFB and GTFA of L. reuteri 121, 46% identity and 61% similarity in 1,683 amino acids) but also clear and characteristic differences (Fig. 2). The three catalytic residues present (D1024, E1061, and D1133 in L. reuteri 121 GTFA [this numbering is used throughout unless indicated otherwise]) in glucansucrases are also present in the group of 4,6-α-glucanotransferases (D1015, E1053, and D1125 [L. reuteri 121 GTFB numbering]). Nevertheless, a large number of amino acid residues conserved in regions I, II, III, and IV of glucansucrases are absent in the 4,6-α-glucanotransferase group of enzymes (Fig. 2). In region II (encompassing the putative nucleophilic residue), the conserved residue V1026 (Pro in GTFA and GTFO) is replaced by Ala in the 4,6-α-glucanotransferases. Region III, the region downstream of the putative acid/base catalyst E1061, is completely different in glucansucrases versus 4,6-α-glucanotransferases. The conserved residue W1063 is replaced by a Tyr residue in the 4,6-α-glucanotransferases (Fig. 2). Also, region IV, with the transition state-stabilizing D1133 residue, is very different in glucansucrases versus 4,6-α-glucanotransferases. The GTFB homologs contain a gap immediately upstream of the location of the Q1137 residue, and all have a Lys residue instead of this Gln residue. In conserved region I, the consensus sequence DWVPDQ, present in most glucansucrases, differs from the D(I/L)VMNQ motif present in the 4,6-α-glucanotransferases (Fig. 2).
Fig. 2.
Amino acid sequence (http://www.cazy.org) alignment of conserved regions (II, III, IV, and I) in the catalytic domains of (putative) 4,6-α-glucanotransferase enzymes (A), DSRE (5) and DSRP, glucansucrase enzymes containing two catalytic domains (CD1 and CD2) (B), and dextran-, mutan-, alternan-, and reuteransucrase enzymes of lactic acid bacteria (C) (see also references 17 and 23). The seven strictly conserved amino acid residues (indicated by the numbers 1 to 7 above the sequences; underlined and lightly shaded in L. reuteri 121 GTFA and GTFB), with important contributions to the −1 and +1 subsites in glucansucrase enzymes, are also conserved in the 4,6-α-glucanotransferase enzymes. GTFB amino acid D1015 (the putative nucleophilic residue), targeted in this study, is shown in boldface. Dark shading indicates changes in conserved amino acid residues between 4,6-α-glucanotransferases and glucansucrases; the corresponding amino acid numbering is indicated. *, low activity.
Cloning and expression of the gtfB gene.
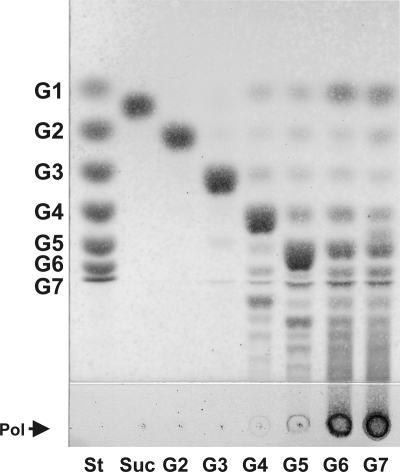
Full-length GTFB, the N-terminally truncated version, and the putative nucleophilic mutant D1015N GTFB were constructed and expressed successfully in E. coli. Following their purification, both the full-length enzyme (Fig. 3) and the N-terminally truncated variant (GTFB-ΔN) (data not shown) showed clear activity on MOS G4 to G7. The GTFB-ΔN variant was not expressed as efficiently as full-length GTFB, and therefore, all further experiments were performed using full-length GTFB. No background activity was detected in E. coli itself, as was evident from a control experiment with an empty pET15b plasmid, which demonstrated that after the His tag purification step, no activity on MOS (maltose to maltoheptaose [G2 to G7]) was detected in the samples obtained (data not shown). The purified full-length D1015N (putative) nucleophilic GTFB mutant also showed no activity on MOS G2 to G7 (data not shown).
Fig. 3.
TLC analysis of the reaction products of 90 nM GTFB incubated for 13 h in 50 mM sodium acetate buffer, pH 4.7, containing 1 mM CaCl2 with 25 mM sucrose or 25 mM maltooligosaccharides. St, standard; Suc, sucrose; G1, glucose; G2, maltose; G3, maltotriose; G4, maltotetraose; G5, maltopentaose; G6, maltohexaose; G7, maltoheptaose; Pol, polymer.
