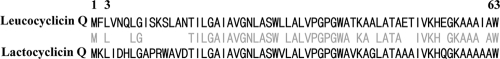Abstract
The culture supernatant of Leuconostoc mesenteroides TK41401, isolated from Japanese pickles, possessed antimicrobial activity against broad range of a bacterial genera and particularly strong activity against Bacillus coagulans, the major contaminant of pickles. An antimicrobial peptide was purified in three chromatographic steps, and its molecular mass was determined to be 6,115.59 Da by electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS). The primary structure of this peptide was determined by amino acid and DNA sequencing, and these analyses revealed that it was translated as a 63-residue precursor. This precursor showed high similarity to the precursor of lactocyclicin Q, a cyclic bacteriocin produced by Lactococcus sp. strain QU 12. The molecular weight calculated after cyclization, which was presumed to involve the same process as in lactocyclicin Q (between L3 and W63), agreed with that estimated by ESI-TOF MS. This peptide was proved to be a novel cyclic bacteriocin, and it was termed leucocyclicin Q. The antimicrobial spectrum of this bacteriocin clearly differed from that of lactocyclicin Q, even though their primary structures were quite similar. This is the first report of a cyclic bacteriocin produced by a strain of the genus Leuconostoc.
INTRODUCTION
Fermented foods represent one of the most common examples of biopreservation. Many microorganisms contribute to the taste, flavor, and preservation of these foods. Lactic acid bacteria (LAB) are known to play important roles in preservation by producing various antimicrobial substances that eliminate other competing microorganisms during the fermentation process (6). Some LAB produce different types of bacteriocins, which are antimicrobial peptides exhibiting bactericidal or bacteriostatic effects on Gram-positive bacteria, including food-borne pathogens, but not against eukaryotic cells (13, 37). In general, LAB bacteriocins are highly resistant to high temperatures and low pH but are easily digested by human proteolytic enzymes. These beneficial characteristics have led to the utilization of bacteriocins as biopreservatives and bacteriocin-producing LAB as starter cultures for fermented foods, such as pickles (10, 22).
LAB bacteriocins are mainly grouped into two classes according to the classification approach of Cotter et al. (13). Class I bacteriocins, so-called lantibiotics, are heat-stable posttranslationally modified peptides containing multiple rings that are bridged by lanthionine or 3-methyl lanthionine residues (30). Class II bacteriocins are small, heat-stable nonlantibiotic peptides and are further divided into four subgroups (13, 31, 32). Class IIa bacteriocins are Listeria-active peptides containing the consensus sequence YGNGVXC at the N terminus (18, 20). Class IIb bacteriocins comprise two peptides, both of which are required for complete antimicrobial activity (33). Class IIc bacteriocins are cyclic bacteriocins, such as enterocin AS-48 (27) and lactocyclicin Q (36). Class IId bacteriocins are the other class II bacteriocins, including enterocin P, which is processed and secreted by the sec pathway (9), and lacticin Q, which is secreted without a leader sequence (21, 40).
Nisin is the most typical bacteriocin and is used as a food preservative around the world (26). Nisin possesses strong, wide-spectrum antimicrobial activity, but some bacteriocins are known to show higher activity against particular species than nisin, such as class IIa bacteriocins against Listeria. Many potent LAB bacteriocins have therefore been studied, and their use as biopreservatives is anticipated. When nisin is added to certain fermented foods or when a nisin-producing strain is used as a starter culture, it sometimes eliminates not only harmful or pathogenic bacteria but also the bacteria responsible for the food's original taste, thereby triggering a decline in food quality. On the other hand, if bacteriocin-producing inhabitant LAB or the bacteriocins can be utilized, they may be able to control bacteria in the fermented foods more effectively without affecting any essential bacteria and their flavors.
Recently, some reports on LAB cyclic bacteriocins (28), such as enterocin AS-48 (27), gassericin A (24, 25), reutericin 6 (23), uberolysin (39), and lactocyclicin Q (36), have been published. These bacteriocins are known to be translated as linear precursors with N-terminal leader sequences of various lengths and then mature by removal of the leader sequences and formation of peptide bonds between the N-terminal and C-terminal residues. In general, cyclic bacteriocins show much higher stability when exposed to pH stress, high temperature, and proteolytic digestion (5, 29, 36). Because of these characteristics, LAB cyclic bacteriocins are expected to emerge as novel biopreservatives succeeding nisin. Enterocin AS-48 is the most extensively studied LAB cyclic bacteriocin, in terms of both its biosynthesis and its application as a biopreservative (8, 16, 19, 35).
In this report, we describe the identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401, isolated from Japanese pickles. Leucocyclicin Q has a cyclic structure in which N and C termini are bound to each other and comprises 61 amino acid residues that show high homology to lactocyclicin Q, a cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Leucocyclicin Q shows a unique antimicrobial spectrum that differs from that of lactocyclicin Q and shows high stability against several bacterial proteases.
MATERIALS AND METHODS
Bacterial strains and media.
