Abstract
We investigated the effects of rare earth elements on enzyme production and secondary metabolism in Bacillus subtilis. Addition of scandium to the growth medium stimulated the production of both amylase and bacilysin at the transcriptional level, thus showing scandium to have a remarkable impact in B. subtilis.
TEXT
The rare earth elements (REEs) are 17 elements consisting of scandium, yttrium, and the 15 lanthanides. Although these elements have been widely used in industry, there have been few reports regarding their significance in biology. Previously, we reported that REEs, especially scandium, enhance antibiotic production in several Streptomyces species (7, 10). These results suggest that REEs are useful for activate bacterial capabilities.
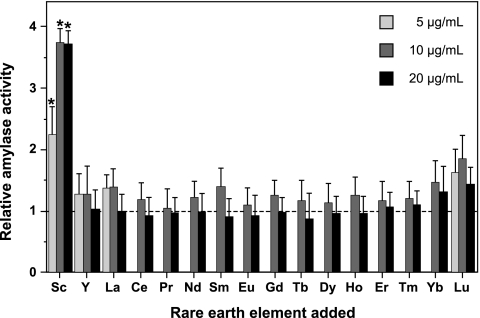
The Gram-positive model bacterium Bacillus subtilis produces not only commercially important extracellular enzymes, such as amylase and protease, but also a variety of antibiotics. To investigate the effects of REEs in B. subtilis, we first examined whether REEs stimulate amylase production. B. subtilis strain 168 (trpC2) was cultivated at 30°C for 48 h in NG medium (9) with or without each REE. All REEs except promethium (all chloride salts; purchased from Wako Pure Chemical) were employed in this study. Amylase activity was measured using the method of Fuwa (2) with slight modifications. Samples of 50 μl were mixed with 0.1 ml of soluble starch solution (50 mM Tris-HCl [pH 6.8], 0.5% soluble starch) and incubated at 40°C. A 4-μl volume of the reaction mixture was added to 0.1 ml of Lugol solution (MP Biomedicals LLC), and the absorbance at 700 nm (A700) was then measured. Specific activity was expressed as (Ablank − Asample) · Ablank−1 · t−1 · 100, where t indicates the time (min) of the reaction and Ablank and Asample refer to the A700 values of the blank (water) and the sample, respectively. Strikingly, B. subtilis amylase production was markedly stimulated by adding scandium chloride to the medium (Fig. 1). When scandium was added to the medium at a concentration of 10 or 20 μg/ml (38.5 or 77 μM, respectively), the amylase activity reached the maximum level (approximately 3.5-fold higher than that in scandium-free medium). At higher concentrations (>50 μg/ml), scandium inhibited the growth of B. subtilis, resulting in a decrease in amylase production (data not shown). Other REEs appeared to have no significant effect on amylase production in B. subtilis, although lutetium had a slight stimulatory effect (Fig. 1). Therefore, we further analyzed the effects of scandium on amylase production. Time course analysis indicated the stimulation of amylase production from 40 h onwards (Fig. 2A). Similar results were observed even when scandium was added at different time points (12 and 24 h after the start of cultivation) (data not shown). In addition, scandium had no effect on amylase activity per se (data not shown). These results suggest that cell response is necessary to exert the stimulatory effect.
Fig. 1.
Effects of REEs on B. subtilis amylase production. B. subtilis strain 168 was cultivated at 30°C for 48 h in NG medium with or without each REE (chloride salt) added at a concentration of 5 (light gray thick bars), 10 (gray thick bars), or 20 (black thick bars) μg/ml. The culture supernatants obtained after centrifugation were used for the amylase assay. Relative amylase activity was calculated by dividing the activity obtained when cells were grown in REE-supplemented medium by that obtained when cells were grown in the control medium (no addition of REE). The average values for three independent experiments are shown. Error bars represent the standard deviations. Asterisks denote significant differences (P < 0.05). Abbreviations: Sc, scandium; Y, yttrium; La, lanthanum; Ce, cerium; Pr, praseodymium; Nd, neodymium; Sm, samarium; Eu, europium; Gd, gadolinium; Tb, terbium; Dy, dysprosium; Ho, holmium; Er, erbium; Tm, thulium; Yb, ytterbium; Lu, lutetium.
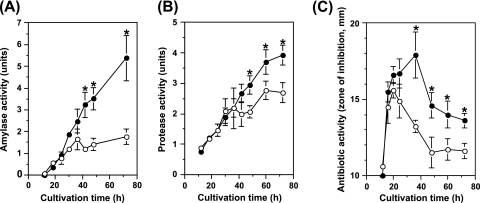
Fig. 2.
Effects of scandium on amylase, protease, and antibiotic production. B. subtilis strain 168 was cultivated at 30°C in NG medium without (open circles) or with (closed circles) scandium chloride at a concentration of 10 μg/ml. Culture samples were withdrawn at the indicated time points and assayed for the activities of amylase (A), protease (B), and antibiotic (C). Asterisks denote significant differences (P < 0.05).