GTFB enzyme characteristics.
The optimal temperature and pH for GTFB activity with maltotetraose (G4) as a substrate were 30 to 37°C and pH 4 to 5, respectively (data not shown). Assays with various combinations of temperatures and buffers yielded the highest activity levels at 37°C and pH 4.7; these values were used in all subsequent assays.
GTFB donor substrates.
GTFB was unable to use sucrose as a donor substrate (Fig. 3, TLC analysis) (15) and was also inactive with the sucrose analogs turanose and palatinose, with raffinose, with the DP5 and DP6 isomaltooligosaccharides (IMO), with panose, and with an oligosaccharide mixture obtained by hydrolysis of a partially purified reuteran (produced by the L. reuteri 121 GTFA enzyme) (data not shown). Clear activity, however, was observed after relatively short incubation times with MOS; i.e., G4 and larger MOS yielded a range of different oligosaccharide products (Fig. 3). With substrates of DP6 or larger, polymeric material as well as oligosaccharides started to accumulate (Fig. 3). Under the incubation conditions tested, virtually no activity was observed on maltose or maltotriose (Fig. 3). Only low activity was observed with amylose-V, resulting mainly in G1 and G2 release (see below).
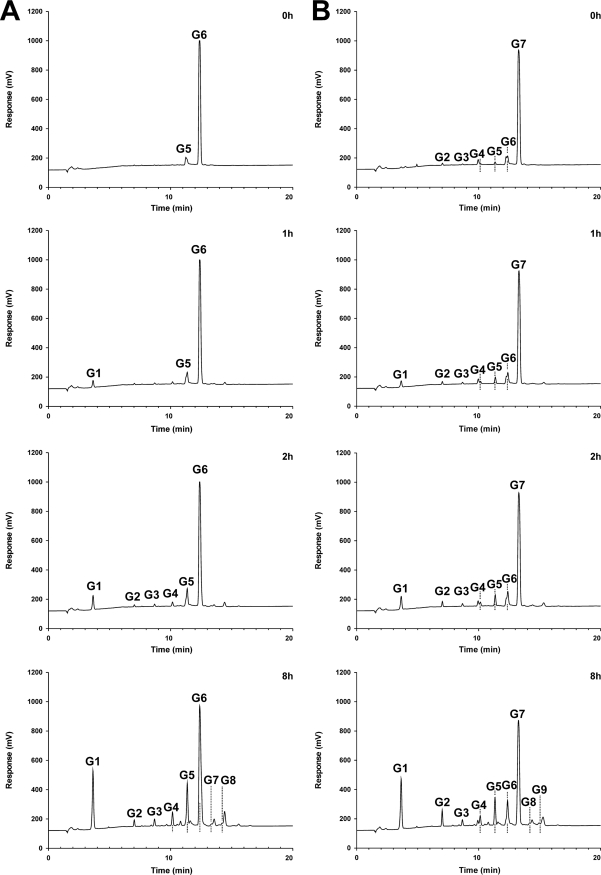
GTFB products from G6 and G7 accumulating in time.
By HPAEC analysis, the first clear reaction products detectable after 1 h with G6 (slightly contaminated with G5) as the substrate were G1 (glucose) and G5 (maltopentaose) (Fig. 4A). Similarly, with G7 (slightly contaminated with oligosaccharides of shorter retention times) as the substrate, the first saccharide products released were G1 (glucose) and G6 (maltohexaose) (Fig. 4B). Later on, incubations with G6 or G7 yielded peaks at retention times of MOS with lower DPs than that of the starting donor, but also peaks that did not fit with the MOS retention times, especially products with longer retention times than that of the starting donor.
Fig. 4.
HPAEC analysis of the reaction products of 90 nM GTFB incubated for 0 to 8 h in 50 mM sodium acetate buffer, pH 4.7, containing 1 mM CaCl2 with 25 mM maltohexaose (A) or 25 mM maltoheptaose (B).
GTFB donor and acceptor substrate studies.