L. mesenteroides TK41401, a bacteriocin-producing strain, was isolated from pickled red turnips and identified by 16S rRNA gene sequence analysis. The strain was stored at −80°C in de Man, Rogosa, and Sharpe (MRS) medium (Oxoid, Basingstoke, United Kingdom) with 15% glycerol and propagated in MRS medium at 30°C for 18 h before use. Indicator strains for the determination of antimicrobial activities were propagated at the respective appropriate temperatures (30°C or 37°C) for 18 h before use. LAB strains were grown in MRS medium, and the other Gram-positive indicator strains were grown in tryptic soy broth (BD, Sparks, MD) supplemented with 0.6% yeast extract (Nacalai Tesque, Kyoto, Japan). Escherichia coli strains DH5α and JM109 were grown in Luria-Bertani (LB) medium (BD), and transformants of E. coli DH5α were selected on LB agar plates containing 50 mg/liter isopropyl-β-d-thiogalactopyranoside and 30 mg/liter ampicillin.
Determination of bacteriocin activity.
Bacteriocin activity was determined by the spot-on-lawn method as previously described (21). Bacillus coagulans JCM 2257T was used as an indicator strain unless otherwise stated. Briefly, 10 μl of a bacteriocin preparation was spotted onto a double-layered agar plate containing 5 ml of Lactobacilli agar AOAC (BD) inoculated with an overnight culture of an indicator strain as the upper layer and 10 ml of MRS medium with 1.2% agar as the bottom layer. After overnight incubation at the temperatures appropriate for the indicator strains, bacterial lawns were analyzed for inhibition zones. In each purification step, the activity titer was defined as the reciprocal of the highest dilution that yielded a clear zone of growth inhibition in the indicator lawn; this value was expressed in arbitrary activity units (AU) per milliliter of bacteriocin preparation. Purified bacteriocin was diluted with 0.1% (vol/vol) Tween 80 solution to the appropriate concentrations and tested for antimicrobial activity against various indicator strains as described above. The MIC was defined as the minimum bacteriocin concentration that yielded clear zones of growth inhibition in the indicator lawn. All activity tests were performed in duplicate.
Purification procedure for bacteriocin.
Bacteriocin purification was carried out in a three-step procedure using the supernatant of a 1-liter culture of L. mesenteroides TK41401 grown for 20 h in MRS broth at 30°C. The cell-free supernatant was mixed with 25 g of Amberlite XAD-16 (Sigma-Aldrich, St. Louis, MO), a synthetic hydrophobic resin previously activated with 50% (vol/vol) isopropanol, and equilibrated with distilled water. The mixture was gently shaken at 4°C overnight and transferred to a column (10-mm internal diameter, 200-mm length). The mixture was washed with distilled water and 40% ethanol, followed by elution of the bacteriocin with 200 ml of 80% isopropanol containing 0.1% trifluoroacetic acid (TFA). The active eluted solution was placed in a rotary evaporator (Tokyo Rikakikai, Tokyo, Japan) to remove the isopropanol. The resulting solution was then diluted with 50 mM sodium phosphate buffer (PB; pH 5.7) to 100 ml and loaded onto an SP-Sepharose Fast Flow cation-exchange column (15-mm internal diameter, 100-mm length; GE Healthcare, Uppsala, Sweden) pre-equilibrated with PB. The column was then subjected to serial washes with 100 ml PB. The bacteriocin was eluted with 40 ml of PB containing 0.25 M and 0.5 M NaCl. For further purification, the active eluted solution was applied to a reverse-phase column (Resource RPC 3-ml; GE Healthcare) incorporated in the LC-2000Plus high-performance liquid chromatography (HPLC) system (Jasco, Tokyo, Japan), and eluted with a gradient of MilliQ water-acetonitrile containing 0.1% TFA at a flow rate of 1 ml/min as follows: 0 to 10 min, 30% (vol/vol); 10 to 40 min, 30 to 60% (vol/vol); and 40 to 45 min, 60 to 100% (vol/vol) acetonitrile. The active fraction was subjected to RP-HPLC again, followed by thorough removal of acetonitrile in a Speed-Vac concentrator (Savants, Farmingdale, NY). The purified bacteriocin solution was used for MIC determination, and further characterization, such as amino acid sequence analysis and stability assessment, is described below. The antimicrobial activity of the fractions was determined as described above for each purification step. The protein concentration (mg/ml) of each fraction was estimated from the A280 by using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) according to the manufacturer's instructions. The concentration of purified bacteriocin was also determined using a Pierce BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA). These concentrations were significantly equivalent, and the former was used to determine the MICs.
Sensitivity analyses against peptidases.
The peptidase sensitivity of leucocyclicin Q was determined for pepsin, trypsin, α-chymotrypsin, proteinase K, carboxypeptidase Y, and Asp-N peptidase (all from Sigma-Aldrich). All enzymes were dissolved in the respective appropriate buffers with optimal pH, and the solutions were filter sterilized and added to the purified leucocyclicin Q at an enzyme-to-peptide molar ratio of 1:100. Following incubation at 37°C for 4 h, the reaction samples were heated at 100°C for 5 min to denature the enzymes. The residual antimicrobial activities were determined by agar diffusion assay as described in a previous study (3) by using Lactobacillus sakei subsp. sakei JCM 1157T as an indicator strain. A sample without enzyme treatments was used as the negative control. All sensitivity tests were performed in duplicate.