We next examined whether scandium can also stimulate protease and antibiotic production. Protease activity was measured by a method similar to that described by Geele et al. (3). The culture supernatants (20 μl) obtained after centrifugation were mixed with reaction buffer consisting of 1.25% azocasein, 2 mM CaC12, and 50 mM Tris-HCl (pH 7.5) and incubated at 37°C for 20 min. Reactions were then stopped by adding 0.2 ml of 10% trichloroacetic acid. After centrifugation, the absorbance at 335 nm (A335) of the supernatant fraction was measured. Specific activity was expressed as (Asample − Ablank) · t−1 · 100, where t indicates the time of reaction and Ablank and Asample refer to the A335 values of the blank (water) and the sample, respectively. Antibiotic activity was determined by a paper disk-agar diffusion assay as described previously (5). Staphylococcus aureus 209P was used as the test organism. The results indicated that scandium had a slight stimulatory effect on protease production at late stationary phase (Fig. 2B). The effect of scandium was more pronounced on antibiotic than on protease production (Fig. 2C). In the scandium-free medium, antibiotic production reached the maximum level at 20 h and decreased abruptly during late stationary phase. In contrast, in the presence of scandium, antibiotic production continued during late stationary phase. B. subtilis is known to produce the dipeptide antibiotic bacilysin (11). As no antibiotic activity was detected in the culture broth of a mutant lacking bacilysin synthetase (alanine-anticapsin ligase) activity (data not shown), it was concluded that scandium stimulated bacilysin production in late stationary phase.
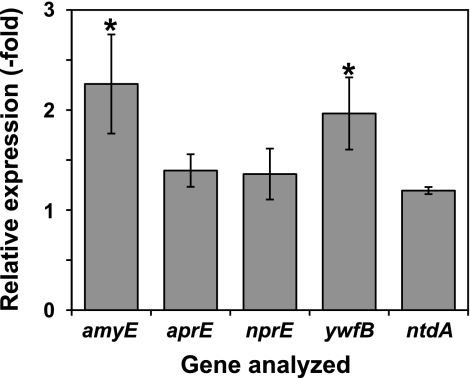
To examine whether the observed effects of scandium were exerted at the transcription level, we analyzed the gene expression of amylase (amyE), alkaline protease (aprE), neutral protease (nprE), bacilysin (ywfB, which is the first gene of the bacilysin biosynthetic operon) (4), and the other antibiotic neotrehalosadiamine (ntdA, which is the first gene of the neotrehalosadiamine biosynthetic operon) (5) by reverse transcription quantitative PCR (RT-qPCR) analysis. The oligonucleotides used for RT-qPCR analysis are listed in Table 1. In cells in early stationary phase (24 h of cultivation), no significant increases in gene expression were observed (data not shown). In contrast, in cells at late stationary phase (48 h of cultivation), the relative expression levels of amyE and ywfB in cells grown in scandium-supplemented medium were approximately 2-fold higher than those in cells grown in scandium-free medium (Fig. 3). Scandium had no or only a slight effect on aprE, nprE, and ntdA expression, in agreement with the results from the production experiments.
Table 1.
Oligonucleotides used for RT-qPCR
| Primer | Oligonucleotide sequence (5′→3′) |
|---|---|
| RT-qPCR 16S-F | ATCTTCCGCAATGGACGA |
| RT-qPCR 16S-R | GCCGTGGCTTTCTGGTTA |
| RT-qPCR amyE-F | CCACCAGTGATTATGCCG |
| RT-qPCR amyE-R | TCTGCGTGACATCCCATC |
| RT-qPCR aprE-F | CCGCAGCAACATTGGATG |
| RT-qPCR aprE-R | TTGAGAGTGAAGAGCCGG |
| RT-qPCR nprE-F | AAAAAGCGCTGGCACTCG |
| RT-qPCR nprE-R | CGTCACGTCGTAAGCAAG |
| RT-qPCR ywfB-F | AGGGGACAAGCAGTGAGT |
| RT-qPCR ywfB-R | TATTCCAGCGGGCTTTGG |
| RT-qPCR ntdA-F | GCAGTGGAACCGATGCTA |
| RT-qPCR ntdA-R | ATAGGAACGCCTCCCGAA |
Fig. 3.
Effects of scandium on the transcription of various genes. B. subtilis strain 168 was cultivated at 30°C for 48 h in NG medium with or without scandium chloride at a concentration of 10 μg/ml. Total RNAs were extracted and used for RT-qPCR analysis. The levels of amyE, aprE, nprE, ywfB, and ntdA transcription were normalized relative to the amount of 16S rRNA in each RNA sample. The relative expression of each gene was calculated by dividing the relative amount of each mRNA in cells grown in scandium-supplemented medium by that in cells grown in scandium-free medium. The average values (with standard deviations) for three independent experiments are shown. The asterisks denote significant differences (P < 0.05).