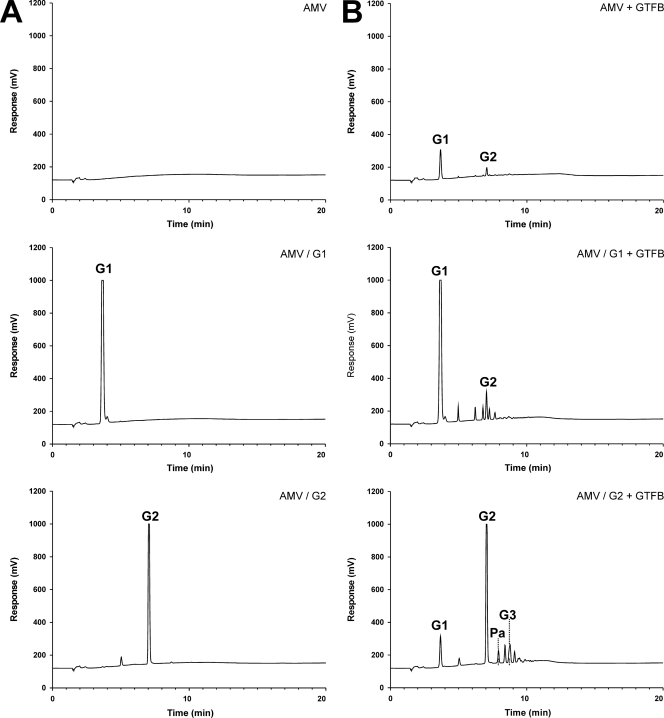
As indicated by HPAEC analysis, incubation of amylose-V with GTFB yielded some glucose (G1) and maltose (G2) (compare Fig. 5A and B). When amylose-V as a donor substrate was incubated with GTFB and glucose as an acceptor substrate, a range of oligosaccharides was synthesized (Fig. 5); also, larger amounts of maltose were detected than for the incubation with amylose-V alone (Fig. 5). Incubation of amylose-V with GTFB and maltose as the acceptor substrate yielded, among others, HPAEC peaks at the positions of panose [Glc-(α1→6)-maltose] and maltotriose (G3) (Fig. 5).
Fig. 5.
HPAEC analysis of samples with 0.25% amylose-V (AMV) alone (donor substrate) or amylose-V with 25 mM glucose or 25 mM maltose (acceptor substrates), either without the GTFB enzyme (A) or with GTFB (90 nM) (B) incubated overnight at 37°C in 25 mM sodium acetate buffer, pH 4.7, containing 1 mM CaCl2. Pa, panose.
Characterization of the GTFB products with maltoheptaose (G7).
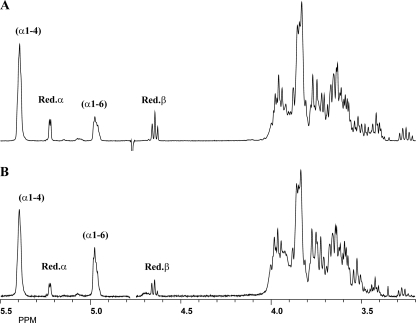
MALDI-TOF MS analysis of the 150 mM G7 incubation sample showed visible [M + Na]+ peaks in the region of DP3 (m/z 527) up to DP10 (m/z 1,661), with DP5 as the major peak (100%), followed by DP6 (48%) (Table 1). It should be noted that m/z values below 500 were not recorded due to matrix-related noise in that area. The 1-dimensional 1H NMR spectrum (Fig. 6A) revealed, besides the anomeric signals at δ 5.42 to 5.37 (indicative of α1→4 linkages, in the absence of α1→3 linkages), δ 5.225 [reducing -(1→4)-α-d-Glcp unit and α-anomer of free glucose], δ 4.653 [reducing -(1→4)-β-d-Glcp unit; H-2 at δ 3.271], and δ 4.634 (β-anomer of free glucose; H-2 at δ 3.237), an additional broad anomeric peak at δ 5.00 to 4.95, indicative of α1→6 linkages (35, 37). The molar ratio of the α1→4-linked, α1→6-linked, and reducing unit glucose residues was 60:18:22. Ethanol treatment of the G7 incubation sample yielded a supernatant fraction (O1) and a precipitate fraction (P2).
Table 1.