Mass spectrometry and amino acid sequencing.
Molecular mass analyses of the purified bacteriocin and fragments were conducted by electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS) using a JMS-T100LC mass spectrometer (JEOL, Tokyo, Japan). The N-terminal amino acid sequences of the purified bacteriocin and fragments were determined by Edman degradation using a PPSQ-21 gas-phase automatic protein sequencer (Shimadzu, Kyoto, Japan). Fragmentation with 2-(2′-nitrophenylsulfenyl)-3-methyl-3 bromoindolenine (BNPS-skatole) was performed as previously described (36) to obtain a part of the amino acid sequence. All fragments were fractionated by HPLC as described above and used for subsequent analyses.
Analysis of the genes encoding the bacteriocins.
DNA manipulations were performed as described in a previous report (21). DNA polymerases, restriction enzymes, and other DNA-modifying enzymes were used according to the manufacturers' instructions. The total DNA of the strain TK41041 was extracted from cells treated with lysozyme (Seikagaku, Tokyo, Japan) and cetyltrimethylammonium bromide (Wako, Osaka, Japan) according to the procedures described previously (21) and used as a template for PCR and subsequent procedures. The total DNA of strain TK4101 was digested by the restriction enzymes BamHI, EcoRI, SacI, KpnI, and XbaI, and digested DNA fragments were subsequently ligated into the pUC18 cloning vector (Toyobo, Osaka, Japan) or self-ligated for use as templates in nested anchor rapid PCR (NAR-PCR) and inverse PCR. The oligonucleotide primers used in this study are listed in Table 1. The degenerate primers dAKA-F1, F2, R2, and R3, designed from the amino acid sequence obtained for the bacteriocin, were used for NAR-PCR as anchor primers. NAR-PCRs were performed using Taq DNA polymerase (Promega, Madison, WI) under the following conditions: denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at the optimal temperature for each primer for 30 s, and polymerization at 72°C for 2 min. The PCR fragments were cloned into the pGEM-T vector (Promega) and introduced into E. coli DH5α by electroporation in a MicroPulser apparatus (Bio-Rad Laboratories, Hercules, CA) according to the procedure recommended by the manufacturer. DNA sequencing was carried out by FASMAC Co., Ltd. (Kanagawa, Japan). To obtain the upstream and downstream sequences of the bacteriocin structural gene, inverse PCR was performed with Taq DNA polymerase by using the primers aka-F1, F2, R1, and R3, designed from the DNA sequence obtained by NAR-PCR, under the following conditions: denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at the optimal temperature for each primer for 30 s, and polymerization at 72°C for 2 min. The products obtained by inverse PCR were cloned and sequenced as described above. On the basis of sequences obtained from the inverse PCR products, the new specific primers Lcy-F and Lcy-R were synthesized and used to confirm the DNA sequence around the structural gene by using the procedures described above.
Table 1.
Oligonucleotide primers used to obtain the structural gene of leucocyclicin Q
| Primer name | Sequence (5′–3′)a |
|---|---|
| dAKA-F1 | TGGGCNACNAARGCNGC |
| dAKA-F2 | AARCAYGARGGNAARGC |
| dAKA-R2 | GCYTTNCCYTCRTGYTT |
| dAKA-R3 | CCANGCDATNGCNGC |
| 1stMup13-f | TTAACTATGCGGCATCAGA |
| 1stMup13-r | TAATGTGAGTTAGCTCACTC |
| Mup13-f | AAGGCGATTAAGTTGGGTA |
| Mup13-r | GTATGTTGTGTGGAATTGTG |
| s-M13-f | GTAAAACGACGGCCAGT |
| s-M13-r | TTCACACAGGAAACAGG |
| aka-F1 | GATATGCGTTCAGTTAATTTGGC |
| aka-F2 | GAGATTAAACCAATTATTGAGCCTG |
| aka-R1 | CAAATCTTTTTACCACGCAATAGC |
| aka-R3 | CTAATAATCTTTGCTTACGTTCTTC |
| Lcy-F | TTTGTTGTCGTAAAAATCTTATCTT |
| Lcy-R | GACGCAACAATACCAGTAGAAG |
In degenerate primers, N, R, Y, and D indicate A/T/G/C, A/G, T/C, and A/T/G, respectively.
Computer analysis of DNA and amino acid sequences.
The DNA and amino acid sequences obtained were analyzed with GENETYX-WIN software, version 8.0.1 (Genetyx, Tokyo, Japan). Database searches were performed using the BLAST program of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/BLAST/).
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the DDBJ database under the accession no. AB649282.
RESULTS
Purification and molecular mass analysis of leucocyclicin Q.