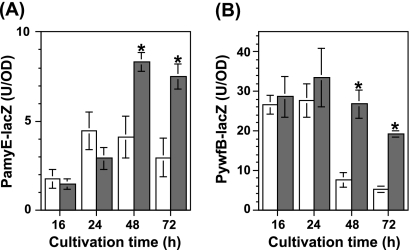
For further investigation, we monitored the effect of scandium on the promoter activity using the lacZ reporter gene. The ywfB reporter strain TI351 [trpC2 amyE::(PywfB-lacZ cat)], carrying the ywfB promoter (192 bp) fused to the lacZ gene at the amyE locus, was constructed previously (6). To construct the amyE reporter strain, the upstream promoter region of amyE (413 bp) was amplified by PCR with the primer pair comprising PamyE-F (5′-GGTAGTGGTGCTTACGATG-3′) and PamyE-R (5′-TCTTGACACTCCTTATTTGATTT-3′). The amplified fragment was cloned into pCR2.1 (Invitrogen). The EcoRI fragment containing the upstream region of amyE was inserted into the EcoRI site of pDL2 (1). The resulting plasmid, pDL2-PamyE, was linearized with PstI and inserted into the amyE locus of the B. subtilis 168 chromosome, with selection for chloramphenicol resistance. The strain thus generated was designated TI416 [trpC2 amyE::(PamyE-lacZ cat)]. These reporter strains, TI351 and TI416, were grown in NG medium with or without scandium at a concentration of 10 μg/ml. As expected, the addition of scandium into the growth medium resulted in elevated expression of PamyE-lacZ and PywfB-lacZ during late stationary phase (48 h and 72 h) but not early stationary phase (16 h and 24 h) (Fig. 4). The amyE promoter in cells grown in the presence of scandium was apparently activated in late stationary phase (48 h and 72 h), whereas it reached the maximum level at 24 h in the scandium-free medium (Fig. 4A). On the other hand, in the presence of scandium, the expression of PywfB-lacZ appeared to be sustained at higher levels for a significantly longer period, resulting in higher levels of expression of PywfB-lacZ in late stationary phase (Fig. 4B). Thus, scandium had a remarkable impact on the expression of these genes during late stationary phase in B. subtilis.
Fig. 4.
Effects of scandium on the expression of PamyE-lacZ and PywfB-lacZ. Strains TI416 [amyE::(PamyE-lacZ cat)] (A) and TI351 [amyE::(PywfB-lacZ cat)] (B) were grown in NG medium without (white thick bars) or with (gray thick bars) scandium chloride at a concentration of 10 μg/ml. Culture samples were withdrawn at the indicated time points, and β-galactosidase activity was determined as described previously (8). The average values (with standard deviations) for three independent experiments are shown. The asterisks denote significant differences (P < 0.05).
In the present study, we demonstrated that scandium has a significant effect on amylase and bacilysin production in B. subtilis. Taken together with previous reports (7, 10), these results indicate that scandium may be a valuable reagent as a component of bacterial growth medium in the biotechnology industry. Scandium has the lowest atomic weight (44.96) among the 17 REEs, and its chemical property resembles that of aluminum. These characteristics may be related, more or less, to the enigmatic results found in this study.
Acknowledgments
This work was supported by a Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science, and Technology (KAKENHI no. 22780078).
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Fukuchi K., et al. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573–1583 [DOI] [PubMed] [Google Scholar]
- 2. Fuwa H. 1954. A new method for microdetermination of amylase activity by the use of amylose as the substrate. J. Biochem. 41:583–603 [Google Scholar]
- 3. Geele G., Garrett E., Hageman J. H. 1975. Effect of benzeneboronic acids on sporulation and on production of serine preteases in Bacillus subtilis cells, p. 391–396 In Gerhardt P., Costilow R. N., Sadoff H. L. (ed.), Spores VI. American Society for Microbiology, Washington, DC [Google Scholar]
- 4. Inaoka T., Takahashi K., Ohnishi-Kameyama M., Yoshida M., Ochi K. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169–2176 [DOI] [PubMed] [Google Scholar]
- 5. Inaoka T., Takahashi K., Yada H., Yoshida M., Ochi K. 2004. RNA polymerase mutation activates the production of a dormant antibiotic 3,3′-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 279:3885–3892 [DOI] [PubMed] [Google Scholar]
- 6. Inaoka T., Wang G., Ochi K. 2009. ScoC regulates bacilysin production at the transcription level in Bacillus subtilis. J. Bacteriol. 191:7367–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawai K., Wang G., Okamoto S., Ochi K. 2007. The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol. Lett. 274:311–315 [DOI] [PubMed] [Google Scholar]
- 8. Kim J. Y., et al. 2009. Identification and characterization of a novel multidrug resistance operon, mdtRP (yusOP), of Bacillus subtilis. J. Bacteriol. 191:3273–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kurosawa K., Hosaka T., Tamehiro N., Inaoka T., Ochi K. 2006. Improvement of α-amylase production by modulation of ribosomal component protein S12 in Bacillus subtilis 168. Appl. Environ. Microbiol. 72:71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka Y., Hosaka T., Ochi K. 2010. Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2). J. Antibiot. (Tokyo) 63:477–481 [DOI] [PubMed] [Google Scholar]
- 11. Walker J. E., Abraham E. P. 1970. The structure of bacilysin and other products of Bacillus subtilis. Biochem. J. 118:563–570 [DOI] [PMC free article] [PubMed] [Google Scholar]