DP distribution of the glucooligomers, as detected by MALDI-TOF MS analysis, in the incubation mixture of maltoheptaose with GTFB, its ethanol supernatant fraction O1, and its ethanol precipitate fraction P2
| DP | Peak intensity (%) of the glucooligomer in: |
||
|---|---|---|---|
| Incubation mixture | Fraction O1 | Fraction P2 | |
| 3 | 11 | 8 | 12 |
| 4 | 12 | 10 | 14 |
| 5 | 100 | 100 | 100 |
| 6 | 48 | 67 | 77 |
| 7 | 17 | 17 | 46 |
| 8 | 8 | 10 | 35 |
| 9 | 7 | 9 | 31 |
| 10 | 6 | 2 | 28 |
| 11 | NDa | ND | 19 |
| 12 | ND | ND | 8 |
| 13 | ND | ND | 4 |
| 14 | ND | ND | 3 |
| 15 | ND | ND | 2 |
| 16 | ND | ND | 1 |
| 17 | ND | ND | ND |
ND, not detected under the conditions used.
Fig. 6.
Five hundred-megahertz 1-dimensional 1H NMR analysis of maltoheptaose (DP7) incubated with 90 nM GTFB for 7 days in 50 mM sodium acetate buffer, pH 4.7, containing 1 mM CaCl2. (A) Incubation sample; (B) ethanol-precipitated fraction (P2) from the incubation sample. Red., reducing glucose unit.
MALDI-TOF MS analysis of fraction O1 showed evidence of the same DP range as that in the incubation sample, with DP5 as the major component(s) (100%), followed by DP6 (67%) (Table 1). Methylation (linkage) analysis of fraction O1 showed the presence of 4-substituted, 6-substituted, and terminal glucose residues at a molar ratio of 63, 17, and 20%, indicating the presence of linear products only (no branched glucose residues).
MALDI-TOF MS analysis of fraction P2 showed visible [M + Na]+ peaks in the region of DP3 (m/z 527) up to DP16 (m/z 2633), with DP5 as the major peak (100%), followed by DP6 (77%). Compared with those in the incubation mixture and fraction O1, the peak intensities of DP7 to DP11 glucooligomers were significantly increased. Methylation (linkage) analysis of fraction P2 showed 4-substituted, 6-substituted, and terminal glucose residues at a molar ratio of 52, 35, and 13%, indicating the presence of linear products only. In the 1-dimensional 1H NMR spectrum of fraction P2 (Fig. 6B), the areas of the anomeric signals at δ 5.42 to 5.37 (α1→4 linkages [36, 37]), δ 5.226 and 4.655 (α and β configuration, respectively, of the reducing glucose unit [37]), and δ 5.00 to 4.95 (α1→6 linkages [35, 37]) occurred at a molar ratio of 55, 33, and 12%, indicating a significant increase in the percentage of α1→6-linked glucose residues over those in the incubation sample. This suggests that the α1→6 linkages are specifically included in the higher-DP linear chains.
DISCUSSION
This paper reports the identification and characterization of a new reaction specificity within the GH70 family, namely, the disproportionation of maltooligosaccharides (MOS), also introducing α1→6 glycosidic linkages into the products formed.
L. reuteri 121 GTFB is the first member of this group of enzymes that has been characterized in detail. Surprisingly, GTFB has virtually no activity with sucrose. Incubation of L. reuteri 121 GTFB with MOS of different DPs produced saccharides with α1→4 glycosidic linkages alone and saccharides that also contained α1→6 glycosidic linkages. Incubation of GTFB with amylose as a donor substrate and glucose or maltose as an acceptor substrate confirmed the ability of the enzyme to synthesize α1→4 glycosidic linkages also. In particular, with maltose as an acceptor, among other unidentified products, panose [glucosyl-(α1→6)-maltose] and G3 [glucosyl-(α1→4)-maltose] were detected. Preliminary investigation of the products generated when GTFB was incubated with maltoheptaose (DP7) by methylation (linkage) analysis (GLC-EI-MS), 1H NMR analysis, and MALDI-TOF MS revealed the presence of only linear oligosaccharides containing α1→4 and α1→6 glycosidic linkages. There was a range of glucooligosaccharides from DP3 to DP16, with an overall content of around 18% α1→6 glycosidic bonds, and the percentage of α1→6 glycosidic linkages was higher in the higher-DP products (33% in the ethanol-precipitated fraction). Based on these findings, we have designated the GTFB enzymes (1→4)-α-d-glucan:(1→4)(1→6)-α-d-glucan α-glucanotransferases (in short, 4,6-α-glucanotransferases).