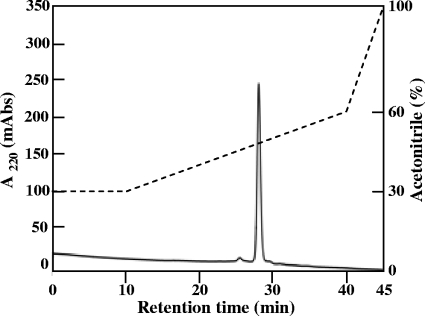
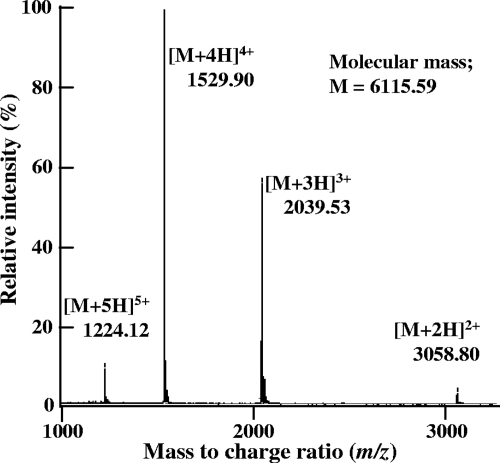
The bacteriocin produced by L. mesenteroides TK41401 was purified by a three-step procedure, which included hydrophobic interaction, cation-exchange chromatography, and reverse-phase HPLC. Most of the activity in the culture supernatant was recovered by hydrophobic interaction chromatography, and the bacteriocin activity was primarily recovered in the 0.5 M NaCl fraction of the cation-exchange chromatography. This fraction was subsequently subjected to reverse-phase HPLC. One peak with antimicrobial activity was obtained, and the active fraction was subjected to RP-HPLC again (Fig. 1). The final yield of the peptide obtained in these purification steps was 25.6% from culture supernatant activity; the details are summarized in Table 2. ESI-TOF MS analysis was performed using the purified peptide, and the molecular mass of this peptide was determined to be 6,115.59 Da (Fig. 2). This molecular mass was not identical to that of any known bacteriocin. Therefore, we concluded that this peptide was a novel bacteriocin and designated it as leucocyclicin Q.
Fig. 1.
Second reverse-phase HPLC of a bacteriocin from L. mesenteroides TK41401. The broken line indicates the acetonitrile gradient.
Table 2.
Purification of leucocyclicin Q produced by L. mesenteroides TK41401
| Step | Vol (ml) | Total activity (AU)a | Yield (%) | Sp act (AU/mg) | Purification (fold) |
|---|---|---|---|---|---|
| Supernatant | 1,000 | 1.60 × 106 | 100 | 8.85 × 10 | 1.00 |
| Amberlite XAD-16 | 200 | 1.28 × 106 | 80 | 6.88 × 103 | 77.8 |
| SP-Sepharose | 100 | 1.28 × 106 | 80 | 1.66 × 104 | 188 |
| RP-HPLC | 2 | 4.10 × 105 | 25.6 | 7.40 × 104 | 836 |
Antimicrobial activity (in arbitrary units [AU]) was assayed by the spot-on-lawn method using B. coagulans JCM 2257T as an indicator strain.
Fig. 2.
ESI-TOF mass spectrum of purified leucocyclicin Q. Multiple charged molecular ions are indicated. The molecular mass was calculated from the values of the major peak of tetravalent ions.
Amino acid sequencing analysis of leucocyclicin Q.
Amino acid sequencing of purified leucocyclicin Q was performed, but the Edman degradation did not proceed. This suggested that either the N-terminal amino acid is blocked by some modification or that leucocyclicin Q possesses cyclic structure binding between the N and C termini. To obtain partial amino acid sequences, purified leucocyclicin Q was fragmented by BNPS-skatole treatment, and the fragments were purified by HPLC. One fragment (2,437.24 Da) was subjected to amino acid sequencing, and a 23-residue amino acid sequence of leucocyclicin Q was obtained. C-terminal tryptophan residues were cut by BNPS-skatole treatment. Considering tryptophan residues at both the N and C termini of the fragment, a 25-residue amino acid sequence was identified as (W)ATKAALATAETIVKHEGKAAAIA(W). Based on this partial sequence, degenerate primers (Table 1) were designed and used for NAR-PCR.
DNA sequencing analysis to elucidate the structure gene.
NAR-PCRs were performed using degenerate primers as anchor primers, and the downstream DNA sequence of the structure gene of leucocyclicin Q was successfully obtained. Based on this sequence, specific primers were constructed for inverse PCR, and a DNA sequence of around 700 bp was obtained using these primers. Sequencing analysis revealed some open reading frames, including lcyQ, encoding the precursor of leucocyclicin Q (Fig. 3), which consisted of 63 amino acid residues. The calculated mass of this precursor was 6,411.50 Da, which is higher than the mass of purified leucocyclicin Q determined by ESI-TOF MS, suggesting that it contained a leader sequence at the N terminus.
Fig. 3.
Nucleotide sequence of the region encoding leucocyclicin Q and the deduced amino acid sequence. The putative ribosome binding site is underlined. The structural gene is in italics, and the asterisk indicates the stop codon.
The database search we performed showed that leucocyclicin Q was a novel bacteriocin, and its precursor showed high and characteristic homology (72%) with that of lactocyclicin Q, a known cyclic bacteriocin produced by Lactococcus sp. strain QU 12 (Fig. 4). The precursor of lactocyclicin Q contains an N-terminal leader sequence of two amino acid residues, and cyclization occurs between L3 and 63W with cleavage of the leader sequence (36). When this process was applied to leucocyclicin Q, the calculated mass after considering the cleavage of the leader sequence (MF) and cyclization (−18 Da) was 6,115.13 Da, which agreed with that obtained by ESI-TOF MS. These results suggest that leucocyclicin Q is a novel cyclic bacteriocin.