Phylogenetic analysis shows that the 4,6-α-glucanotransferases form a separate group within the glucansucrases (Fig. 1). The three amino acids crucial for catalysis in glucansucrase enzymes (D1024, E1061, and D1133 [L. reuteri 121 GTFA numbering]) (18) are also conserved in the 4,6-α-glucanotransferase group of enzymes (D1015, E1053, and D1125 [L. reuteri 121 GTFB numbering]) (Fig. 2). The recent elucidation of the first 3D structure of the N-terminally truncated glucansucrase from L. reuteri 180 showed that glucansucrase enzymes contain 5 domains (A, B, C, IV, and V) and that the peptide chain follows a “U” path. The A, B, and C domains are similar and can be superimposed on the A, B, and C domains present in Bacillus licheniformis α-amylase. Domains IV and V are unique to glucansucrase enzymes (25, 38). A domain organization similar to that in glucansucrase enzymes is also found in the 4,6-α-glucanotransferases. These 4,6-α-glucanotransferase enzymes thus represent structural and functional evolutionary intermediates between family GH13 and GH70 enzymes.
Mutation of the GTFB putative nucleophile amino acid residue (D1015N) resulted in an inactive enzyme, confirming the crucial importance of this residue in catalysis. The other six conserved residues in GH70 enzymes, present in regions I, II, III, and IV, are also conserved in 4,6-α-glucanotransferase enzymes (Fig. 2). Interestingly, a large number of amino acid residues conserved in regions II, III, IV, and I in the glucansucrase type of enzymes are absent or differ in the 4,6-α-glucanotransferase group of enzymes (Fig. 2). Amino acid residues in regions I to IV contribute to the −1, +1, and +2 donor/acceptor substrate binding subsites and therefore are important in determining substrate specificity (38). Differences in amino acid residues contributing to these donor and acceptor substrate binding subsites may explain the differences in substrate specificity between glucansucrases and 4,6-α-glucanotransferases. The same applies to differences in glycosidic bond specificity between the products synthesized (8, 19).
Although the active sites of glucansucrases and 4,6-α-glucanotransferases show clear similarities, there are some differences. Questions arise about the active-site organization in 4,6-α-glucanotransferases, in particular the number of donor substrate binding subsites present. Only one donor subsite is present in glucansucrase enzymes, whereas other amylolytic enzymes, such as amylases and amylomaltases, have more than one donor subsite. In order to address this question, GTFB enzyme incubations with maltohexaose (G6) or maltoheptaose (G7) as the substrate were followed over time using HPAEC. The first reaction products detectable with G6 were G1 (glucose) and G5 (maltopentaose) (Fig. 4A). Similarly, with G7, the first products released were G1 (glucose) and G6 (maltohexaose) (Fig. 4B). With both G6 and G7, other MOS of lower DPs also accumulated, and at later times, unknown saccharides of higher DPs, which, besides α1→4 glycosidic linkages, were likely to contain other linkages (in fact, α1→6 glycosidic linkages, as demonstrated by methylation and 1H NMR analysis), were also present. The data thus suggest that 4,6-α-glucanotransferases have an active-site architecture similar to that of glucansucrases, both containing only a single donor substrate binding subsite (−1).
The enzymes from glycoside hydrolase families GH13, GH70, and GH77 together form clan GH-H (www.cazy.org). All GH-H members employ similar catalytic mechanisms involving a covalent glucosyl-enzyme intermediate and retention of the α-anomeric configuration of the product upon hydrolysis (13, 32). Amylomaltases (also known as 4-α-glucanotransferases) from GH77 are capable of synthesizing large cyclic glucans and disproportionating MOS. Notably, amylomaltases almost exclusively catalyze transglycosylation reactions and only cleave and synthesize α1→4 glycosidic bonds (3). The GH13 enzyme family, acting mainly on starch-like substrates and displaying a large variety of different reaction specificities, is the largest of the GH families at present, organized in different subfamilies in the CAZy database (30). Amylosucrase is the only enzyme from family GH13 that uses sucrose as a substrate to synthesize an amylose type of α-glucan polymer. Amylosucrase enzymes thus have the GH13 type of domain architecture and the GH70 type of glucansucrase activity (24, 29).
All α-amylases have similar domain organizations (see above), even those of archaeal origin (20). Glucansucrases are found only in Bacteria, suggesting that the precursor amylase enzyme from which GH70 glucansucrases and present-day amylases have evolved had a similar domain organization. In addition, it is most likely that amylolytic activity, the main activity of α-amylases, emerged first, and that the α-glucanotransferase and sucrase types of activities evolved from the precursor amylase. It appears less likely that a family GH77 enzyme is an intermediate, since these proteins possess various additional domains (B1, partly present in α-amylase members; B2 and B3, unique to amylomaltases); in addition, they lack the C domain (26). Furthermore, GH77 enzymes do not possess α1→6 specificity, in contrast to GH13 and GH70 family members. The GTFB enzyme clearly is a glucansucrase type of protein with regard to its amino acid sequence and domain organization but lacks the ability to act on sucrose. Surprisingly, GTFB acts on MOS, substrates used by various GH13 (and GH77) family members, and is able to cleave α1→4 glycosidic linkages and to synthesize α1→4 and α1→6 glycosidic linkages. GTFB from the GH70 family therefore provides a link between the GH13 α-amylase and GH70 glucansucrase families. The latter use sucrose as a substrate to synthesize linear as well as branched α-glucan polymers, differing in the type of glycosidic linkages (reuteran, α1→4; dextran, α1→6; alternan, α1→3/α1→6; mutan, α1→3), the degree and type of branching, the length of the glucan chains, molecular mass, and the conformation of the polymers (34).
Glucansucrase enzymes also use MOS as acceptor substrates with sucrose as donor substrate. This results in the synthesis of a range of oligosaccharides, e.g., a maltose extended with a series of glucose units bound via α1→6 linkages in the case of dextransucrase. In this case, however, the α1→4 linkages in MOS substrates are not cleaved; MOS are used only as acceptor substrates (6, 12). This is a major difference from the GFTB enzyme, which fails to act on sucrose and instead uses MOS as donor and acceptor substrates, cleaving α1→4 linkages and introducing new α1→4 and α1→6 linkages. GTFB thus appears to be an evolutionary precursor for the glucansucrase type of enzymes with the GH70 domain architecture and the GH13 amylolytic activity (38).
Although 4,6-α-glucanotransferases and glucansucrase enzymes are commonly arranged in tandem on the genome (10, 15), the exact in vivo role of GTFB-like enzymes remains unknown. They may scavenge and modify the oligosaccharides formed by glucansucrase enzymes as substrates for the synthesis of larger saccharides, which are inaccessible to other microbes. They may also play a role in the modification of the glucan synthesized by the glucansucrase enzyme of the microorganism. Interestingly, the genome of L. reuteri DSM 20016 contains only a GTFB-like enzyme and no glucansucrase enzyme. This Lactobacillus strain may be the key to answering questions about the in vivo role of 4,6-α-glucanotransferase enzymes.
Conclusions.
Based on the enzymatic activity of GTFB, we propose to give this group of enzymes a separate EC number, different from EC 2.4.1.5 (for common glucansucrase enzymes) or EC 2.4.1.140 (for alternansucrase). We propose to name these enzymes (1→4)-α-d-glucan:(1→4),(1→6)-α-d-glucan α-glucanotransferase enzymes (in short 4,6-α-glucanotransferases). We also propose to divide GH70 family enzymes into two subfamilies in the CAZy database, as has been done for GH13 enzymes (30), namely, glucansucrase enzymes acting on sucrose or on MOS.
The precise in vivo reaction and the physiological function of the GTFB type of enzymes remain to be determined. Clearly, this new group of α-glucanotransferase enzymes provides a valuable asset in the toolbox for saccharide synthesis. The linear oligosaccharides synthesized by GTFB containing α1→4 and α1→6 glycosidic linkages may have interesting physicochemical properties, which remain to be determined, and may have potentially new applications in the food, cosmetics, and/or pharmaceutical industries. In the future, we will characterize the saccharides synthesized by GTFB in more detail, as well as the characteristic properties of other members of this enzyme group.
ACKNOWLEDGMENTS
We thank Peter Sanders (TNO Quality of Life) for anion-exchange (Dionex) analysis and Rolf Boelens (Bijvoet Center, Department of NMR Spectroscopy, Utrecht University, Utrecht, The Netherlands) for providing us with measuring time on the 500-MHz NMR instrument.