Fig. 4.
Alignment of leucocyclicin Q and lactocyclicin Q. The middle line indicates identical amino acid residues.
Antimicrobial spectrum of leucocyclicin Q.
Purified leucocyclicin Q was tested for antimicrobial activity against various indicator strains by using the spot-on-lawn method (Table 3). As described above, although the leucocyclicin Q structure showed very high similarity to lactocyclicin Q, their antimicrobial spectra were very dissimilar. Both bacteriocins showed quite strong activities against Lactobacillus sakei and Bacillus coagulans and weak activities against Gram-negative bacteria. In addition, leucocyclicin Q possessed higher activity against a broad range of Gram-positive bacteria, such as Lactococcus, Weissella paramesenteroides, Pediococcus dextrinicus, Enterococcus, Streptococcus, and Leuconostoc, than lactocyclicin Q. On the other hand, lactocyclicin Q possessed higher activity against other Gram-positive bacteria, such as Weissella cibaria, Pediococcus pentosaceus, and Bacillus subtilis. Interestingly, both producer strains showed quite higher tolerance to each bacteriocin than their related species.
Table 3.
Antimicrobial spectra of leucocyclicin Q and lactocyclicin Q
| Indicator straina | MIC (μM)b |
|
|---|---|---|
| Leucocyclicin Q | Lactocyclicin Q | |
| Lactococcus lactis subsp. lactis JCM 7638 | 0.038 | 0.141 |
| L. lactis subsp. lactis ATCC 19435T | 0.038 | 0.141 |
| Lactococcus sp. QU 12 | 1.203 | 22.90 |
| Lactobacillus sakei subsp. sakei JCM 1157T | 0.038 | 0.015 |
| Weissella cibaria JCM 12495T | 2.405 | 0.710 |
| W. paramesenteroides JCM 9890T | 0.075 | ND |
| Pediococcus pentosaceus JCM 5885 | 2.405 | 0.550 |
| P. dextrinicus JCM 5887T | 0.038 | 0.141 |
| Enterococcus faecium JCM 5804T | 0.038 | 0.710 |
| E. faecalis JCM 5803T | 0.150 | 0.260 |
| Streptococcus salivarius JCM 5707T | 2.405 | 22.90 |
| S. bovis JCM 5802T | 0.601 | 22.90 |
| S. mutans JCM 5705T | 19.25 | NA |
| Bacillus coagulans JCM 2257T | 0.075 | 0.015 |
| B. subtilis subsp. subtilis JCM 1465T | 0.601 | 0.064 |
| B. cereus JCM 2152T | 2.405 | 11.40 |
| Kocuria rhizophila NBRC 12708 | 9.623 | 5.70 |
| Listeria innocua ATCC 33090T | 0.601 | 1.03 |
| Leuconostoc mesenteroides subsp. mesenteroides JCM 6124T | 0.300 | 1.03 |
| L. mesenteroides TK41401 | NA | 22.90 |
| Escherichia coli JM109 | 38.49 | 34.30 |
| E. coli NBRC 3301 | 38.49 | 17.30 |
| Staphylococcus aureus subsp. aureus ATCC 12600T | NA | 91.60 |
ATCC, American Type Culture Collection, Rockville, MD,; JCM, Japan Collection of Microorganisms, Saitama, Japan: NBRC, NITE Biological Resource Center, Chiba, Japan.
The antimicrobial spectrum of lactocyclicin Q was drawn from the previous report (36). Only the MIC against L. mesenteroides TK41401 was determined in this study. ND, not determined; NA, no activity even with maximum concentrations of leucocyclicin Q and lactocyclicin Q (38.49 and 91.60 μM, respectively).
Sensitivity against peptidases.
As shown in Table 4, leucocyclicin Q was sensitive to digestion by human digestive enzymes such as pepsin and α-chymotrypsin but was tolerant to trypsin. On the other hand, leucocyclicin Q showed high stability against bacterial peptidases, with the exception of proteinase K. It is noteworthy that leucocyclicin Q showed no susceptibility to carboxypeptidase Y, which digests peptides from C-terminal residues.
Table 4.
Peptidase sensitivity of leucocyclicin Q
| Peptidase | Diam (mm)a |
|---|---|
| Control (no peptidase) | 23 |
| Trypsin | 23 |
| Pepsin | 0 |
| α-Chymotrypsin | 0 |
| Proteinase K | 0 |
| Carboxypeptidase Y | 23 |
| Asp-N peptidase | 23 |
Enzyme treatments were performed at 37°C for 4 h, and L. sakei subsp. sakei JCM 1157T was used as the indicator strain. Diameters were based on the clear zones of inhibition.