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Argüello-Morales M. A., et al. 2000. Sequence analysis of the gene encoding alternansucrase, a sucrose glucosyltransferase from Leuconostoc mesenteroides NRRL B-1355. FEMS Microbiol. Lett. 182:81–85 [DOI] [PubMed] [Google Scholar]
- 2. Ausubel F. M., et al. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 3. Barends T. R. M., et al. 2007. Three-way stabilization of the covalent intermediate in amylomaltase, an α-amylase-like transglycosylase. J. Biol. Chem. 282:17242–17249 [DOI] [PubMed] [Google Scholar]
- 4. Devulapalle K. S., Goodman S. D., Gao Q., Hemsley A., Mooser G. 1997. Knowledge-based model of a glucosyltransferase from the oral bacterial group of mutans streptococci. Protein Sci. 6:2489–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fabre E., et al. 2005. Role of the two catalytic domains of DSR-E dextransucrase and their involvement in the formation of highly α-1,2 branched dextran. J. Bacteriol. 187:296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fu D., Robyt J. F. 1991. Maltodextrin acceptor reactions of Streptococcus mutans 6715 glucosyltransferases. Carbohydr. Res. 217:201–211 [DOI] [PubMed] [Google Scholar]
- 7. Hård K., van Zadelhoff G., Moonen P., Kamerling J. P., Vliegenthart J. F. G. 1992. The Asn-linked carbohydrate chains of human Tamm-Horsfall glycoprotein of one male. Novel sulfated and novel N-acetylgalactosamine-containing N-linked carbohydrate chains. Eur. J. Biochem. 209:895–915 [DOI] [PubMed] [Google Scholar]
- 8. Hellmuth H., et al. 2008. Engineering the glucansucrase GTFR enzyme reaction and glycosidic bond specificity: toward tailor-made polymer and oligosaccharide products. Biochemistry 47:6678–6684 [DOI] [PubMed] [Google Scholar]
- 9. Ito K., et al. 2011. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J. Mol. Biol. 408:177–186 [DOI] [PubMed] [Google Scholar]
- 10. Kaditzky S. B., et al. 2008. Influence of pH on the formation of glucan by Lactobacillus reuteri TMW 1.106 exerting a protective function against extreme pH values. Food Biotechnol. 22:398–418 [Google Scholar]
- 11. Kamerling J. P., Vliegenthart J. F. G. 1989. Carbohydrates, p. 176–263 In Lawson A. M. (ed.), Clinbiochemistry: principles, methods, applications, vol. 1. mass spectrometry. Walter de Gruyter, Berlin, Germany [Google Scholar]
- 12. Kang H. K., Oh J.-S., Kim D. 2009. Molecular characterization and expression analysis of the glucansucrase DSRWC from Weissella cibaria synthesizing a α(1→6) glucan. FEMS Microbiol. Lett. 292:33–41 [DOI] [PubMed] [Google Scholar]
- 13. Kelly R. M., Dijkhuizen L., Leemhuis H. 2009. Starch and α-glucan acting enzymes, modulating their properties by directed evolution. J. Biotechnol. 140:184–193 [DOI] [PubMed] [Google Scholar]
- 14. Kralj S., Stripling E., Sanders P., van Geel-Schutten G. H., Dijkhuizen L. 2005. Highly hydrolytic reuteransucrase from probiotic Lactobacillus reuteri strain ATCC 55730. Appl. Environ. Microbiol. 71:3942–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kralj S., et al. 2004. Glucan synthesis in the genus Lactobacillus: isolation and characterization of glucansucrase genes, enzymes and glucan products from six different strains. Microbiology 150:3681–3690 [DOI] [PubMed] [Google Scholar]
- 16. Kralj S., et al. 2002. Molecular characterization of a novel glucosyltransferase from Lactobacillus reuteri strain 121 synthesizing a unique, highly branched glucan with α-(1→4) and α-(1→6) glucosidic bonds. Appl. Environ. Microbiol. 68:4283–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kralj S., van Geel-Schutten G. H., van der Maarel M. J. E. C., Dijkhuizen L. 2003. Efficient screening methods for glucosyltransferase genes in Lactobacillus strains. Biocatal. Biotransform. 21:181–187 [Google Scholar]
- 18. Kralj S., van Geel-Schutten G. H., van der Maarel M. J. E. C., Dijkhuizen L. 2004. Biochemical and molecular characterization of Lactobacillus reuteri 121 reuteransucrase. Microbiology 150:2099–2112 [DOI] [PubMed] [Google Scholar]
- 19. Kralj S., van Geel-Schutten I. G. H., Faber E. J., van der Maarel M. J. E. C., Dijkhuizen L. 2005. Rational transformation of Lactobacillus reuteri 121 reuteransucrase into a dextransucrase. Biochemistry 44:9206–9216 [DOI] [PubMed] [Google Scholar]
- 20. Linden A., Mayans O., Meyer-Klaucke W., Antranikian G., Wilmanns M. 2003. Differential regulation of a hyperthermophilic α-amylase with a novel (Ca,Zn) two-metal center by zinc. J. Biol. Chem. 278:9875–9884 [DOI] [PubMed] [Google Scholar]
- 21. MacGregor E. A., Jespersen H. M., Svensson B. 1996. A circularly permuted α-amylase-type α/β-barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 378:263–266 [DOI] [PubMed] [Google Scholar]
- 22. Malik A., Radji M., Kralj S., Dijkhuizen L. 2009. Screening of lactic acid bacteria from Indonesia reveals glucansucrase and fructansucrase genes in two different Weissella confusa strains from soya. FEMS Microbiol. Lett. 300:131–138 [DOI] [PubMed] [Google Scholar]
- 23. Monchois V., Willemot R.-M., Monsan P. 1999. Glucansucrases: mechanism of action and structure-function relationships. FEMS Microbiol. Rev. 23:131–151 [DOI] [PubMed] [Google Scholar]
- 24. Moulis C., et al. 2006. Understanding the polymerization mechanism of glycoside-hydrolase family 70 glucansucrases. J. Biol. Chem. 281:31254–31267 [DOI] [PubMed] [Google Scholar]
- 25. Pijning T., et al. 2008. Biochemical and crystallographic characterization of a glucansucrase from Lactobacillus reuteri 180. Biocatal. Biotransform. 26:12–17 [Google Scholar]
- 26. Przylas I., et al. 2000. Crystal structure of amylomaltase from Thermus aquaticus, a glycosyltransferase catalysing the production of large cyclic glucans. J. Mol. Biol. 296:873–886 [DOI] [PubMed] [Google Scholar]
- 27. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Sarkar G., Sommer S. S. 1990. The “megaprimer” method of site-directed mutagenesis. Biotechniques 8:404–407 [PubMed] [Google Scholar]
- 29. Skov L. K., et al. 2001. Amylosucrase, a glucan-synthesizing enzyme from the α-amylase family. J. Biol. Chem. 276:25273–25278 [DOI] [PubMed] [Google Scholar]
- 30. Stam M. R., Danchin E. G. J., Rancurel C., Coutinho P. M., Henrissat B. 2006. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng. Des. Sel. 19:555–562 [DOI] [PubMed] [Google Scholar]
- 31. Svensson B. 1994. Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol. Biol. 25:141–157 [DOI] [PubMed] [Google Scholar]
- 32. Uitdehaag J. C. M., et al. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat. Struct. Biol. 6:432–436 [DOI] [PubMed] [Google Scholar]
- 33. van Geel-Schutten G. H., et al. 1999. Biochemical and structural characterization of the glucan and fructan exopolysaccharides synthesized by the Lactobacillus reuteri wild-type strain and by mutant strains. Appl. Environ. Microbiol. 65:3008–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Hijum S. A. F. T., Kralj S., Ozimek L. K., Dijkhuizen L., van Geel-Schutten I. G. 2006. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol. Mol. Biol. Rev. 70:157–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Leeuwen S. S., et al. 2008. Structural analysis of the α-d-glucan (EPS180) produced by the Lactobacillus reuteri strain 180 glucansucrase GTF180 enzyme. Carbohydr. Res. 343:1237–1250 [DOI] [PubMed] [Google Scholar]
- 36. van Leeuwen S. S., et al. 2008. Structural analysis of the α-d-glucan (EPS35-5) produced by the Lactobacillus reuteri strain 35-5 glucansucrase GTFA enzyme. Carbohydr. Res. 343:1251–1265 [DOI] [PubMed] [Google Scholar]
- 37. van Leeuwen S. S., Leeflang B. R., Gerwig G. J., Kamerling J. P. 2008. Development of a 1H NMR structural-reporter-group concept for the primary structural characterisation of α-d-glucans. Carbohydr. Res. 343:1114–1119 [DOI] [PubMed] [Google Scholar]
- 38. Vujičić-Žagar A., et al. 2010. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc. Natl. Acad. Sci. U. S. A. 107:21406–21411 [DOI] [PMC free article] [PubMed] [Google Scholar]