DISCUSSION
In recent years, LAB cyclic bacteriocins have been carefully evaluated worldwide as the next generation of antimicrobial compounds because of their unusual characteristics. The search for novel LAB cyclic bacteriocins has resulted in discoveries of several bacteriocins produced by different genera, such as enterocin AS-48 from Enterococcus, uberolysin from Streptococcus, gassericin A, acidocin B, and reutericin 6 from Lactobacillus, carnocyclin A from Carnobacterium (29), and lactocyclicin Q and garvicin ML (5) from Lactococcus. To the best of our knowledge, there are no reports to date on cyclic bacteriocins produced by members of the genus Leuconostoc. This study therefore presents the first report on cyclic bacteriocins produced by a member of this genus.
Leucocyclicin Q was purified from the culture supernatant of L. mesenteroides TK41401 isolated from Japanese pickles by a three-step purification method and subsequently subjected to structure analyses. Edman degradation analysis, NAR-PCR, and inverse PCR were performed to obtain the structural gene of leucocyclicin Q (Fig. 3). The amino acid sequence of its precursor was deduced from this DNA information, and it showed very high similarity to that of lactocyclicin Q (Fig. 4). Lactocyclicin Q contains a leader sequence of two amino acids, and cyclization occurs between L3 and W63 with concomitant cleavage of the leader sequence. Leucocyclicin Q also contains a two-amino-acid leader sequence. In addition, the N and C termini of the cleavage peptide are flanked by leucine and tryptophan, respectively, the same amino acids flanking the termini of lactocyclicin Q. The calculated mass of mature leucocyclicin Q after cyclization and cleavage of the leader sequence agreed with the molecular mass obtained by ESI-TOF MS. These results indicate that leucocyclicin Q is a novel cyclic bacteriocin. In addition to lactocyclicin Q and leucocyclicin Q, circularin A (24) and uberolysin (39) are known to possess short leader sequences, consisting of three and six residues, respectively. Other cyclic bacteriocins such as enterocin AS-48 possess long leader sequences, more than 20 amino acid residues (28). Regardless of the length of the leader sequences, there are certain differences in their secretion and cyclization processes, although little information is available regarding the cleavage reaction for maturation and function of these sequences. As predicted in a previous report on lactocyclicin Q (36), leucocyclicin Q possesses a KXXXXXW sequence conserved at the C termini of many LAB cyclic bacteriocins (Fig. 4), which may be linked to the cyclization process. In addition, a leucine residue constitutes the N terminus of cleaved leucocyclicin Q, which is the same in most cyclic bacteriocins. The structure of leucocyclicin Q also supported the prediction that these N and C termini might be important factors for secretion and cyclization and may serve as recognition sites for the cyclization enzyme.
The cyclization mechanisms of cyclic peptides, including those of LAB bacteriocins, are not well characterized. The enzymes McjB and McjC were recently shown to be essential for the cyclization of microcin J25 (17), a cyclic bacteriocin produced by E. coli AY 25 (4, 38). In contrast to most LAB cyclic bacteriocins, microcin J25 does not possess aromatic amino acids in the N or C termini, indicating that LAB cyclic bacteriocins possess an alternative cyclization mechanism. Eukaryotic animals and plants are also known to produce cyclic antibiotic peptides, such as defensin in animals and cyclotide in plants (7, 12, 14). These eukaryotic cyclic peptides include both N-terminal and C-terminal extensions in their linear precursor peptides, and both extensions are thought to function as processing sites and/or recognition sites in ligation reactions (15). There have been no reports on LAB cyclic bacteriocins with C-terminal extensions, and it is certain that the cyclization mechanism differs from that of eukaryotic cyclic peptides, while the N-terminal leader sequences are linked to their individual cyclization processes. In general, cyclotides possess longer N-terminal extensions, such as those in some LAB cyclic bacteriocins, and this region is possibly processed twice, once before and once during cyclization (1, 2, 15). Therefore, long leader sequences of cyclic bacteriocins can be considered tags that are recognized by a certain carrier that shuttles precursors to a cyclization factory. After cleavage of this tag, the remaining short leader sequence plays an important role in cyclization. On the other hand, bacteriocins with short leader sequences may use different systems to transfer the precursor to the appropriate cyclization position, because they do not possess long leader sequences that can act as tags.
Lactocyclicin Q shows high antimicrobial activity against Bacillus and Lactococcus, and other cyclic bacteriocins show different spectra of activity. Enterocin AS-48 exerts strong antimicrobial activity against some Gram-positive bacteria, such as Bacillus, Staphylococcus, and Listeria, and also against some Gram-negative bacteria, such as Salmonella and E. coli (1, 2). The spectrum of leucocyclicin Q differed from the spectra of these previously reported cyclic bacteriocins, exerting high activity against a very broad range of Gram-positive bacteria, with especially strong activity against L. lactis, L. sakei, W. paramesenteroides, P. dextrinicus, Enterococcus faecium, and B. coagulans but no activity against Staphylococcus aureus. Leucocyclicin Q also exerted significant activity against Gram-negative bacteria when employed in high concentrations (Table 3). Although the primary structure of leucocyclicin Q was similar to that of lactocyclicin Q, the antimicrobial spectrum was quite different. In addition, the predicted secondary structure of leucocyclicin Q included four α-helices, a structure similar to that of lactocyclicin Q. The antimicrobial mechanism of cyclic bacteriocins has not yet been completely characterized. Further elucidation of the mode of action of these bacteriocins and structure analyses will help further our knowledge of LAB cyclic bacteriocin.
As shown in Table 3, L. mesenteroides TK41401 showed significantly high cross-immunity against lactocyclicin Q, to the same extent as the original producer strain, Lactococcus sp. strain QU 12. This cross-immunity must depend on the leucocyclicin Q immunity system, because another strain, L. mesenteroides subsp. mesenteroides JCM 6124T, a non-bacteriocin-producing strain, showed quite high sensitivity. On the other hand, Lactococcus sp. strain QU 12 did not exhibit any immunity against leucocyclicin Q significantly higher than that of strains of other genera; instead, it was approximately 40-fold more tolerant than other Lactococcus strains, including a nisin-producing strain (JCM 7638). These results suggest that the self-immunity systems of these strains employ a common mechanism that influences their comparatively lower susceptibilities to both cyclic bacteriocins.
In general, cyclic bacteriocins are known to possess extremely high stability against several stresses (11, 24, 27). For instance, lactocyclicin Q maintains full activity after pH changes ranging from pH 3.0 to 9.0 and after exposure to 121°C for 15 min under acidic conditions. In addition, lactocyclicin Q possesses high resistance against digestive enzymes, such as α-chymotrypsin, trypsin, pepsin, and proteinase K. When leucocyclicin Q was exposed to stresses such as acidic, basic, or heat stress, it also exhibited remarkable stability (data not shown). Moreover, as shown in Table 4, leucocyclicin Q exhibited noteworthy resistance against digestive enzymes and against bacterial peptidases. In contrast to lactocyclicin Q (36), leucocyclicin Q was not resistant to α-chymotrypsin or pepsin but exhibited significant resistance to trypsin.
Leuconostoc is one of the major LAB and can be isolated from a variety of fermented vegetables, such as Japanese pickles and sauerkraut (34). L. mesenteroides TK41401 produces a promising novel cyclic bacteriocin, leucocyclicin Q, which exhibits remarkable antimicrobial activity and great stability against several stresses but is at the same time safely degradable by human digestive enzymes. Because of this useful bacteriocin production, it is expected that L. mesenteroides TK41401 will be used as a novel starter culture for fermented vegetables, and leucocyclicin Q itself also holds enormous potential as a biopreservative. In addition, leucocyclicin Q is a significantly potent research subject not only in applied food microbiology but also for the study of LAB cyclic bacteriocin biosynthesis, which is still not well characterized. Future studies will open a new platform for the application of LAB cyclic bacteriocins.
ACKNOWLEDGMENTS
This work was partially supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Abriouel H., Valdivia E., Gálvez A., Maqueda M. 2001. Influence of physico-chemical factors on the oligomerization and biological activity of bacteriocin AS-48. Curr. Microbiol. 42:89–95 [DOI] [PubMed] [Google Scholar]
- 2. Abriouel H., Valdivia E., Gálvez A., Maqueda M. 1998. Response of Salmonella choleraesuis LT2 spheroplasts and permeabilized cells to the bacteriocin AS-48. Appl. Environ. Microbiol. 64:4623–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basanta A., et al. 2008. Antimicrobial activity of Enterococcus faecium L50, a strain producing enterocins L50 (L50A and L50B), P and Q., against beer-spoilage lactic acid bacteria in broth, wort (hopped and unhopped), and alcoholic and non-alcoholic lager beers. Int. J. Food Microbiol. 125:293–307 [DOI] [PubMed] [Google Scholar]
- 4. Blond A., et al. 2002. Thermolysin-linearized microcin J25 retains the structured core of the native macrocyclic peptide and displays antimicrobial activity. Eur. J. Biochem. 269:6212–6222 [DOI] [PubMed] [Google Scholar]
- 5. Borrero J., et al. 2011. Characterization of garvicin ML, a novel circular bacteriocin produced by Lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos). Appl. Environ. Microbiol. 77:369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caplice E., Fitzgerald G. F. 1999. Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol. 50:131–149 [DOI] [PubMed] [Google Scholar]
- 7. Cascales L., Craik D. J. 2010. Naturally occurring circular proteins: distribution, biosynthesis and evolution. Org. Biomol. Chem. 8:5035–5047 [DOI] [PubMed] [Google Scholar]
- 8. Cebrián R., et al. 2010. Insights into the functionality of the putative residues involved in enterocin AS-48 maturation. Appl. Environ. Microbiol. 76:7268–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cintas L. M., Casaus P., Håvarstein L. S., Hernández P. E., Nes I. F. 1997. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63:4321–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cleveland J., Montville T. J., Nes I. F., Chikindas M. L. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1–20 [DOI] [PubMed] [Google Scholar]
- 11. Cobos E. S., et al. 2001. AS-48: a circular protein with an extremely stable globular structure. FEBS Lett. 505:379–382 [DOI] [PubMed] [Google Scholar]
- 12. Conlan B. F., Gillon A. D., Craik D. J., Anderson M. A. 2010. Circular proteins and mechanisms of cyclization. Biopolymers 94:573–583 [DOI] [PubMed] [Google Scholar]
- 13. Cotter P. D., Hill C., Ross R. P. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788 [DOI] [PubMed] [Google Scholar]
- 14. Craik D. J., Cemazar M., Daly N. L. 2006. The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Dev. 9:251–260 [PubMed] [Google Scholar]
- 15. Daly N. L., Rosengren K. J., Craik D. J. 2009. Discovery, structure and biological activities of cyclotides. Adv. Drug Deliv. Rev. 61:918–930 [DOI] [PubMed] [Google Scholar]
- 16. Diaz M., et al. 2003. Characterization of a new operon, as-48EFGH, from the as-48 gene cluster involved in immunity to enterocin AS-48. Appl. Environ. Microbiol. 69:1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duquesne S., et al. 2007. Two enzymes catalyze the maturation of a lasso peptide in Escherichia coli. Chem. Biol. 14:793–803 [DOI] [PubMed] [Google Scholar]
- 18. Ennahar S., Sashihara T., Sonomoto K., Ishizaki A. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85–106 [DOI] [PubMed] [Google Scholar]
- 19. Fernández M., Martínez-Bueno M., Martin M. C., Valdivia E., Maqueda M. 2007. Heterologous expression of enterocin AS-48 in several strains of lactic acid bacteria. J. Appl. Microbiol. 102:1350–1361 [DOI] [PubMed] [Google Scholar]
- 20. Fimland G., Johnsen L., Dalhus B., Nissen-Meyer J. 2005. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J. Pept. Sci. 11:688–696 [DOI] [PubMed] [Google Scholar]
- 21. Fujita K., et al. 2007. Structural analysis and characterization of lacticin Q., a novel bacteriocin belonging to a new family of unmodified bacteriocins of gram-positive bacteria. Appl. Environ. Microbiol. 73:2871–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gálvez A., Abriouel H., López R. L., Ben Omar N. 2007. Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 120:51–70 [DOI] [PubMed] [Google Scholar]
- 23. Kawai Y., et al. 2004. Structural and functional differences in two cyclic bacteriocins with the same sequences produced by lactobacilli. Appl. Environ. Microbiol. 70:2906–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawai Y., Kemperman R., Kok J., Saito T. 2004. The circular bacteriocins gassericin A and circularin A. Curr. Protein Pept. Sci. 5:393–398 [DOI] [PubMed] [Google Scholar]
- 25. Kawai Y., Saito T., Kitazawa H., Itoh T. 1998. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci. Biotechnol. Biochem. 62:2438–2440 [DOI] [PubMed] [Google Scholar]
- 26. Lubelski J., Rink R., Khusainov R., Moll G. N., Kuipers O. P. 2008. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 65:455–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maqueda M., et al. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr. Protein Pept. Sci. 5:399–416 [DOI] [PubMed] [Google Scholar]
- 28. Maqueda M., et al. 2008. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol. Rev. 32:2–22 [DOI] [PubMed] [Google Scholar]
- 29. Martin-Visscher L. A., et al. 2008. Isolation and characterization of carnocyclin A, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 74:4756–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McAuliffe O., Ross R. P., Hill C. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285–308 [DOI] [PubMed] [Google Scholar]
- 31. Nes I. F., et al. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70:113–128 [DOI] [PubMed] [Google Scholar]
- 32. Nes I. F., Holo H. 2000. Class II antimicrobial peptides from lactic acid bacteria. Biopolymers 55:50–61 [DOI] [PubMed] [Google Scholar]
- 33. Nissen-Meyer J., Oppegard C., Rogne P., Haugen H. S., Kristiansen P. E. 2010. Structure and mode-of-action of the two-peptide (class-IIb) bacteriocins. Probiotics Antimicrob. Proteins 2:52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ogier J. C., Casalta E., Farrokh C., Saihi A. 2008. Safety assessment of dairy microorganisms: the Leuconostoc genus. Int. J. Food Microbiol. 126:286–290 [DOI] [PubMed] [Google Scholar]
- 35. Sánchez-Hidalgo M., et al. 2010. Conformational stability and activity of circular Enterocin AS-48 derivatives. Protein Pept. Lett. 17:708–714 [DOI] [PubMed] [Google Scholar]
- 36. Sawa N., et al. 2009. Identification and characterization of lactocyclicin Q., a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Appl. Environ. Microbiol. 75:1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scheirlinck I., et al. 2007. Influence of geographical origin and flour type on diversity of lactic acid bacteria in traditional Belgian sourdoughs. Appl. Environ. Microbiol. 73:6262–6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Solbiati J. O., et al. 1999. Sequence analysis of the four plasmid genes required to produce the circular peptide antibiotic microcin J25. J. Bacteriol. 181:2659–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wirawan R. E., Swanson K. M., Kleffmann T., Jack R. W., Tagg J. R. 2007. Uberolysin: a novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 153:1619–1630 [DOI] [PubMed] [Google Scholar]
- 40. Yoneyama F., et al. 2009. Lacticin Q., a lactococcal bacteriocin, causes high-level membrane permeability in the absence of specific receptors. Appl. Environ. Microbiol. 75:538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]